Genotypic Characterisation and Antimicrobial Resistance of Extended-Spectrum β-Lactamase-Producing Escherichia coli in Humans, Animals, and the Environment from Lusaka, Zambia: Public Health Implications and One Health Surveillance
Abstract
:1. Introduction
2. Results
2.1. Prevalence of Antimicrobial Resistance
2.2. Distribution of MLST Sequence Type and O-Antigen Serotype
2.2.1. Prevalence of Resistance Genes for Trimethoprim, Sulphonamides, Tetracyclines, and Acquired Quinolone Resistance
2.2.2. Prevalence of β-Lactam AMR Genes in in ESBL-Producing E. coli Strains
3. Discussion
4. Materials and Methods
4.1. Study Site and Sampling
4.2. E. coli Isolation and Identification
4.3. Antimicrobial Susceptibility Testing
4.4. Whole-Genome Sequencing and Bioinformatics Analysis
4.5. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Authority, European Food Safety; European Centre for Disease Prevention and Control (ECDC). The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2020/2021. EFSA J. 2023, 21, e07867. [Google Scholar] [CrossRef]
- Ayukekbong, J.A.; Ntemgwa, M.; Atabe, A.N. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. Control 2017, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Mudenda, S.; Chomba, M.; Chabalenge, B.; Hikaambo, C.N.; Banda, M.; Daka, V.; Zulu, A.; Mukesela, A.; Kasonde, M.; Lukonde, P.; et al. Antibiotic Prescribing Patterns in Adult Patients According to the WHO AWaRe Classification: A Multi-Facility Cross-Sectional Study in Primary Healthcare Hospitals in Lusaka, Zambia. Pharmacol. Pharm. 2022, 13, 379–392. [Google Scholar] [CrossRef]
- Kasanga, M.; Shempela, D.M.; Daka, V.; Mwikisa, M.J.; Sikalima, J.; Chanda, D.; Mudenda, S. Antimicrobial resistance profiles of Escherichia coli isolated from clinical and environmental samples: Findings and implications. JAC Antimicrob. Resist. 2024, 6, dlae061. [Google Scholar] [CrossRef]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef] [PubMed]
- Odey, T.O.J.; Tanimowo, W.O.; Afolabi, K.O.; Jahid, I.K.; Reuben, R.C. Antimicrobial use and resistance in food animal production: Food safety and associated concerns in Sub-Saharan Africa. Int. Microbiol. 2024, 27, 1–23. [Google Scholar] [CrossRef]
- Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: 2022. Available online: https://www.who.int/publications/i/item/9789240062702 (accessed on 15 June 2024).
- Lipworth, S.; Vihta, K.-D.; Chau, K.; Barker, L.; George, S.; Kavanagh, J.; Davies, T.; Vaughan, A.; Andersson, M.; Jeffery, K.; et al. Molecular epidemiology of Escherichia coli and Klebsiella species bloodstream infections in Oxfordshire (UK) 2008–2018. medRxiv 2021. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Kariuki, S.; Kering, K.; Wairimu, C.; Onsare, R.; Mbae, C. Antimicrobial Resistance Rates and Surveillance in Sub-Saharan Africa: Where Are We Now? Infect. Drug Resist. 2022, 15, 3589–3609. [Google Scholar] [CrossRef]
- Antimicrobial Resistance (AMR). Available online: https://www.healthdata.org/research-analysis/health-risks-issues/antimicrobial-resistance-amr (accessed on 13 August 2024).
- Ture, Z.; Güner, R.; Alp, E. Antimicrobial stewardship in the intensive care unit. J. Intensive Med. 2023, 3, 244–253. [Google Scholar] [CrossRef]
- Moniruzzaman, M.; Hussain, M.T.; Ali, S.; Hossain, M.; Hossain, S.; Alam, M.A.U.; Galib, F.C.; Islam, T.; Paul, P.; Islam, S.; et al. Multidrug-resistant Escherichia coli isolated from patients and surrounding hospital environments in Bangladesh: A molecular approach for the determination of pathogenicity and resistance. Heliyon 2023, 9, e22109. [Google Scholar] [CrossRef]
- Girijan, S.K.; Pillai, D. Genetic diversity and prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in aquatic environments receiving untreated hospital effluents. J. Water Health 2023, 21, 66–80. [Google Scholar] [CrossRef]
- Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef]
