Abstract
Objective: Multidrug-resistant, highly pathogenic Escherichia coli strains are the primary causative agents of intestinal and extraintestinal human diseases. The extensive utilization of antibiotics for farm animals has been identified as a contributing factor to the emergence and dissemination of E. coli strains that exhibit multidrug resistance and possess high pathogenic potential. Consequently, a significant research objective is to examine the genetic diversity of pathogenic E. coli strains and to identify those that may pose a threat to human health. Methods: In this study, we present the results of genome sequencing and analysis, as well as the physiological characterization of E. coli strain APEC 36, which was isolated from the liver of a broiler chicken with generalized colibacillosis. Results: We found that APEC 36 possess a number of mechanisms of antibiotic resistance, including antibiotic efflux, antibiotic inactivation, and antibiotic target alteration/replacement/protection. The most widely represented group among these mechanisms was that of antibiotic efflux. This finding is consistent with the strain’s documented resistance to multiple antibiotics. APEC 36 has an extremely rare variant of the beta-lactamase CTX-M-169. Notwithstanding the multitude of systems for interfering with foreign DNA present in the strain, seven plasmids have been identified, three of which may possess novel replication origins. Additionally, qnrS1, which confers resistance to fluoroquinolones, was found to be encoded in the genome rather than in the plasmid. This suggests that the determinants of antibiotic resistance may be captured in the genome and stably transmitted from generation to generation. Conclusions: The APEC 36 strain has genes for toxins, adhesins, protectins, and an iron uptake system. The obtained set of genetic and physiological characteristics allowed us to assume that this strain has a high pathogenic potential for humans.
1. Introduction
The uncontrolled use of antibiotics in medicine and agriculture over an extended period has resulted in the proliferation of antimicrobial-resistant (AMR) strains of microorganisms. Consequently, the World Health Organization (WHO) has indicated that drug-resistant microorganisms are responsible for the deaths of more than 1.2 million individuals annually, with this figure projected to reach 10 million by 2050 [1,2]. Furthermore, the World Bank projects that AMR could result in an additional USD 1 trillion in healthcare expenditures by 2050. At present, the most common AMR bacterial causative agent is Escherichia coli, accounting for 43% of cases [2]. The WHO has identified antibiotic resistance as one of the most significant public health threats. Given that farm animals are considered a primary source of human pathogens including E. coli, it is essential to monitor the actual pathogens of farm animals for the presence of antibiotic resistance and virulence genes.
Commensal non-pathogenic E. coli are normal microbiota of the gastrointestinal tract of humans and animals. However, some representatives of this species of bacteria have the potential to cause diseases of the digestive system and generalized systemic infections [3]. Cattle are recognized as the principal reservoir for diarrheagenic Escherichia coli (DEC). Concurrently, poultry represents a significant source of extraintestinal pathogenic Escherichia coli (ExPEC) strains [4,5,6,7]. The virulence potential of E. coli is attributable to the presence of pathogenicity genes that encode adhesins, invasins, toxins, resistance factors to immune system, and iron uptake system proteins [8]. The intraspecific heterogeneity of this microorganism, which is due to the possibility of horizontal transfer of genetic determinants associated with virulence and antibiotic resistance, provides a high degree of plasticity and adaptability of bacteria in the changing conditions of pathogen habitats, including those in the closed circuit of agro-industrial complexes [9,10]. Consequently, hybrid strains of E. coli emerge, exhibiting a combination of genetic traits from diverse pathotypes of E. coli and a multidrug-resistant (MDR) phenotype [11].
The development of bacterial resistance to antibacterial drugs represents a significant challenge in the context of livestock production, particularly given the potential for agricultural businesses to serve as hotspots for the emergence and dissemination of antibiotic-resistant pathogens [12,13]. The transmission of antibiotic-resistant microorganisms from animals to humans and vice versa is a documented phenomenon. The dissemination of these bacteria into the environment is also facilitated by the use of organic fertilizers and farm waste products [14]. Modern genomic technologies permit the tracking of the representation of antibiotic resistance genes and the genetic “organization” of the resistome (e.g., the identification of mobile genetic elements, including plasmids, transposons, and integrons) within and between microbial populations. Furthermore, the implementation of these techniques allows for the tracking of the sources of antibiotic-resistant pathogens and the modeling of the evolutionary relationships and transmission of antimicrobial resistance determinants [15].
Therefore, the most crucial research objective at this juncture is to examine the genetic diversity of pathogenic E. coli strains and to ascertain those strains that are potentially pathogenic to humans. The present study is focused on the bioinformatic analysis of the genome and partial physiological characterization of an E. coli MDR strain isolated from the liver of a broiler chicken with generalized colibacillosis.
2. Results
2.1. Genome Sequencing-Based General Characterization and Identification of E. coli Strain APEC 36
An analysis conducted using the Quast program (version 5.2.0, [16]) which evaluates the quality of genome assembly, revealed that the results of sequencing and primary data processing yielded eight circular contigs, each exceeding1000 bp in length, with a total length of 4,979,540 bp. The length of the longest contig 5 was 4,629,530 bp, which is presumed to be the genome. The N50 and N90 values were both 4,629,530, while the L50 and L90 values were both 1. The coverage value was 135. The proportion of GC pairs was determined to be 50.74%. The data obtained indicate that the genome assembly is of a high quality. As indicated by the data from the BUSCO program BUSCO program (version 5.5.0 [17]), the completeness of the strain genome assembly, estimated from characterized genomes of the order Enterobacterales, is 98.9%. A total of 435 complete gene copies and five fragmented genes were identified from 440 gene groups. The level of contamination, as determined by the MiGA online server [18], was found to be 0.9%. Therefore, the obtained quality of the sequenced genome of strain APEC 36 is sufficient for further more detailed analysis.
The gene annotation program Prokka [19] identified 4762 protein-coding genes in the genome, of which 1009 are hypothetical and 3753 have functional annotation. Additionally, the genome contains 22 rRNA genes, one of which is duplicated, 88 tRNA genes, and one tmRNA gene. An analysis conducted using the RASTtk program [20] identified 4989 protein-coding genes (of which 591 genes are hypothetical and 4398 have functional annotation), 86 tRNA genes, and 21 rRNA genes.
As indicated in the PubMLST database [21], the ribosomal MLST profile is identical to rST 113079, which corresponds to the E. coli species (taxon identifier in NCBI: 1452441). The MLST scheme for the identification of strains of E. coli employs the analysis of seven housekeeping genes: adenylate kinase adk, class II fumarate hydratase fumC, DNA gyrase beta subunit gyrB, isocitrate dehydrogenase icd, malate dehydrogenase mdh, phosphoribosylaminoimidazolecarboxamidformyltransferase pur, and recombinase recA [21]. The combination of alleles observed in the strain under consideration corresponds to the ST-695 sequence type, as detailed in (Table 1).

Table 1.
Results of MLST analysis of E. coli strain APEC 36.
A phylogenetic analysis conducted using the BV-BRC database, which stores information on human microbial pathogens [22], revealed that E. coli strain APEC 36 is closely related to E. coli strain O157:H7 Sakai [23] (Figure 1). E. coli O157:H7 is a significant human pathogen that causes hemorrhagic colitis and the hemolytic-uremic syndrome. It has the potential to cause severe illness and large-scale outbreaks globally [24]. These findings suggest that E. coli APEC 36 may also have a high pathogenic potential for humans.
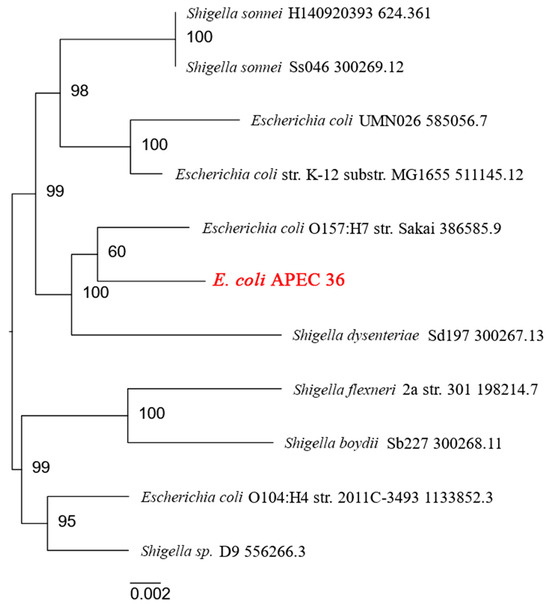
Figure 1.
Human pathogenic strains close to E. coli APEC 36 according to the BV-BRC database.
PlasmidFinder 2.1 [25] was employed to identify plasmids in the APEC 36 strain. The results are summarized in Table 2. Of the seven circular contigs of putative plasmids, the program identified four plasmids. The remaining three contigs (3 (3028 bp), 4 (5879 bp), and 7 (101,931 bp)) are presumed to represent three plasmids with novel types of replication origins.

Table 2.
Plasmids identified in E. coli strain APEC 36 using PlasmidFinder 2.1.
As indicated by the findings of the PADLOC web server [26], the strain APEC 36 has been observed to possess a number of antiviral defense systems. The strain possesses a CRISPR/Cas9 type I-E system, a VSPR, a PDC, a PD-Lambda-1, an ietAS, a gabija, a Mokosh Type II, a restriction-modification type II, a DMS, and a tmn (see Supplementary Figure S1). The majority of these defense systems’ operons are located in the genome, and only the tmn is located on the plasmid (contig_7).
Table 3 provides a summary of the results of database searches for virulence factor genes as well as genes associated with antibiotic resistance [27]. This includes transporter genes [28,29], as well as drug targets [30]. Our results indicate that APEC 36 has several hundred genes associated with antibiotic resistance and virulence, which suggests a high pathogenic potential.

Table 3.
General statistics on the number of genes characterizing the pathogenic potential of strain APEC 36.
2.2. Genes and Potential Mechanisms of Antibiotic Resistance of E. coli Strain APEC 36
Subsequently, a comprehensive search for genes linked to antibiotic resistance mechanisms was conducted using the assembled genome of the APEC 36 strain and the RGI service in the CARD database [31]. The results of this search are presented in Table 4. The results indicate that the E. coli strain under study has the potential to reduce the efficacy of antibacterial drugs by inactivating the antibiotic, altering its target, replacing the target, expelling the antibiotic from the cell, and reducing the permeability of the cell wall. Transcriptional regulators and signal transduction systems may be involved in the activation of these cell-protecting processes.

Table 4.
Genes associated with antibiotic resistance found in the genome of APEC 36 strain.
Specifically, among the resistance genes identified by CARD RGI, more than half of the resistance genes function through the antibiotic efflux mechanism. These include genes that encode efflux pumps as well as regulators of their expression. Notably, all of these genes, with the exception qacL, are located on contig 5, which corresponds to the bacterial chromosome. In particular, the majority of the genes encode subunits of ABC transporters that are embedded in the inner membrane of the bacterial cell and belong to two major families: the major facilitator superfamily (MFS) and the resistance-nodulation-cell division (RND) family. Members of these families differ in structure and substrate specificity (MFS is somewhat broader), yet both can confer drug resistance in bacteria. Members of both families often form pores across both membranes and associate with outer membrane proteins such as TolC to form a tripartite complex [32,33].