- Kasanga, M.; Kwenda, G.; Wu, J.; Kasanga, M.; Mwikisa, M.J.; Chanda, R.; Mupila, Z.; Yankonde, B.; Sikazwe, M.; Mwila, E.; et al. Antimicrobial Resistance Patterns and Risk Factors Associated with ESBL-Producing and MDR Escherichia coli in Hospital and Environmental Settings in Lusaka, Zambia: Implications for One Health, Antimicrobial Stewardship and Surveillance Systems. Microorganisms 2023, 11, 1951. [Google Scholar] [CrossRef]
- Hayati, Z.; Rizal, S.; Putri, R. Isolation of Extended-Spectrum B-Lactamase (ESBL) Producing Escherichia coli and Klebsiella pneumiae from Dr. Zainoel Abidin General Hospital, Aceh. Int. J. Trop. Vet. Biomed. Res. 2019, 4, 16–22. [Google Scholar] [CrossRef]
- Nolan, L.K.; Li, G.; Logue, C.M. Origin and Dissemination of Antimicrobial Resistance among Uropathogenic Escherichia coli. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Ndihokubwayo, J.B.; Yahaya, A.A.; Desta, A.T.; Ki-Zerbo, G.; Odei, E.A.; Keita, B.; Pana, A.P.; Nkhoma, W. Antimicrobial resistance in the African Region: Issues, challenges and actions proposed. Afr. Health Monit. 2013, 16, 27–30. [Google Scholar]
- Masich, A.M.; Vega, A.D.; Callahan, P.; Herbert, A.; Fwoloshi, S.; Zulu, P.M.; Chanda, D.; Chola, U.; Mulenga, L.; Hachaambwa, L.; et al. Antimicrobial usage at a large teaching hospital in Lusaka, Zambia. PLoS ONE 2020, 15, e0228555. [Google Scholar] [CrossRef]
- Chirwa, E.; Mulundu, G.; Ndashe, K.; Kanongesha, K.; Simpokolwe, K.; Kachinda, W.; Hangombe, B.M. Antimicrobial Susceptibility Pattern and Detection of Extended-Spectrum Beta-Lactamase (blaCTX-M) Gene in Escherichia coli from Urinary Tract Infections at the University Teaching Hospital in Lusaka, Zambia. medRxiv 2020. [Google Scholar] [CrossRef]
- Aworh, M.K.; Kwaga, J.K.P.; Hendriksen, R.S.; Okolocha, E.C.; Harrell, E.; Thakur, S. Quinolone-resistant Escherichia coli at the interface between humans, poultry and their shared environment- a potential public health risk. One Health Outlook 2023, 5, 2. [Google Scholar] [CrossRef]
- Karanika, S.; Karantanos, T.; Arvanitis, M.; Grigoras, C.; Mylonakis, E. Fecal Colonization with Extended-spectrum Beta-lactamase–Producing Enterobacteriaceae and Risk Factors Among Healthy Individuals: A Systematic Review and Metaanalysis. Clin. Infect. Dis. 2016, 63, 310–318. [Google Scholar] [CrossRef]
- Husna, A.; Rahman, M.M.; Badruzzaman, A.T.M.; Sikder, M.H.; Islam, M.R.; Rahman, M.T.; Alam, J.; Ashour, H.M. Extended-Spectrum β-Lactamases (ESBL): Challenges and Opportunities. Biomedicines 2023, 11, 2937. [Google Scholar] [CrossRef]
- Shawa, M.; Furuta, Y.; Paudel, A.; Kabunda, O.; Mulenga, E.; Mubanga, M.; Kamboyi, H.; Zorigt, T.; Chambaro, H.; Simbotwe, M.; et al. Clonal relationship between multidrug-resistant Escherichia coli ST69 from poultry and humans in Lusaka, Zambia. FEMS Microbiol. Lett. 2022, 368, fnac004. [Google Scholar] [CrossRef]
- Shawa, M.; Furuta, Y.; Mulenga, G.; Mubanga, M.; Mulenga, E.; Zorigt, T.; Kaile, C.; Simbotwe, M.; Paudel, A.; Hang’ombe, B.; et al. Novel chromosomal insertions of ISEcp1-blaCTX-M-15 and diverse antimicrobial resistance genes in Zambian clinical isolates of Enterobacter cloacae and Escherichia coli. Antimicrob. Resist. Infect. Control 2021, 10, 79. [Google Scholar] [CrossRef]
- 27. Arcilla, M.S.; Van Hattem, J.M.; Bootsma, M.C.; van Genderen, P.J.; Goorhuis, A.; Grobusch, M.P.; Klaassen, C.H.; Lashof, A.M.O.; Schultsz, C.; Stobberingh, E.E.; et al. Prevalence and risk factors for carriage of ESBL-producing Enterobacteriaceae in a population of Dutch travellers: A cross-sectional study. Travel Med. Infect. Dis. 2020, 33, 101547. [Google Scholar] [CrossRef]
- Ribeiro, L.F.; Nespolo, N.M.; Rossi, G.A.M.; Fairbrother, J.M. Exploring Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli in Food-Producing Animals and Animal-Derived Foods. Pathogens 2024, 13, 346. [Google Scholar] [CrossRef]
- Mudenda, S.; Malama, S.; Munyeme, M.; Matafwali, S.K.; Kapila, P.; Katemangwe, P.; Mainda, G.; Mukubesa, A.N.; Hadunka, M.A.; Muma, J.B. Antimicrobial resistance profiles of Escherichia coli isolated from laying hens in Zambia: Implications and significance on one health. JAC Antimicrob. Resist 2023, 5, dlad060. [Google Scholar] [CrossRef]