Complexes that can be formed with members of the RND family include AcrAB-TolC, which protects against a wide range of drugs, including beta-lactams, tetracyclines, fluoroquinolones, rifamycin, phenicols, and so forth; AcrAD-TolC, which protects against beta-lactams and tetracyclines; and so on. In addition, the complexes can be formed with members of the RND family, including AcrAB-TolC (which protects against a wide range of drugs, including beta-lactams, tetracyclines, fluoroquinolones, rifamycin, phenicols, etc.), AcrAD-TolC (which protects against beta-lactams, tetracyclines, fluoroquinolones, rifamycin, phenicols, etc.), AcrEF-TolC (which moves beta-lactams and fluoroquinolones), MdtABC-TolC (which moves aminocoumarins), and MdtEF-TolC (which moves macrolides, fluoroquinolones, and penams). The MFS family includes the following complexes: ErmAB-TolC (which excretes fluoroquinolones), ErmKY-TolC (which excretes tetracyclines), MdtNOP-TolC (which excretes nucleoside antibiotics), and so forth [34,35,36]; and some MFS transporters, including Tet(A), which forms homodimers and acts against tetracyclines [37]. MdfA, which facilitates the cell’s clearance of tetracyclines and disinfectants, and CmlA5, which is anti-phenicol, operate in a TolC-independent manner [38]. Other groups include the KpnEF transporter of the small drug resistance (SMR) family, which eliminates macrolides, aminoglycosides, cephalosporins, tetracyclines, peptide antibiotics, rifampin, and other compounds [39].
Furthermore, the E. coli strain under investigation harbors the E448K mutation within the gene encoding GlpT. The mutation impairs the efficiency of fosfomycin antibiotic transport into the cell through the GlpT transporter [40,41]. Since fosfomycin inhibits the cytoplasm-localized MurA enzyme, which catalyzes the initial step of cell wall biosynthesis, namely the reaction of phosphoenolpyruvate addition to UDP-GlcNAc to form UDP-enoylpyruvyl-GlcNAc, the reduced antibiotic transport mitigates the adverse effects of fosfomycin.
With regard to the regulatory proteins involved in the control of antibiotic defense mechanisms, these include EmrR, H-NS, LeuO, RsmA, AcrS, CpxA, BaeR, CRP, GadX, KdpE, MarA, and the EvgA-EvgS system. ErmR is a member of the TetR family of proteins that functions as a negative regulator of the ErmAB operon [42]. AcrS and AcrR have been demonstrated to repress the acrAB operon [43,44], while MarA has been shown to activate the AcrAB-TolC efflux system [45]. The marA gene is a component of the marRAB operon, which is subject to negative regulation by the MarR repressor [45]. The Y137H and G103S mutations were identified in MarR, which is known to result in elevated expression of marRAB and mdtE [46] and, subsequently, the activation of the AcrAB efflux pump [47]. The LysR family transcription factor LeuO has been demonstrated to stimulate the transcription of operons encoding AcrEF, MdtNOP, and other transporters [48]. Conversely, H-NS is a universal silencer and an antagonist of LeuO [49]. EvgAS is a two-component system comprising EvgS, a sensory kinase that detects environmental stress, and EvgA, a response regulator that regulates the activity of numerous proteins [50]. The SoxRS systems is also a two-component system, with the SoxR protein containing [2Fe-2S] clusters. The latter are oxidized by reactive oxygen species and other oxidants that occur under conditions of cellular stress. This results in the activation of SoxR, which induces the expression of soxS. In turn, this increases the expression of transporter genes (such as acrAB), genes involved in DNA repair and oxidative stress control systems, and decreases the expression of genes encoding porins, such as OmpF, in order to reduce cell membrane permeability [51]. The sequenced genome revealed the presence of the A12S mutation in the SoxS protein, which has the potential to enhance the expression of the AcrAB efflux pump genes and result in diminished cell wall permeability due to reduced OmpF porin production. Low antibiotic input and activated drug efflux are hallmarks of multiple drug resistance phenotypes observed in E. coli clinical isolates [52].
It can be reasonably deduced that the upregulation or mutation of efflux pumps plays a significant role in the development of antibiotic and disinfectant resistance in the strain, both under conditions of stress and in the absence of such conditions.
Additionally, the genome of the strain contains multiple genes that confer resistance through the antibiotic target alteration mechanism. These are antibiotic targets that have undergone specific mutations, rendering them insensitive to inhibition. For example, the penicillin-binding protein PBP3, which is an alternative name for transpeptidase, has undergone mutations at residues D350N and S357N, rendering it insensitive to beta-lactam antibiotics [53]. An additional example is the translation elongation factor EF-Tu. Typically, GTP-bound EF-Tu forms a complex with aminoacyl-tRNA. Subsequently, the entire complex attaches to the ribosome, where EF-Tu hydrolyzes GTP and catalyzes the incorporation of the amino acid into the nascent polypeptide chain. Subsequently, the EF-Tu-GDP complex dissociates from the ribosome. With the assistance of another translation factor, EF-Ts, EF-Tu replaces GDP with GTP, thereby initiating another cycle. Elfamycin antibiotics, including pulvomycin and kirromycin, inhibit the activity of EF-Tu. They prevent the aminoacyl-tRNA binding step to the EF-Tu-GTP complex. The R234F mutation, which is present in two copies of the EF-Tu gene in our E. coli strain, should render it insensitive to these antibiotics [54,55].
The E. coli strain under investigation also harbors a number of genes that function by the mechanism of antibiotic inactivation. The genes encode enzymes that facilitate the degradation of the antibiotic or the addition of extra groups to it, which ultimately results in the loss of its activity. Of the 11 such genes that were identified, only four are located on the chromosome, and another four on contig 2, which corresponds to the plasmid, and another three on contig 8, which is also a plasmid. Subsequently, we will discuss the genes in relation to their role in antibiotic resistance mechanisms.
Beta-lactams were the first antibiotics to be discovered and remain the most diverse and widely used group of antibiotics. The mechanism of action of beta-lactams is the binding of the antibiotic to transpeptidase, an enzyme that catalyzes the formation of interpeptide cross-links in the cell wall. This binding occurs due to the molecular similarity of the antibiotic to the dimer of D-alanine, the natural substrate of transpeptidase. Subsequently, the beta-lactam ring is opened, resulting in the formation of a covalent bond with serine within the active site of transpeptidase, which inhibits the enzyme irreversibly. The most prevalent mechanism of resistance to beta-lactams is the production of beta-lactamases. These enzymes belong to the hydrolase class and function by hydrolyzing the amide bond in the beta-lactam ring, thereby rendering the antibiotic inactive. Among the enzymes whose genes were identified in the strain genome through sequencing, five beta-lactamases were detected that confer resistance to beta-lactam antibiotics by cleaving the beta-lactam ring. Two representatives of the TEM family, one enzyme of the CTX-M family (specifically CTX-M-169) and one of the OXA family (OXA-10 according to CARD RGI or OXA-48 according to DFAST), were identified. Additionally, an AmpC beta-lactamase was detected. As annotated by DFAST, the beta-lactamase was identified as CTX-M-169, one of the OXA family (specifically OXA-10 according to CARD RGI or OXA-48 according to DFAST), and one AmpC beta-lactamase. Both the TEM and CTX-M families are classified as class A beta-lactamases according to Ambler’s classification system. These enzymes are frequently classified as extended-spectrum beta-lactamases, which possess the ability to degrade not only penicillins but also cephalosporins and monobactams. It is fortunate that these enzymes are rarely resistant to inhibitors. The OXA family is classified as a class D beta-lactamase. These enzymes are frequently carbapenemases, and their sensitivity to inhibitors is variable. AmpC beta-lactamase belongs to Ambler’s class C, which is distinguished by variable substrate specificity and frequently observed insensitivity to inhibitors [56]. The remaining three enzymes conferred resistance to aminoglycosides, namely two ANT(3′) proteins (encoded by the aadA gene) and one AAC(3) protein (encoded by the AAC(3)-IId gene). AAC(3) utilizes acetyl-CoA to acetylate the 3-amino group of the antibiotic, whereas ANT(3′) transfers the adenyl group to the hydroxyl at the 3′ position in an ATP-dependent manner. Aminoglycoside antibiotics inhibit bacterial translation by binding to the 30S subunit of the ribosome. However, aminoglycosides modified by the enzymes identified in this study exhibit a markedly reduced affinity for the target [57,58,59]. Additionally, genes for three enzymes that similarly modify other groups of antibiotics were identified: chloramphenicol acetyltransferase (catIII gene), streptothricin acetyltransferase (sat2 gene), and rifampin ADP-ribosyltransferase (arr2 gene). Chloramphenicol targets protein synthesis on ribosomes (it binds to the 50S subunit), yet loses its activity after acetylation by the CAT enzyme [60]. Similarly, streptothricin binds to the 30S subunit of the ribosome. Following acetylation by the SAT enzyme, the interaction is no longer possible [61]. Rifampin, in turn, binds to the β-subunit of bacterial RNA polymerase, inhibiting transcription. The enzyme Arr, using NAD+ as a substrate, ADP-ribosylates rifampin, thereby removing the interference with RNA polymerase activity [62].
Some genes confer resistance through the mechanism of “antibiotic target replacement”, whereby they encode alternative enzymes that are not inhibited by the antibiotics. In the case of the strain under consideration, these are alternative enzymes in the folate biosynthesis pathway, which is required for the biosynthesis of purines (which in turn are required for DNA and RNA synthesis and cell proliferation) and a number of other metabolic pathways. The biosynthesis of tetrahydrofolate, which possesses reducing properties and is the active form of folate, in bacteria comprises three steps. Firstly, the enzyme dihydropteroate synthase (DHPS) catalyzes the formation of 7,8-dihydropterolate (DHP) from 6-hydroxymethyl-7,8-dihydropterin diphosphate (DHPP) and p-aminobenzoic acid (pABA). Subsequently, DHP is transformed into 7,8-dihydrofolate (DHF). In the third step, 7,8-dihydrofolate (DHF) is reduced to 5,6,7,8-tetrahydrofolate (THF). This final step is catalyzed by dihydrofolate reductase (DHFR). Sulfonamide antibiotics inhibit DHPS, which catalyzes the initial step of this pathway, and diaminopyrimidines act on DHFR, which is responsible for the final step. The sul genes (comprising two sul2 and one sul3 genes) encode alternative forms of DHPS that are insensitive to the action of sulfonamides. Additionally, dfrA14 encodes DHFR that is not inhibited by diaminopyrimidine antibiotics. Accordingly, the genome sequence data indicated that the sul and dfrA14 genes enable the APEC 36 E. coli strain to synthesize folate in response to both sulfonamides and diaminopyrimidines [63,64]. Furthermore, the qnrS1 gene was identified, which is typically located on plasmids but was observed to be present on the chromosome. This gene confers resistance to fluoroquinolones through the mechanism of antibiotic target protection. As the target of quinolones is DNA gyrase, the action of these antibiotics results in the inability of the bacterial genome to replicate. The QnrS1 protein is capable of binding to the gyrase, thereby competing with the antibiotic. The interaction of QnrS1 with the enzyme does not interfere with the gyrase functioning and allows normal catalysis even in the presence of fluoroquinolones [65].
The drug resistance mechanisms identified for the E. coli APEC 36 strain are presented in Figure 2.
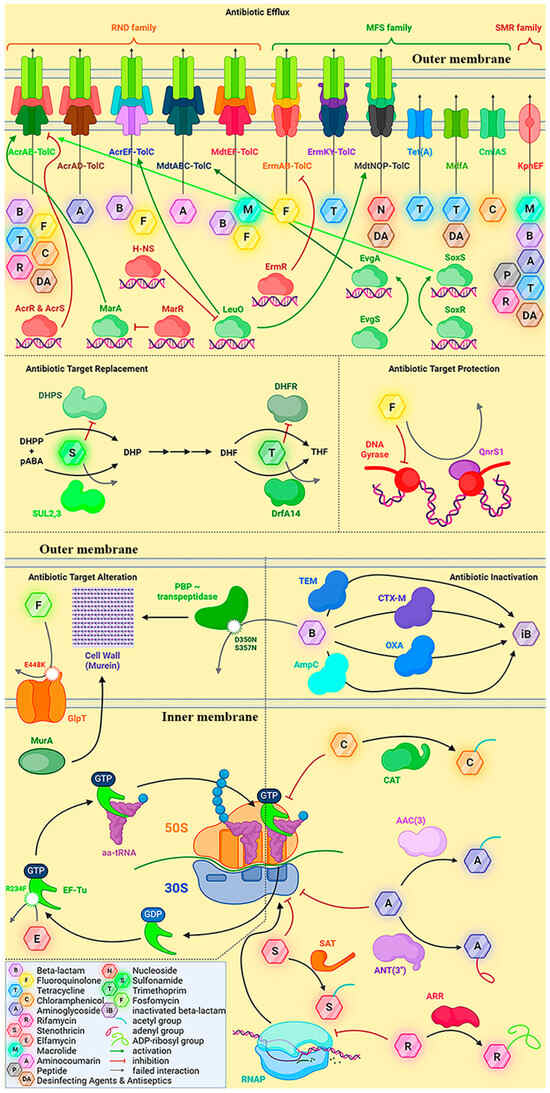
Figure 2.
The antibiotic resistance mechanisms proposed for the E. coli APEC 36 strain, based on the analysis of its sequenced genome.
2.3. PCR Verification of the Presence of Virulence and Antibiotic Resistance Genes
Additionally, the presence of genes associated with virulence and antibiotic resistance was evaluated through analytical PCR (Table 5). The data indicated the presence of specific genes associated with the AREC pathotype, namely hlyF (a gene encoding for a virulence factor known as avian hemolysin) and iutA (a gene encoding for the receptor for the siderophore aerobactin). Among the genes of general pathogenicity, the presence of kpsMT and iroN genes was confirmed. Genes characteristic of the ExPEC group (UPEC) were represented by upaG (mediates adhesion to bladder epithelial cells) and fimH (encodes fimbrial adhesin). Furthermore, specific DEC/IPEC marker genes of the estI, eltA, subA, eastI, and iha group were identified. The presence of genes encoding beta-lactamases belonging to the CTX-M and TEM families was also confirmed by PCR.

Table 5.
Detection of virulence and antibiotic resistance genes by PCR.
2.4. Physiological Characteristics of E. coli Strain APEC 36
It is important to note that the presence of genes associated with pathogenicity and antibiotic resistance does not necessarily indicate the presence of the corresponding phenotype in the strain. This may be attributed to point mutations occurring in the genes themselves and in their regulatory regions or transcription factors. Subsequently, the antibiotic resistance of strain APEC 36 was evaluated to ascertain the functionality of the identified genes (Table 6). The results demonstrated that the strain exhibited resistance to several cephalosporins (cefuroxime, cefotaxime, and cefixime), fluoroquinolones (ciprofloxacin and levofloxacin), tetracyclines, sulfonamides, and chloramphenicol.

Table 6.
Antibiotic resistance profile of E. coli strain APEC 36.
Given that the strain in question exhibited the presence of both iha and fimH genes as determined by PCR (see Table 5), we proceeded to assess its capacity for adhesion to erythrocytes. APEC 36 exhibited minimal adhesion to human red blood cells (IAM = 1.47) and low adhesion to chicken red blood cells (IAM = 2.16) (Figure 3A). Furthermore, the strain was observed to lack hemolytic activity against human blood, yet exhibited the ability to lyse chicken red blood cells (Figure 3B). This phenotype aligns with the absence of the hlyA lysine gene and the presence of the hlyF lysine gene (Table 5).

Figure 3.
Assessment of pathogenicity of E. coli strain APEC 36. (A). An example of specific adhesion of E. coli APEC 36 on chicken red blood cells (arrow), 30 min, staining with gentian violet, 1000×. (B). An example of E. coli growth on chicken blood agar: viewed with incident light–obvious hemolysis. (C). Assessment of the siderophore production level.
The functionality of the iron uptake system was evaluated through the estimation of siderophore production levels. The coefficient was found to be 1.67 (Figure 3C), which lends support to the phenotypic manifestation of the iroN and iutA genes (Table 5).
Subsequently, the strain was examined for the presence of bacteriophages. No lysogenic phages were identified (Figure 4). These findings indicate the functionality of anti-phage defense systems, such as the CRISPR/Cas IE system encoded in the genome of the APEC 36 strain.

Figure 4.
APEC 36 strain does not contain lysogenic prophages. (A) E. coli APEC 36; (B) control lysogenic prophage-carrying strain.
3. Discussion
It is established that healthy farm animals can serve as a source of microorganisms with high virulence potential due to the presence of pathogenicity genes encoding adhesins, invasins, toxins, immune system resistance factors, and proteins of iron uptake systems. The initial characteristic of a strain’s virulence potential is typically regarded as its affiliation with a specific phylogenetic group. The available evidence indicates that APEC representatives are more frequently associated with phylogroup B1 or F [66,67,68], which encompasses a diverse range of pathogens with a multitude of virulence factors. The isolated E. coli APEC 36 strain was found to carry genes for toxins, adhesins, protectins, genes for UPEC-specific proteins, and iron uptake systems (Table 5). Based on this set of genetic characteristics, the strain was classified as a representative of phylogroup B1, since B1 is the predominant group among Shiga toxin-producing Escherichia coli (STEC/EHEC) [69]. APEC36 bears a wide range of virulence determinants and appeared to be close to O157:H7 strain; it seems to have a high pathogenic potential with respect to both poultry and human beings.
Genome analysis revealed the presence of a number of antiphage defense systems, including the CRISPR/Cas9 type I-E system, the VSPR, the PDC, the PD-Lambda-1, the ietAS, the Gabija, the Mokosh Type II, the restriction-modification Type II, the DMS, and the tmn. This raises the question of how plasmids survive the action of these systems.
Plasmids may circumvent the majority of these defensive mechanisms due to their lack of characteristics associated with phage infections. For example, plasmids may not undergo extensive replication or transcription, which precludes their ability to trigger the action of the Gabija defense system. This system is triggered when there is a depletion of NTP or dNTP pools [70]. The action of some other system, such as ietAS or Mokosh, is dependent on specific functional viral proteins, including DNA polymerase, ssDNA binding protein, helicase, terminase, and so forth [71]. Since plasmid replication is dependent on host enzymes, it can avoid the action of such defensive systems.
It seems reasonable to suggest that plasmids are more susceptible to defense systems that are targeted DNA, such as restriction-modification (R-M) systems or CRISPR/Cas systems. In recent work, it has been proposed that plasmids could evade the action of the R-M system in two ways, depending on the size of the plasmid [72]. In the case of relatively small plasmids, the primary mechanism of evasion involves sequence change. In contrast, in the case of relatively large plasmids, the primary mechanism of evasion involves the addition of genes that inactivate the R-M system. Additionally, plasmids can circumvent the action of the functional CRISPR/Cas IE system of E. coli. This is achieved initially via an enhanced replication rate that could be faster than the rate of CRISPR/Cas IE system action [73]. Subsequently, the plasmids accumulate escape mutations in targeted protospacers and become persistent in E. coli strains. It is plausible that plasmids may utilize multiple strategies to evade the defensive systems of E. coli.
The fight against such potentially dangerous strains is often complicated by their ability to survive under antibiotic pressure due to the presence of antibiotic resistance systems, which render the strains more resilient to the effects of antibiotics. This is a direct consequence of the active use of antibacterial drugs throughout the poultry production cycle in the majority of countries worldwide.
The most commonly utilized antibiotics for the prevention and treatment of bacterial infections in poultry within the industrial context are tetracyclines, fluoroquinolones, and sulfonamides. For this reason, a high incidence of strains resistant to these antibiotics has been documented for some time [74,75,76]. It is regrettable that the Food and Drug Administration (FDA) has approved the use of certain antibiotics in both animals and humans, including those classified as Category I antimicrobials, which are considered highly important for the treatment of severe human infections. Such drugs include, in particular, cephalosporins. For example, ceftiofur is employed for the control of omphalitis in broilers. It has been demonstrated that the proportion of E. coli strains exhibiting resistance to this drug was markedly elevated in chickens that received it as a prophylactic supplement [77]. Conversely, on farms where ceftiofur was discontinued, a reduction in the resistance of E. coli to this broad-spectrum cephalosporin was observed in healthy broilers [78].
It is of particular importance to note that resistance to specific antimicrobials can occur in the presence of both related and unrelated antimicrobials. For example, the resistance of E. coli to tetracycline in turkey flocks was found to be increased when not only tetracycline but also injectable aminoglycosides were used. Furthermore, a notable correlation between beta-lactam resistance and the administration of streptogramine in the diet has been documented [79].
According to data from the World Health Organization (WHO), the utilization of antibiotics in veterinary medicine is currently approximately twice the quantity of pharmaceuticals employed in human medicine. In light of these circumstances, efforts are being made to legally restrict the use of antibacterial drugs in agricultural practice with the aim of reducing the probability of the emergence and spread of antibiotic-resistant strains of microorganisms. In 2013, the FDA restricted the use of low-dose antibiotics as growth promoters in livestock, in accordance with the Guidance for Industry (GFI) #213. These antibiotics are important for human medicine [80]. Subsequently, in 2014, a comparable prohibition on select AGPs was enacted in Canada [81]. A number of countries within the Organisation for Economic Co-operation and Development (OECD) have implemented restrictions on the use of AGPs (e.g., Mexico, South Korea, New Zealand), whereas in other countries (e.g., Japan), the use of AGPs remains permitted [82]. The use of AGPs is not prohibited in the majority of non-OECD countries. These include major poultry producers such as China, Brazil, Russia, Argentina, India, Indonesia, the Philippines, and South Africa [82]. Furthermore, Regulation (EU) 2019/6 came into force in European countries on 28 January 2022. This regulation states that antimicrobial medicinal products may not be used for routine prophylaxis, defined as the administration of a medicinal product before the appearance of clinical signs of disease in order to prevent the occurrence of disease or infection. In Russia, the use of antibiotics in agriculture is also subject to regulation [83]. In particular, a list of feed antibiotics that are authorized for use has been established. However, for a variety of reasons, antibacterial drugs are often used in a haphazard manner or in violation of the prescription regime [84].
The E. coli strain APEC 36, as described in this work, contains a wide range of resistance genes to various antibacterial drugs within its genome (Table 4). A detailed examination of the genotype (Table 4) in conjunction with the identified phenotypic parameters of antibiotic resistance (Table 6) has enabled us to ascertain the genetic determinants, the products of which appear to directly contribute to the observed profile of reduced antibiotic sensitivity of the strain.
It is important to note that APEC 36 is sensitive to carbapenems and aminoglycosides (Table 6). Consequently, the genes whose products provide resistance to these drugs are nonfunctional under conditions studied. It is noteworthy that these genes encompass determinants of both specific and multidrug resistance. Consequently, AAC(3)-IId, AadA, ArcD, and KdpE are solely responsible for conferring resistance to aminoglycosides, while BaeR and CpxA are involved in this process for both aminoglycosides and aminocoumarin antibiotics. Additionally, KpnEF, MarA, SoxS, and TolC contribute to the development of resistance to a diverse range of antibacterial drugs.
It is important to note that the SoxS protein does not play a direct role in the cellular defense mechanism against the antibiotic. It functions as a transcriptional regulator, activated by the sensor protein SoxR in response to oxidative stress [85,86]. The activation of the SoxRS regulon results in the expression of genes that encode a variety of multidrug efflux pumps, which facilitate the removal of antibacterial drugs from the cell. It is established that mutations in the SoxR gene result in elevated levels of SoxS expression in the absence of stress. This, in turn, gives rise to the formation of multidrug resistance in clinical strains, thereby ensuring the perpetuation of this trait over multiple generations [87].
A comparable situation arises with the MarRAB operon, which encompasses the transcriptional regulator MarA. MarR binds to the marRAB promoter and functions as a negative regulator of gene expression [88]. In the presence of inducers, MarR binds these inducers and is then unable to interact with DNA [89]. In such a situation, MarA binds the marRAB promoter and acts as an activator of the system [90]. Therefore, if MarA fails to function for any reason, the entire operon becomes non-functional. Consequently, the corresponding proteins can also be excluded from the list of resistance determinants active in the cells of the strain under study.
The TolC protein constitutes a component of the major multidrug efflux pump of Gram-negative bacteria AcrAB-TolC. This pump forms a channel [91] that traverses through the periplasmic space and the outer membrane. The entrance to the channel through the inner membrane on the cytoplasmic side is determined by the AcrZ protein. The functionality of the AcrAB-TolC pump is contingent upon its complete assembly and the optimal operation of all its constituent components. Given that TolC does not afford protection against carbapenems, it would appear that this entire protein complex should be excluded from the list of resistance systems active in the APEC 36 strain. This assertion is corroborated by the observation that the strain under examination is susceptible to inhibitor-protected penicillins. Given that penicillin antibiotics can effectively inhibit the growth of APEC 36 in the presence of clavulanic acid, its resistance to this class of compounds is evidently determined by beta-lactamases rather than by multidrug efflux pumps. Consequently, we have a second line of evidence indicating the non-functionality of the AcrAB-TolC system.
In light of the other resistance determinants whose genes were identified in the genome of the strain under investigation through sequencing, it is notable that AcrR serves as a modulator of AcrAB-TolC pump gene expression [92]. Similarly, AcrS has been demonstrated to act as a repressor of the AcrAB efflux complex, and is associated with the expression of AcrEF. It is postulated that AcrS regulates a switch between AcrAB- and AcrEF-mediated efflux. Given that penicillin resistance has been demonstrated to be independent of efflux pumps (and thus the AcrEFS system, analogous to AcrAB-TolC, is non-functional), it can be concluded that acrR and acrEFS are not significant contributors to resistance.
In addition, the cAMP receptor protein (CRP) has been identified as a potential determinant of resistance for penicillins and fluoroquinolones (Table 6). CRP is one of seven global regulators in E. coli, influencing the expression of nearly 490 genes. These genes are involved in a number of processes, including the regulation of multidrug efflux pumps [93]. The other transcriptional regulator is GadX. This protein is responsible for the activation of the glutamate decarboxylation system, which serves as a defensive mechanism of E. coli against acid stress [94]. GadX has also been demonstrated to activate drug efflux pumps [95], though it is not directly involved in resistance, unlike CRP. In addition to the aforementioned transcription factors, other proteins indirectly involved in antibiotic resistance include the H-NS protein, which is a DNA-binding protein with a central role in gene regulation and nucleoid structuring [96], the RsmA protein, which is RNA-binding protein regulating a set of genes including those of the Type III secretion system [97], and the two-component signal-transduction system EvgAS, which regulates the EmrKY pump [98].
By shifting the focus to an analysis of the factors influencing the resistance of the APEC 36 strain to specific antibacterial drugs, and considering the spectrum of resistance systems that are either dysfunctional or indirectly related to the development of reduced antibiotic sensitivity, we can conclude the following.
It is evident that resistance to sulfonamides (cotrimoxazole) is a consequence of the activity of the sul2 and sul3 gene products, which act by the mechanism of antibiotic target replacement (Figure 2). This ensures the appearance of alternative forms of DHPS in the cell that are not inhibited by the antibacterial drug.
The resistance to amphenicol (chloramphenicol) appears to be due to the proteins CatIII (chloramphenicol acetyltransferase, which ensures inactivation of the antibiotic), CmlA5 (efflux pump), and MdtM (also an antibiotic efflux pump) [99]. Conversely, CatIII and CmlA5 are exclusively specific for amphenicols, while MdtM can also function with antibacterial drugs belonging to the fluoroquinolone group.
It has been proposed that insensitivity to tetracyclines may be related to the EmrKY, MdfA, and Tet(A) systems. All are efflux pumps, with MdfA being largely analogous to the amphenicol-specific CmlA.
With regard to fluoroquinolones, the most critical defense systems of the bacterial cell appear to be EmrRAB, MdtEFHM, and QnrS1. In this instance, MdtEF represents a multidrug efflux pump that may also exhibit activity against penicillins [100]. Given that the APEC 36 strain displays sensitivity to penicillins in the presence of clavulanic acid, it can be concluded that this system is ineffective under the conditions studied. In contrast, MdtM and MdtH may be functional. While the MdtH efflux pump is specific for fluoroquinolones [101], MdtM has broader substrate specificity [99] and may also confer APEC 36 immunity to amphenicols.
Another fluoroquinolone-specific drug efflux pump is EmrRAB [102], and QnrS1 is a specific protein that interacts with regions of gyrase besides the DNA-binding groove, potentially allowing more specific binding and destabilization of the topoisomerase-DNA-quinolone cleavage complex [103]. Therefore, while EmrRAB and MdtHM facilitate the extrusion of fluoroquinolones from the cytoplasm, QnrS1 operates in accordance with the mechanism of antibiotic target protection (Figure 2). It is noteworthy that qnrS1 is typically encoded by plasmids [104]. However, in the APEC 36 strain, this gene is located in the genome, indicating that the determinants of antibiotic resistance may be captured in the genome and stably transmitted from generation to generation.
Upon returning to the subject of beta-lactam antibiotics, it can be stated with a high degree of confidence that the E. coli strain under study possesses at least one beta-lactamase enzyme capable of degrading penicillin antibiotics. However, this enzyme is inhibited by clavulanic acid. The APEC 36 genome encodes four distinct beta-lactamase variants. These are OXA, TEM, CTX-M, and AmpC. AmpC activity has been demonstrated to be independent of clavulanic acid presence [105]. This leads to the conclusion that AmpC is inactive. OXA beta-lactamases confer resistance to ampicillin and oxacillin, although some of them are capable of degrading cefotaxime, cefepime, and ceftazidime molecules. The beta-lactamase OXA-10 detected in the strain belongs to this category and should provide resistance to ceftazidime, cefepime, and aztreonam [105]. The results demonstrate that strain APEC 36 is susceptible to these antibiotics (Table 6), indicating that the beta-lactamase OXA-10, similar to AmpC, is nonfunctional. Therefore, in the context of penicillin antibiotics, the sole determinant of observed antibiotic resistance is the TEM-1 beta-lactamase. The aforementioned conclusion is indirectly corroborated by the previously published paper, wherein the genomic analysis of E. coli strains isolated from diseased chickens in the Czech Republic [106] demonstrated that the multiresistant phenotype observed in the majority of sequenced strains was predominantly based on resistance to β-lactams and quinolones, which were associated with TEM-type beta-lactamase genes and chromosomal gyrA mutations. It is important to note that, despite the existence of at least 19 distinct TEM inhibitor-resistant β-lactamases, the enzyme type identified in this study is clearly a clavulanic acid-sensitive variant.
The protection of the APEC 36 strain against cephalosporins also appears to be determined, at least in part, by the presence of TEM-1 beta-lactamase. An alternative enzyme that may contribute is the CTX-M-169 beta-lactamase. In general, CTX-beta-lactamases readily hydrolyze ceftazidime. However, our strain is sensitive to this drug, suggesting either nonfunctionality of CTX-M-169 (in which case, the resistance to cephalosporins depends entirely on TEM-1) or the inability of this enzyme variant (CTX-M-169) to degrade ceftazidime specifically.
It is noteworthy that the PBP3 protein is listed among the determinants of antibiotic resistance that can influence resistance to beta-lactams (Table 6). The protein is involved in cell wall synthesis and is targeted by beta-lactam antibiotics. Mutations in PBP3 can result in resistance to penicillins and cephalosporins through the mechanism of antibiotic target alteration [107]. However, in the presence of clavulanate, APEC 36 is sensitive to penicillins, indicating that PBP3 does not carry resistance mutations.
4. Materials and Methods
4.1. Isolation and Identification of E. coli Strain APEC 36
From 2016 to 2018, we conducted a post-mortem examination of the organs of broiler chickens (Gallus gallus L.) of the Ross 308 cross that had succumbed to generalized colibacillosis at a large poultry farm in the Perm region of Russia. The E. coli strain APEC 36 was isolated from the liver of a seven-day-old (December 2017) and subsequently deposited in the Ex Culture Collection of the Department of Biology, Faculty of Biotechnology, University of Ljubljana (Univerza v Ljubljani, Slovenia) under the designation L-5866.
The isolation and identification of the strain was conducted in accordance with the bacteriological method recommended by the Order on Unification of Microbiological Research Methods No. 535, 1985. The isolated strain was verified using the diagnostic test system ENTEROtest 16 (Erba Lachema s.r.o., Brno, Czech Republic), which is a Russified version of Microb-2 (Microbiological System Monitoring Microb-2, SMMM-2). The SMMM-2 was employed along with the PCR amplification of the 16S RNA fragment using primers 16s-F: GACCTCGGTTTAGTTCACAGA, 16s-R: CACACGCTGACGCTGACCA [108]. The resulting product was then subjected to Sanger sequencing.
4.2. Antibiotic Sensitivity Test
The determination of antibiotic sensitivity was conducted in accordance with the clinical guidelines “Determination of sensitivity of microorganisms to antimicrobial agents” of the Interregional Association of Clinical Microbiology and Antimicrobial Chemotherapy (MACMAX Version-2018-03). The sensitivity to antibiotics was determined for beta-lactams (ampicillin (10 µg), cefotaxime (5 µg), ceftazidime (10 µg)), as well as for aminoglycosides (amikacin (30 µg), gentamicin (10 µg)), tetracyclines (tetracycline (30 µg)), fluoroquinolones (ciprofloxacin (5 µg), levofloxacin (5 µg)), and sulfonamides (co-trimoxazole (trimethoprim/sulfamethoxazole, 1. The minimum inhibitory concentrations (MICs) of 25/23.75 µg were determined by the disk-diffusion method (MACMAX, version 2015-02, EUCAST, version 11.0, valid from 1 January 2021). The production of extended-spectrum beta-lactamases (ESBLs) was determined by the double-disc method.
4.3. The Hemolysis Test
The culture’s hemolytic ability was determined by sowing it on 5% blood agar using human or chicken erythrocytes. The presence of hemolysis was assessed visually by observing the formation of a clear zone around the streaks after 24 h of culture incubation at 37 °C.
4.4. Iron Uptake Test
The level of siderophore production was tested as previously described [109]. It was estimated semi-quantitatively by applying a coefficient defined as the ratio of the diameter (in mm) of the yellow-orange zone on the agar plate to the diameter of the bacterial colony.
4.5. Lysogenic Phage Detection
The presence of bacteriophages within cells was determined through the use of the ultraviolet radiation induction method. A liquid culture of the strain under study, with an optical density of 2.0 according to McFarland, was distributed across the surface of Petri dishes. Subsequently, the dishes were irradiated with a DB-30-1 arc bactericidal lamp with a power of 30 W (wavelength 253.7 nm, UV-C spectrum) at a distance of 1 m from the dishes for 70 s. Following this, the dishes were covered with lids and incubated for 1 h at 37 °C. The resulting suspension was mixed with a culture of the sensitive strain E. coli DH5α, added to melted 0.6% agar (46 °C), mixed and layered on pre-prepared dishes with agarized Luria Bertani (LB) medium. The dishes were then incubated for 24 h at 37 °C. The formation of lysis zones in the sensitive strain was documented. In parallel, dishes with a negative control, lacking any cultures, were prepared.
4.6. Bacterial Adhesion to Red Blood Cells
The study of bacterial-specific adhesion to red blood cells was conducted in accordance with the Brilis method in Eppendorf tubes [110]. To account for the adhesive properties of bacteria, human red blood cells (type O, Rh+) and chicken erythrocytes were utilized. The erythrocytes were washed in saline phosphate buffer (PBS), then diluted to 108 cells/mL. The bacteria were cultivated overnight, rinsed with a phosphate buffer, and resuspended to a concentration of 108 cells/mL. Subsequently, a bacterial suspension was combined with an erythrocyte mass in a 1:1 ratio, and incubated at 37 °C with agitation at 120 rpm for 30 min. Blood smears were prepared and stained with a 0.5% solution of gentian violet. In examining the preparations under optical microscopy, three key indicators were considered: the average adhesion index (AAI), which represents the mean number of microorganisms attached to the surface of a single red blood cell; the adhesion coefficient (AC), which denotes the percentage of red blood cells with bacteria on their surface; and the index of adhesiveness of microorganism (IAM), which is the ratio of AAI and AC. Counting was conducted on 100 cells, with the entire glass slide being examined. Based on the IAM values, the microorganisms were classified as follows: non-adhesive (IAM < 1.75), low-adhesive (IAM = 1.76–2.49), medium-adhesive (IAM = 2.50–3.99), and highly adhesive (IAM > 4.0).
4.7. Phylogenetic Group Determination
The phylogenetic group was determined using multiplex polymerase chain reaction (quadruplex PCR) with primers according to Clermont O. et al. (2013) [111]. The results were interpreted using the key for determining the phylogenetic group. As a control, E. coli strains B2, BJ30, BJ32, and BJ33 of phylogroup B2 from the collection of the Faculty of Biotechnology, University of Ljubljana (Univerza v Ljubljani, Slovenia) were used.
4.8. PCR Screening for Virulence and Beta-Lactamase Genes
The following gene fragments were amplified: those encoding toxins (cnf1, east1, ehxA, estI, estII, eltA, hlyA, hlyF, stx1, and stx2), adhesins (fimH, papC, sfaDE, afa/draBC, iha and flu), protectins (ompT, kpsMTII, and iss), iron uptake system proteins (iroN and iutA), UPEC-specific protein (usp), and beta-lactamase genes (blaTEM, blaSHV, blaOXA, and blaCTX-M) using the primers listed in Table 7. The reaction mixtures were prepared using reagents produced by Syntol LLC (Moscow, Russia). The reactions were conducted on a DNA Engine Dyad thermocycler (Bio-Rad, Foster City, CA, USA) under the conditions previously described in the relevant literature sources, which also provided the sequences of the oligonucleotides. PCR products were separated by electrophoresis in 1% agarose gel in the presence of ethidium bromide. The bands were then visualized using a Gel-Doc XR gel documentation system (Bio-Rad, Foster City, CA, USA).

Table 7.
Oligonucleotides used in the work.
4.9. Genomic DNA Isolation and Assessment of Its Quality and Quantity
Genomic DNA from E. coli strain APEC 36 was isolated from overnight culture grown in LB using GeneJET Genomic DNA purification kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. DNA quality was assessed by electrophoresis in 1% agarose gel and using a Nanodrop spectrophotometer (Thermo Fisher Scientific). The 260/280 nm absorbance ratio demonstrated a value of 1.96, 260/230 nm, and an absorbance ratio of 2.21. According to the manufacturer’s recommendations, DNA quality is considered good for nanopore sequencing if the A260/A280 ratio is 1.8–2.0 and A260/A230 ratio is 2.0–2.2. Precise DNA concentration was determined using a Qubit 3.0 fluorimeter (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). Sample preparation for fluorimetry was performed using the Qudye dsDNA HS Assay kit (Lumiprobe RUS, Moscow, Russia). The measured DNA concentration of APEC 36 strain was 209.86 ng/μL.
4.10. DNA Library Preparation for Nanopore Sequencing
Genomic libraries were prepared from 400 ng of genome DNA using SQK-LSK109 ligation kits (Oxford Nanopore, Oxford, UK) and additional modules and enzymes NEBNext Ultra II End repair/dA-tailing Module (NEB, cat # E7546), NEBNext Quick Ligation Module (NEB, cat # E6056), NEB Blunt/TA Ligase Master Mix (NEB, cat # M0367), and NEBNext FFPE Repair Mix (NEB, M6630) according to the manufacturer’s protocol (Ligation sequencing gDNA Native Barcoding Kit 24 V14 SQK-NBD114. 24 NBE, Oxford Nanopore). The first step involved DNA repair and preparation of the ends for ligation of native adaptors, in which 1 μL of diluted control DNA (DCS) was added to the reaction mixture according to the manufacturer’s protocol. The second step was to ligate the native barcode. Adapters were ligated for 20 min at room temperature, followed by overnight at 40 °C. In the third step, adaptors were ligated for 60 min at room temperature. Reaction mixtures were stirred on a Hula mixer (Elmi, Riga, Latvia) to avoid fragmentation of long DNA strands. Purification at all steps from reaction impurities, unligated adapters, and short fragments was performed using AMPure XP magnetic particles (Beckman Coulter Brea, CA, USA), after which DNA was washed with freshly prepared 70% ethanol. The library was eluted with nuclease-free water. DNA concentration was measured using a fluorimeter.
4.11. DNA Sequencing Using Nanopore Technology
The sequencing cell of the SpotON FlowCell R10.4.1 was loaded with 20 femtomoles of a pre-prepared DNA library. At the outset of the sequencing process, the cell was loaded with 900 active pores. Sequencing was conducted on a MinION instrument (Oxford Nanopore, Oxford, UK). The Guppy basecaller 6.5.7 software was employed to convert raw data into pod5 format and generate fastq basecalls. Debarcoding of samples was accomplished using the aforementioned software in conjunction with the basecalling procedure (high accuracy model). All reads with quality Q < 8 were excluded from subsequent data analysis.
4.12. Genome Assembly and Annotation
The prior genome assembly reads were trimmed using the Chopper software (version 0.6.0) with a minimum read length of 500 and a minimum Phred average quality score of 13. A de novo genome assembly was conducted using the Flye program (version 2.9.2.) in ONT regular reads mode with two rounds of polishing [127]. The quality of the assembly was evaluated using the programs Quast (version 5.2.0, [16]) and BUSCO (version 5.5.0, [17]). Genome annotation and strain identification were conducted using Prokka (version 1.14.6, [19]) with the default settings. The BV-BRC database services [22], including RASTtk [20], the Resistance Gene Identifier (RGI, version 6.0.3) service of the Comprehensive Antibiotic Resistance Database (CARD, version 3.2.9) [31], the PubMLST database [21], and the MiGA online server [18] were also utilized.
4.13. Phylogenetic Analysis
A phylogenetic analysis of the E. coli APEC 36 strain was conducted using the comprehensive genome analysis algorithm, accessible via the BV-BCR website [22]. It comprised a number of distinct phases. First, a set of reference and representative genomes that were most similar to the query genome were identified by Mash/MinHash [128], using a pre-formed database provided by PATRIC. Secondly, PGFams [129] were selected from all the aforementioned genomes, including the query genome, and aligned using MUSCLE [130]. Subsequently, the nucleotides from the genomes were mapped to this alignment. Ultimately, the entirety of the data was consolidated into a data matrix, which was then subjected to analysis by RaxML [131]. The tree’s support values were generated with the assistance of fast bootstrapping [132].
5. Conclusions
Summarizing the information obtained by sequencing the genome of strain APEC 36 and evaluating its phenotypic properties, it can be noted that the described E. coli strain exhibits a high degree of pathogenic potential. The genome contains genes for toxins and protectins, as well as functional adhesins, hemolysins, and proteins involved in iron uptake. The presence of multiple determinants of antibiotic resistance establishes the strain’s insensitivity to a wide range of antibacterial drugs. Conversely, the potential for antibiotic resistance remains unfulfilled, with numerous resistance determinants remaining unexpressed phenotypically. Notably, this is the case with the multidrug efflux systems AcrAB-TolC and MdtEF, as well as the beta-lactamases AmpC and OXA.
This correlation of phenotypic and genotypic traits suggests a probable history of the strain’s emergence. It is plausible that the strain was formed in an environment with some level of antibacterial drug pressure, which resulted in the acquisition of the corresponding resistance genes. However, as the influence of antibiotics waned and the corresponding resistance determinants were “canned,” the cell “turned them off” to enhance fitness by redirecting resources toward growth and reproduction. Consequently, the resistance genes are present within the cells, yet they are not functional. This fact serves to increase the clinical danger of the APEC 36 isolate. It would appear that relatively short-term exposure to moderate doses of antibiotics can result in the activation of previously dormant resistance determinants, rendering the cells sensitive to a significantly reduced spectrum of antibacterial drugs and complicating the treatment of infection. This prospect is particularly concerning given the inactivity of multidrug efflux pumps, including the AcrAB-TolC system, which is the primary one for Gram-negative bacteria.
In light of the considerable diversity of biological characteristics, virulence factors, and determinants of antibiotic resistance observed among strains of agricultural origin, a comprehensive study of these factors is of paramount importance for the effective control of epidemic and epizootic situations in agricultural enterprises and the prevention of the environmental dissemination of potentially pathogenic microorganisms. A comprehensive understanding of the biology (resistance determinants, transmissible potential) of antibiotic resistance development in the main representatives of the microbiocenosis of farm animals and poultry is essential for the effective development of measures and technologies to combat antimicrobial resistance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics13100945/s1, Figure S1: Antiphage defensive systems of the E. coli APEC 36 strain.
Author Contributions
Conceptualization, D.S.K., M.V.K. and A.V.G.; methodology, M.S.S. and I.A.T.; software, E.M.K., T.K. and I.A.T.; validation, D.S.K., M.A.K. and M.V.K.; formal analysis, D.S.K., M.A.K. and E.M.K.; investigation, D.S.K., E.M.K., T.K., M.S.S., P.J.O. and M.V.K.; resources, M.V.K.; data curation, E.M.K. and T.K.; writing—original draft preparation, D.S.K., M.S.S., E.M.K. and M.V.K.; writing—review and editing, D.S.K., M.S.S., S.V.P., M.V.K., I.A.T., V.A.M. and A.V.G.; visualization, M.A.K.; supervision, D.S.K., I.A.T., S.V.P., M.V.K. and A.V.G.; project administration, A.V.G.; funding acquisition, V.A.M. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by the Ministry of Science and Higher Education of the Russian Federation (Contract in the electronic budget system no. 075-10-2021-113, project ID: RF----193021X0001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Antimicrobial Resistance, C. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- World Health Organization. Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 7 August 2024).
- Yu, D.; Banting, G.; Neumann, N.F. A review of the taxonomy, genetics, and biology of the genus Escherichia and the type species Escherichia coli. Can. J. Microbiol. 2021, 67, 553–571. [Google Scholar] [CrossRef] [PubMed]
- Belanger, L.; Garenaux, A.; Harel, J.; Boulianne, M.; Nadeau, E.; Dozois, C.M. Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic E. coli. FEMS Immunol. Med. Microbiol. 2011, 62, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mora, A.; Viso, S.; Lopez, C.; Alonso, M.P.; Garcia-Garrote, F.; Dabhi, G.; Mamani, R.; Herrera, A.; Marzoa, J.; Blanco, M.; et al. Poultry as reservoir for extraintestinal pathogenic Escherichia coli O45:K1:H7-B2-ST95 in humans. Vet. Microbiol. 2013, 167, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R. Escherichia coli and urinary tract infections: The role of poultry-meat. Clin. Microbiol. Infect. 2016, 22, 122–129. [Google Scholar] [CrossRef]
- Mageiros, L.; Meric, G.; Bayliss, S.C.; Pensar, J.; Pascoe, B.; Mourkas, E.; Calland, J.K.; Yahara, K.; Murray, S.; Wilkinson, T.S.; et al. Genome evolution and the emergence of pathogenicity in avian Escherichia coli. Nat. Commun. 2021, 12, 765. [Google Scholar] [CrossRef]
- Sora, V.M.; Meroni, G.; Martino, P.A.; Soggiu, A.; Bonizzi, L.; Zecconi, A. Extraintestinal Pathogenic Escherichia coli: Virulence Factors and Antibiotic Resistance. Pathogens 2021, 10, 1355. [Google Scholar] [CrossRef]
- Jalil, A.; Gul, S.; Bhatti, M.F.; Siddiqui, M.F.; Adnan, F. High Occurrence of Multidrug-Resistant Escherichia coli Strains in Bovine Fecal Samples from Healthy Cows Serves as Rich Reservoir for AMR Transmission. Antibiotics 2022, 12, 37. [Google Scholar] [CrossRef]
- Stanley, D.; Batacan, R., Jr.; Bajagai, Y.S. Rapid growth of antimicrobial resistance: The role of agriculture in the problem and the solutions. Appl. Microbiol. Biotechnol. 2022, 106, 6953–6962. [Google Scholar] [CrossRef]
- Santos, A.C.M.; Santos, F.F.; Silva, R.M.; Gomes, T.A.T. Diversity of Hybrid- and Hetero-Pathogenic Escherichia coli and Their Potential Implication in More Severe Diseases. Front. Cell. Infect. Microbiol. 2020, 10, 339. [Google Scholar] [CrossRef]
- Cao, H.; Bougouffa, S.; Park, T.J.; Lau, A.; Tong, M.K.; Chow, K.H.; Ho, P.L. Sharing of Antimicrobial Resistance Genes between Humans and Food Animals. mSystems 2022, 7, e0077522. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Gao, M.; Wang, X.; Qiu, T.; Guo, Y.; Zhang, L. Animal farms are hot spots for airborne antimicrobial resistance. Sci. Total Environ. 2022, 851, 158050. [Google Scholar] [CrossRef]
- Fatoba, D.O.; Amoako, D.G.; Abia, A.L.K.; Essack, S.Y. Transmission of Antibiotic-Resistant Escherichia coli from Chicken Litter to Agricultural Soil. Front. Environ. Sci. 2022, 9, 751732. [Google Scholar] [CrossRef]
- Djordjevic, S.P.; Jarocki, V.M.; Seemann, T.; Cummins, M.L.; Watt, A.E.; Drigo, B.; Wyrsch, E.R.; Reid, C.J.; Donner, E.; Howden, B.P. Genomic surveillance for antimicrobial resistance—A One Health perspective. Nat. Rev. Genet. 2024, 25, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Simao, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.L.; Gunturu, S.; Harvey, W.T.; Rossello-Mora, R.; Tiedje, J.M.; Cole, J.R.; Konstantinidis, K.T. The Microbial Genomes Atlas (MiGA) webserver: Taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acids Res. 2018, 46, W282–W288. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef] [PubMed]
- Kailasan Vanaja, S.; Bergholz, T.M.; Whittam, T.S. Characterization of the Escherichia coli O157:H7 Sakai GadE regulon. J. Bacteriol. 2009, 191, 1868–1877. [Google Scholar] [CrossRef] [PubMed]
- Gambushe, S.M.; Zishiri, O.T.; El Zowalaty, M.E. Review of Escherichia coli O157:H7 Prevalence, Pathogenicity, Heavy Metal and Antimicrobial Resistance, African Perspective. Infect. Drug Resist. 2022, 15, 4645–4673. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Hasman, H. PlasmidFinder and In Silico pMLST: Identification and Typing of Plasmid Replicons in Whole-Genome Sequencing (WGS). Methods Mol. Biol. 2020, 2075, 285–294. [Google Scholar] [CrossRef]
- Payne, L.J.; Meaden, S.; Mestre, M.R.; Palmer, C.; Toro, N.; Fineran, P.C.; Jackson, S.A. PADLOC: A web server for the identification of antiviral defence systems in microbial genomes. Nucleic Acids Res. 2022, 50, W541–W550. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Saier, M.H.; Reddy, V.S.; Moreno-Hagelsieb, G.; Hendargo, K.J.; Zhang, Y.; Iddamsetty, V.; Lam, K.J.K.; Tian, N.; Russum, S.; Wang, J.; et al. The Transporter Classification Database (TCDB): 2021 update. Nucleic Acids Res. 2021, 49, D461–D467. [Google Scholar] [CrossRef]
- Mao, C.; Abraham, D.; Wattam, A.R.; Wilson, M.J.; Shukla, M.; Yoo, H.S.; Sobral, B.W. Curation, integration and visualization of bacterial virulence factors in PATRIC. Bioinformatics 2015, 31, 252–258. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Yang, X.; Ye, W.; Qi, Y.; Ying, Y.; Xia, Z. Overcoming Multidrug Resistance in Bacteria Through Antibiotics Delivery in Surface-Engineered Nano-Cargos: Recent Developments for Future Nano-Antibiotics. Front. Bioeng. Biotechnol. 2021, 9, 696514. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Wang-Kan, X.; Neuberger, A.; van Veen, H.W.; Pos, K.M.; Piddock, L.J.V.; Luisi, B.F. Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Horiyama, T.; Nishino, K. AcrB, AcrD, and MdtABC multidrug efflux systems are involved in enterobactin export in Escherichia coli. PLoS ONE 2014, 9, e108642. [Google Scholar] [CrossRef] [PubMed]
- Anes, J.; McCusker, M.P.; Fanning, S.; Martins, M. The ins and outs of RND efflux pumps in Escherichia coli. Front. Microbiol. 2015, 6, 587. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.L.; He, G.-X.; Kakarla, P.; KC, R.; Kumar, S.; Lakra, W.S.; Mukherjee, M.M.; Ranaweera, I.; Shrestha, U.; Tran, T.; et al. Multidrug efflux pumps from Enterobacteriaceae, Vibrio cholerae and Staphylococcus aureus bacterial food pathogens. Int. J. Environ. Res. Public Health 2015, 12, 1487–1547. [Google Scholar] [CrossRef]
- Moller, T.S.; Overgaard, M.; Nielsen, S.S.; Bortolaia, V.; Sommer, M.O.; Guardabassi, L.; Olsen, J.E. Relation between tetR and tetA expression in tetracycline resistant Escherichia coli. BMC Microbiol. 2016, 16, 39. [Google Scholar] [CrossRef]
- Slipski, C.J.; Zhanel, G.G.; Bay, D.C. Biocide Selective TolC-Independent Efflux Pumps in Enterobacteriaceae. J. Membr. Biol. 2018, 251, 15–33. [Google Scholar] [CrossRef]
- Srinivasan, V.B.; Rajamohan, G. KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob. Agents Chemother. 2013, 57, 4449–4462. [Google Scholar] [CrossRef]
- Cao, Y.; Peng, Q.; Li, S.; Deng, Z.; Gao, J. The intriguing biology and chemistry of fosfomycin: The only marketed phosphonate antibiotic. RSC Adv. 2019, 9, 42204–42218. [Google Scholar] [CrossRef]
- Takahata, S.; Ida, T.; Hiraishi, T.; Sakakibara, S.; Maebashi, K.; Terada, S.; Muratani, T.; Matsumoto, T.; Nakahama, C.; Tomono, K. Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int. J. Antimicrob. Agents 2010, 35, 333–337. [Google Scholar] [CrossRef]
- Rossbach, S.; Kunze, K.; Albert, S.; Zehner, S.; Gottfert, M. The Sinorhizobium meliloti EmrAB efflux system is regulated by flavonoids through a TetR-like regulator (EmrR). Mol. Plant-Microbe Interact. 2014, 27, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Takumi-Kobayashi, A.; Theisen, U.; Hirata, T.; Nishino, K.; Yamaguchi, A. AcrS/EnvR represses expression of the acrAB multidrug efflux genes in Escherichia coli. J. Bacteriol. 2008, 190, 6276–6279. [Google Scholar] [CrossRef] [PubMed]
- Subhadra, B.; Kim, J.; Kim, D.H.; Woo, K.; Oh, M.H.; Choi, C.H. Local Repressor AcrR Regulates AcrAB Efflux Pump Required for Biofilm Formation and Virulence in Acinetobacter nosocomialis. Front. Cell. Infect. Microbiol. 2018, 8, 270. [Google Scholar] [CrossRef] [PubMed]
- Praski Alzrigat, L.; Huseby, D.L.; Brandis, G.; Hughes, D. Resistance/fitness trade-off is a barrier to the evolution of MarR inactivation mutants in Escherichia coli. J. Antimicrob. Chemother. 2021, 76, 77–83. [Google Scholar] [CrossRef]
- Vinue, L.; Hooper, D.C.; Jacoby, G.A. Chromosomal mutations that accompany qnr in clinical isolates of Escherichia coli. Int. J. Antimicrob. Agents 2018, 51, 479–483. [Google Scholar] [CrossRef]
- Alekshun, M.N.; Kim, Y.S.; Levy, S.B. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol. Microbiol. 2000, 35, 1394–1404. [Google Scholar] [CrossRef]
- Shimada, T.; Yamamoto, K.; Ishihama, A. Involvement of the leucine response transcription factor LeuO in regulation of the genes for sulfa drug efflux. J. Bacteriol. 2009, 191, 4562–4571. [Google Scholar] [CrossRef]
- Shimada, T.; Bridier, A.; Briandet, R.; Ishihama, A. Novel roles of LeuO in transcription regulation of E. coli genome: Antagonistic interplay with the universal silencer H-NS. Mol. Microbiol. 2011, 82, 378–397. [Google Scholar] [CrossRef]
- Nishino, K.; Yamaguchi, A. Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J. Bacteriol. 2001, 183, 1455–1458. [Google Scholar] [CrossRef]
- Pomposiello, P.J.; Demple, B. Redox-operated genetic switches: The SoxR and OxyR transcription factors. Trends Biotechnol. 2001, 19, 109–114. [Google Scholar] [CrossRef]
- Aly, S.A.; Boothe, D.M.; Suh, S.J. A novel alanine to serine substitution mutation in SoxS induces overexpression of efflux pumps and contributes to multidrug resistance in clinical Escherichia coli isolates. J. Antimicrob. Chemother. 2015, 70, 2228–2233. [Google Scholar] [CrossRef] [PubMed]
- Misawa, K.; Tarumoto, N.; Tamura, S.; Osa, M.; Hamamoto, T.; Yuki, A.; Kouzaki, Y.; Imai, K.; Ronald, R.L.; Yamaguchi, T.; et al. Single nucleotide polymorphisms in genes encoding penicillin-binding proteins in beta-lactamase-negative ampicillin-resistant Haemophilus influenzae in Japan. BMC Res. Notes 2018, 11, 53. [Google Scholar] [CrossRef] [PubMed]
- Prezioso, S.M.; Brown, N.E.; Goldberg, J.B. Elfamycins: Inhibitors of elongation factor-Tu. Mol. Microbiol. 2017, 106, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Zeef, L.A.; Bosch, L.; Anborgh, P.H.; Cetin, R.; Parmeggiani, A.; Hilgenfeld, R. Pulvomycin-resistant mutants of E. coli elongation factor Tu. EMBO J. 1994, 13, 5113–5120. [Google Scholar] [CrossRef] [PubMed]
- Philippon, A.; Arlet, G.; Labia, R.; Iorga, B.I. Class C beta-Lactamases: Molecular Characteristics. Clin. Microbiol. Rev. 2022, 35, e0015021. [Google Scholar] [CrossRef]
- Zarate, S.G.; De la Cruz Claure, M.L.; Benito-Arenas, R.; Revuelta, J.; Santana, A.G.; Bastida, A. Overcoming Aminoglycoside Enzymatic Resistance: Design of Novel Antibiotics and Inhibitors. Molecules 2018, 23, 284. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside modifying enzymes. Drug Resist. Updat. 2010, 13, 151–171. [Google Scholar] [CrossRef]
- Takahashi, Y.; Igarashi, M. Destination of aminoglycoside antibiotics in the ‘post-antibiotic era’. J. Antibiot. 2017, 71, 4–14. [Google Scholar] [CrossRef]
- Ghafoori, S.M.; Robles, A.M.; Arada, A.M.; Shirmast, P.; Dranow, D.M.; Mayclin, S.J.; Lorimer, D.D.; Myler, P.J.; Edwards, T.E.; Kuhn, M.L.; et al. Structural characterization of a Type B chloramphenicol acetyltransferase from the emerging pathogen Elizabethkingia anophelis NUHP1. Sci. Rep. 2021, 11, 9453. [Google Scholar] [CrossRef]
- Franck, E.; Crofts, T.S. History of the streptothricin antibiotics and evidence for the neglect of the streptothricin resistome. npj Antimicrob. Resist. 2024, 2, 3. [Google Scholar] [CrossRef]
- Harbottle, J.; Mosaei, H.; Allenby, N.; Zenkin, N. Kanglemycin A Can Overcome Rifamycin Resistance Caused by ADP-Ribosylation by Arr Protein. Antimicrob. Agents Chemother. 2021, 65, e0086421. [Google Scholar] [CrossRef] [PubMed]
- Venkatesan, M.; Fruci, M.; Verellen, L.A.; Skarina, T.; Mesa, N.; Flick, R.; Pham, C.; Mahadevan, R.; Stogios, P.J.; Savchenko, A. Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics. Nat. Commun. 2023, 14, 4031. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Qiao, W.; An, Q.; Yang, T.; Luo, Y. Dihydrofolate reductase inhibitors for use as antimicrobial agents. Eur. J. Med. Chem. 2020, 195, 112268. [Google Scholar] [CrossRef] [PubMed]
- Miranda, C.D.; Concha, C.; Godoy, F.A.; Lee, M.R. Aquatic Environments as Hotspots of Transferable Low-Level Quinolone Resistance and Their Potential Contribution to High-Level Quinolone Resistance. Antibiotics 2022, 11, 1487. [Google Scholar] [CrossRef]
- Borzi, M.M.; Cardozo, M.V.; Oliveira, E.S.; Pollo, A.S.; Guastalli, E.A.L.; Santos, L.F.D.; Avila, F.A. Characterization of avian pathogenic Escherichia coli isolated from free-range helmeted guineafowl. Braz. J. Microbiol. 2018, 49 (Suppl. S1), 107–112. [Google Scholar] [CrossRef]
- Logue, C.M.; Wannemuehler, Y.; Nicholson, B.A.; Doetkott, C.; Barbieri, N.L.; Nolan, L.K. Comparative Analysis of Phylogenetic Assignment of Human and Avian ExPEC and Fecal Commensal Escherichia coli Using the (Previous and Revised) Clermont Phylogenetic Typing Methods and its Impact on Avian Pathogenic Escherichia coli (APEC) Classification. Front. Microbiol. 2017, 8, 283. [Google Scholar] [CrossRef]
- Oliveira, E.S.; Cardozo, M.V.; Borzi, M.M.; Borges, C.A.; Guastalli, E.A.L.; Ávila, F.A. Highly Pathogenic and Multidrug Resistant Avian Pathogenic Escherichia coli in Free-Range Chickens from Brazil. Braz. J. Poult. Sci. 2019, 21, eRBCA-2019-0876. [Google Scholar] [CrossRef]
- van Overbeek, L.S.; Wichers, J.H.; van Amerongen, A.; van Roermund, H.J.W.; van der Zouwen, P.; Willemsen, P.T.J. Circulation of Shiga Toxin-Producing Escherichia coli Phylogenetic Group B1 Strains Between Calve Stable Manure and Pasture Land With Grazing Heifers. Front. Microbiol. 2020, 11, 1355. [Google Scholar] [CrossRef]
- Cheng, R.; Huang, F.; Wu, H.; Lu, X.; Yan, Y.; Yu, B.; Wang, X.; Zhu, B. A nucleotide-sensing endonuclease from the Gabija bacterial defense system. Nucleic Acids Res. 2021, 49, 5216–5229. [Google Scholar] [CrossRef]
- Stokar-Avihail, A.; Fedorenko, T.; Hor, J.; Garb, J.; Leavitt, A.; Millman, A.; Shulman, G.; Wojtania, N.; Melamed, S.; Amitai, G.; et al. Discovery of phage determinants that confer sensitivity to bacterial immune systems. Cell 2023, 186, 1863–1876.e16. [Google Scholar] [CrossRef]
- Shaw, L.P.; Rocha, E.P.C.; MacLean, R.C. Restriction-modification systems have shaped the evolution and distribution of plasmids across bacteria. Nucleic Acids Res. 2023, 51, 6806–6818. [Google Scholar] [CrossRef] [PubMed]
- Mamontov, V.; Martynov, A.; Morozova, N.; Bukatin, A.; Staroverov, D.B.; Lukyanov, K.A.; Ispolatov, Y.; Semenova, E.; Severinov, K. Persistence of plasmids targeted by CRISPR interference in bacterial populations. Proc. Natl. Acad. Sci. USA 2022, 119, e2114905119. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Maurer, J.J.; Hubert, S.; De Villena, J.F.; McDermott, P.F.; Meng, J.; Ayers, S.; English, L.; White, D.G. Antimicrobial susceptibility and molecular characterization of avian pathogenic Escherichia coli isolates. Vet. Microbiol. 2005, 107, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Koga, V.L.; Rodrigues, G.R.; Scandorieiro, S.; Vespero, E.C.; Oba, A.; de Brito, B.G.; de Brito, K.C.; Nakazato, G.; Kobayashi, R.K. Evaluation of the Antibiotic Resistance and Virulence of Escherichia coli Strains Isolated from Chicken Carcasses in 2007 and 2013 from Parana, Brazil. Foodborne Pathog. Dis. 2015, 12, 479–485. [Google Scholar] [CrossRef]
- Wibisono, F.J.; Effendi, M.H.; Wibisono, F.M. Occurrence, antimicrobial resistance, and potential zoonosis risk of avian pathogenic Escherichia coli in Indonesia: A review. Int. J. One Health 2022, 8, 76–85. [Google Scholar] [CrossRef]
- Boulianne, M.; Arsenault, J.; Daignault, D.; Archambault, M.; Letellier, A.; Dutil, L. Drug use and antimicrobial resistance among Escherichia coli and Enterococcus spp. isolates from chicken and turkey flocks slaughtered in Quebec, Canada. Can. J. Vet. Res. 2016, 80, 49–59. [Google Scholar]
- Hiki, M.; Kawanishi, M.; Abo, H.; Kojima, A.; Koike, R.; Hamamoto, S.; Asai, T. Decreased Resistance to Broad-Spectrum Cephalosporin in Escherichia coli from Healthy Broilers at Farms in Japan After Voluntary Withdrawal of Ceftiofur. Foodborne Pathog. Dis. 2015, 12, 639–643. [Google Scholar] [CrossRef]
- Shrestha, R.D.; Agunos, A.; Gow, S.P.; Deckert, A.E.; Varga, C. Associations between antimicrobial resistance in fecal Escherichia coli isolates and antimicrobial use in Canadian turkey flocks. Front. Microbiol. 2022, 13, 954123. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. CVM GFI #213 New Animal Drugs and New Animal Drug Combination Products Administered in or on Medicated Feed or Drinking Water of Food-Producing Animals: Recommendations for Drug Sponsors for Voluntarily Aligning Product Use Conditions with GFI #209. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/cvm-gfi-213-new-animal-drugs-and-new-animal-drug-combination-products-administered-or-medicated-feed (accessed on 21 August 2024).
- Bean-Hodgins, L.; Kiarie, E.G. Mandated restrictions on the use of medically important antibiotics in broiler chicken production in Canada: Implications, emerging challenges, and opportunities for bolstering gastrointestinal function and health—A review. Can. J. Anim. Sci. 2021, 101, 602–629. [Google Scholar] [CrossRef]
- Hedman, H.D.; Vasco, K.A.; Zhang, L. A Review of Antimicrobial Resistance in Poultry Farming within Low-Resource Settings. Animals 2020, 10, 1264. [Google Scholar] [CrossRef]
- Schmerold, I.; van Geijlswijk, I.; Gehring, R. European regulations on the use of antibiotics in veterinary medicine. Eur. J. Pharm. Sci. 2023, 189, 106473. [Google Scholar] [CrossRef] [PubMed]
- Shchepetkina, S.V. Antibiotics in poultry farming: To prohibit cannot be rationed. Effic. Livest. Breed. 2019, 4, 80–84. [Google Scholar] [CrossRef]
- Koo, M.S.; Lee, J.H.; Rah, S.Y.; Yeo, W.S.; Lee, J.W.; Lee, K.L.; Koh, Y.S.; Kang, S.O.; Roe, J.H. A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO J. 2003, 22, 2614–2622. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Demple, B. SoxS, an activator of superoxide stress genes in Escherichia coli. Purification and interaction with DNA. J. Biol. Chem. 1994, 269, 18371–18377. [Google Scholar] [CrossRef]
- Koutsolioutsou, A.; Pena-Llopis, S.; Demple, B. Constitutive soxR mutations contribute to multiple-antibiotic resistance in clinical Escherichia coli isolates. Antimicrob. Agents Chemother. 2005, 49, 2746–2752. [Google Scholar] [CrossRef]
- Martin, R.G.; Rosner, J.L. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl. Acad. Sci. USA 1995, 92, 5456–5460. [Google Scholar] [CrossRef]
- Alekshun, M.N.; Levy, S.B. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J. Bacteriol. 1999, 181, 4669–4672. [Google Scholar] [CrossRef]
- Martin, R.G.; Jair, K.W.; Wolf, R.E., Jr.; Rosner, J.L. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J. Bacteriol. 1996, 178, 2216–2223. [Google Scholar] [CrossRef][Green Version]
- Du, D.; Wang, Z.; James, N.R.; Voss, J.E.; Klimont, E.; Ohene-Agyei, T.; Venter, H.; Chiu, W.; Luisi, B.F. Structure of the AcrAB-TolC multidrug efflux pump. Nature 2014, 509, 512–515. [Google Scholar] [CrossRef]
- Ma, D.; Alberti, M.; Lynch, C.; Nikaido, H.; Hearst, J.E. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 1996, 19, 101–112. [Google Scholar] [CrossRef]
- Geng, H.; Jiang, R. cAMP receptor protein (CRP)-mediated resistance/tolerance in bacteria: Mechanism and utilization in biotechnology. Appl. Microbiol. Biotechnol. 2015, 99, 4533–4543. [Google Scholar] [CrossRef] [PubMed]
- Tramonti, A.; Visca, P.; De Canio, M.; Falconi, M.; De Biase, D. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 2002, 184, 2603–2613. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Senda, Y.; Yamaguchi, A. The AraC-family regulator GadX enhances multidrug resistance in Escherichia coli by activating expression of mdtEF multidrug efflux genes. J. Infect. Chemother. 2008, 14, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Dorman, C.J. H-NS: A universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2004, 2, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Marden, J.N.; Diaz, M.R.; Walton, W.G.; Gode, C.J.; Betts, L.; Urbanowski, M.L.; Redinbo, M.R.; Yahr, T.L.; Wolfgang, M.C. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2013, 110, 15055–15060. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Ohnishi, H.; Yamamoto, K.; Furuta, E.; Tanabe, H.; Utsumi, R. Transcription of emrKY is regulated by the EvgA-EvgS two-component system in Escherichia coli K-12. Biosci. Biotechnol. Biochem. 2000, 64, 1203–1209. [Google Scholar] [CrossRef]
- Law, C.J.; Alegre, K.O. Clamping down on drugs: The Escherichia coli multidrug efflux protein MdtM. Res. Microbiol. 2018, 169, 461–467. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, M.; Horiyama, T.; Zhang, Y.; Li, X.; Nishino, K.; Yan, A. The multidrug efflux pump MdtEF protects against nitrosative damage during the anaerobic respiration in Escherichia coli. J. Biol. Chem. 2011, 286, 26576–26584. [Google Scholar] [CrossRef]
- Li, X.Z.; Nikaido, H. Efflux-mediated drug resistance in bacteria: An update. Drugs 2009, 69, 1555–1623. [Google Scholar] [CrossRef]
- Lomovskaya, O.; Lewis, K.; Matin, A. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 1995, 177, 2328–2334. [Google Scholar] [CrossRef]
- Tavio, M.M.; Jacoby, G.A.; Hooper, D.C. QnrS1 structure-activity relationships. J. Antimicrob. Chemother. 2014, 69, 2102–2109. [Google Scholar] [CrossRef] [PubMed]
- Juraschek, K.; Kasbohrer, A.; Malorny, B.; Schwarz, S.; Meemken, D.; Hammerl, J.A. Dissection of Highly Prevalent qnrS1-Carrying IncX Plasmid Types in Commensal Escherichia coli from German Food and Livestock. Antibiotics 2021, 10, 1236. [Google Scholar] [CrossRef] [PubMed]
- Nasrollahian, S.; Graham, J.P.; Halaji, M. A review of the mechanisms that confer antibiotic resistance in pathotypes of E. coli. Front. Cell. Infect. Microbiol. 2024, 14, 1387497. [Google Scholar] [CrossRef] [PubMed]
- Papouskova, A.; Masarikova, M.; Valcek, A.; Senk, D.; Cejkova, D.; Jahodarova, E.; Cizek, A. Genomic analysis of Escherichia coli strains isolated from diseased chicken in the Czech Republic. BMC Vet. Res. 2020, 16, 189. [Google Scholar] [CrossRef]
- Sun, S.; Selmer, M.; Andersson, D.I. Resistance to beta-lactam antibiotics conferred by point mutations in penicillin-binding proteins PBP3, PBP4 and PBP6 in Salmonella enterica. PLoS ONE 2014, 9, e97202. [Google Scholar] [CrossRef]
- Hossain, M.K.; Rahman, M.; Nahar, A.; Khair, A.; Alam, M.M. Isolation and identification of diarrheagenic Escherichia coli causing colibacillosis in calf in selective areas of Bangladesh. Bangl. J. Vet. Med. 2013, 11, 145–149. [Google Scholar] [CrossRef]
- Alexander, D.B.; Zuberer, D.A. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 1991, 12, 39–45. [Google Scholar] [CrossRef]
- Brilis, V.I.; Brilene, T.A.; Lentsner Kh, P.; Lentsner, A.A. Method of studying the adhesive process of microorganisms. Lab. Delo 1986, 4, 210–212. [Google Scholar]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Chapman, T.A.; Wu, X.Y.; Barchia, I.; Bettelheim, K.A.; Driesen, S.; Trott, D.; Wilson, M.; Chin, J.J. Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl. Environ. Microbiol. 2006, 72, 4782–4795. [Google Scholar] [CrossRef]
- Kuhar, I.; Grabnar, M.; Zgur-Bertok, D. Virulence determinants of uropathogenic Escherichia coli in fecal strains from intestinal infections and healthy individuals. FEMS Microbiol. Lett. 1998, 164, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Kerenyi, M.; Allison, H.E.; Batai, I.; Sonnevend, A.; Emody, L.; Plaveczky, N.; Pal, T. Occurrence of hlyA and sheA genes in extraintestinal Escherichia coli strains. J. Clin. Microbiol. 2005, 43, 2965–2968. [Google Scholar] [CrossRef] [PubMed]
- Moulin-Schouleur, M.; Reperant, M.; Laurent, S.; Bree, A.; Mignon-Grasteau, S.; Germon, P.; Rasschaert, D.; Schouler, C. Extraintestinal pathogenic Escherichia coli strains of avian and human origin: Link between phylogenetic relationships and common virulence patterns. J. Clin. Microbiol. 2007, 45, 3366–3376. [Google Scholar] [CrossRef]
- Johnson, J.R.; Stell, A.L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 2000, 181, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Guiral, E.; Bosch, J.; Vila, J.; Soto, S.M. Prevalence of Escherichia coli among samples collected from the genital tract in pregnant and nonpregnant women: Relationship with virulence. FEMS Microbiol. Lett. 2011, 314, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Ulett, G.C.; Valle, J.; Beloin, C.; Sherlock, O.; Ghigo, J.M.; Schembri, M.A. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect. Immun. 2007, 75, 3233–3244. [Google Scholar] [CrossRef] [PubMed]
- Le Bouguenec, C.; Archambaud, M.; Labigne, A. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J. Clin. Microbiol. 1992, 30, 1189–1193. [Google Scholar] [CrossRef]
- Johnson, T.J.; Wannemuehler, Y.; Doetkott, C.; Johnson, S.J.; Rosenberger, S.C.; Nolan, L.K. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J. Clin. Microbiol. 2008, 46, 3987–3996. [Google Scholar] [CrossRef] [PubMed]
- Subedi, M.; Luitel, H.; Devkota, B.; Bhattarai, R.K.; Phuyal, S.; Panthi, P.; Shrestha, A.; Chaudhary, D.K. Antibiotic resistance pattern and virulence genes content in avian pathogenic Escherichia coli (APEC) from broiler chickens in Chitwan, Nepal. BMC Vet. Res. 2018, 14, 113. [Google Scholar] [CrossRef]
- Nakano, M.; Yamamoto, S.; Terai, A.; Ogawa, O.; Makino, S.I.; Hayashi, H.; Nair, G.B.; Kurazono, H. Structural and sequence diversity of the pathogenicity island of uropathogenic Escherichia coli which encodes the USP protein. FEMS Microbiol. Lett. 2001, 205, 71–76. [Google Scholar] [CrossRef]
- O’Hara, R.W.; Jenks, P.J.; Emery, M.; Upton, M. Rapid detection of extra-intestinal pathogenic Escherichia coli multi-locus sequence type 127 using a specific PCR assay. J. Med. Microbiol. 2019, 68, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Naas, T.; Le Thomas, I.; Karim, A.; Bingen, E.; Nordmann, P. CTX-M-type extended-spectrum beta-lactamase that hydrolyzes ceftazidime through a single amino acid substitution in the omega loop. Antimicrob. Agents Chemother. 2001, 45, 3355–3361. [Google Scholar] [CrossRef] [PubMed]
- Kotsakis, S.D.; Flach, C.F.; Razavi, M.; Larsson, D.G.J. Characterization of the First OXA-10 Natural Variant with Increased Carbapenemase Activity. Antimicrob. Agents Chemother. 2019, 63, e01817-18. [Google Scholar] [CrossRef] [PubMed]
- Colom, K.; Perez, J.; Alonso, R.; Fernandez-Aranguiz, A.; Larino, E.; Cisterna, R. Simple and reliable multiplex PCR assay for detection of blaTEM, bla(SHV) and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol. Lett. 2003, 223, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Davis, J.J.; Gerdes, S.; Olsen, G.J.; Olson, R.; Pusch, G.D.; Shukla, M.; Vonstein, V.; Wattam, A.R.; Yoo, H. PATtyFams: Protein Families for the Microbial Genomes in the PATRIC Database. Front. Microbiol. 2016, 7, 118. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).