- Shawa, M.; Paudel, A.; Chambaro, H.; Kamboyi, H.; Nakazwe, R.; Alutuli, L.; Zorigt, T.; Sinyawa, T.; Samutela, M.; Chizimu, J.; et al. Trends, patterns and relationship of antimicrobial use and resistance in bacterial isolates tested between 2015–2020 in a national referral hospital of Zambia. PLoS ONE 2024, 19, e0302053. [Google Scholar] [CrossRef]
- Alharazi, T.; Alhoot, M.A.; Alzubiery, T.K.; Aldarhami, A.; Bazaid, A.S.; Qanash, H.; Alcantara, J.C.; Gattan, H.S.; Alsumairy, H. Increasing Resistance of Nosocomial and Community-Acquired Escherichia coli in Clinical Samples from Hospitals and Clinics in Sana’a City. J. Pure Appl. Microbiol. 2024, 18, 1741–1751. [Google Scholar] [CrossRef]
- Alamri, A.; Hassan, B.; Hamid, M.E. Susceptibility of hospital-acquired uropathogens to first-line antimicrobial agents at a tertiary health-care hospital, Saudi Arabia. Urol. Ann. 2021, 13, 166. [Google Scholar] [CrossRef]
- Demir, M.; Kazanasmaz, H. Uropathogens and antibiotic resistance in the community and hospital-induced urinary tract infected children. J. Glob. Antimicrob. Resist. 2020, 20, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Lu, Z.; Hu, X.; Su, T.; Su, L.; Pu, H. Clinical significance of YAP1 and TAZ in esophageal squamous cell carcinoma. Medicine 2021, 100, e26597. [Google Scholar] [CrossRef]
- Sintondji, K.; Fabiyi, K.; Hougbenou, J.; Koudokpon, H.; Lègba, B.; Amoussou, H.; Haukka, K.; Dougnon, V. Prevalence and characterization of ESBL-producing Escherichia coli in healthy pregnant women and hospital environments in Benin: An approach based on Tricycle. Front. Public Health 2023, 11, 1227000. [Google Scholar] [CrossRef]
- Mudenda, S.; Malama, S.; Munyeme, M.; Hang’ombe, B.M.; Mainda, G.; Kapona, O.; Mukosha, M.; Yamba, K.; Bumbangi, F.N.; Mfune, R.L.; et al. Awareness of Antimicrobial Resistance and Associated Factors among Layer Poultry Farmers in Zambia: Implications for Surveillance and Antimicrobial Stewardship Programs. Antibiotics 2022, 11, 383. [Google Scholar] [CrossRef]
- Fenollar-Penadés, A.; Catalá-Gregori, P.; Tallá-Ferrer, V.; Castillo, M.Á.; García-Ferrús, M.; Jiménez-Belenguer, A. Evolution of the Antibiotic Resistance Levels, Multi-Resistance Patterns, and Presence of Antibiotic Resistance Genes in E. coli Isolates from the Feces of Breeding Hens during the Rearing Period. Antibiotics 2024, 13, 753. [Google Scholar] [CrossRef]
- Mohanty, D.; Das, B.K.; Kumari, P.; Dey, S.; Bera, A.K.; Sahoo, A.K.; Dasgupta, S.; Roy, S. Prevalence of Extended-Spectrum β-Lactamases (ESBLs) Producing Aeromonas spp. Isolated from Lamellidens marginalis (Lamark, 1819) of Sewage-Fed Wetland: A Phenotypic and Genotypic Approach. Microorganisms 2024, 12, 723. [Google Scholar] [CrossRef]
- Mshana, S.E.; Falgenhauer, L.; Mirambo, M.M.; Mushi, M.F.; Moremi, N.; Julius, R.; Seni, J.; Imirzalioglu, C.; Matee, M.; Chakraborty, T. Predictors of blaCTX-M-15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania. BMC Infect. Dis. 2016, 16, 187. [Google Scholar] [CrossRef]
- Mwakyoma, A.A.; Kidenya, B.R.; Minja, C.A.; Mushi, M.F.; Sandeman, A.; Sabiti, W.; Holden, M.T.G.; Mshana, S.E. Allele distribution and phenotypic resistance to ciprofloxacin and gentamicin among extended-spectrum β-lactamase-producing Escherichia coli isolated from the urine, stool, animals, and environments of patients with presumptive urinary tract infection in Tanzania. Front. Antibiot. 2023, 2, 1164016. [Google Scholar] [CrossRef]
- Silago, V.; Kovacs, D.; Samson, H.; Seni, J.; Matthews, L.; Oravcová, K.; Lupindu, A.M.; Hoza, A.S.; Mshana, S.E. Existence of Multiple ESBL Genes among Phenotypically Confirmed ESBL Producing Klebsiella pneumoniae and Escherichia coli Concurrently Isolated from Clinical, Colonization and Contamination Samples from Neonatal Units at Bugando Medical Center, Mwanza, Tanzania. Antibiotics 2021, 10, 476. [Google Scholar] [CrossRef]
- Islam, M.S.; Sobur, M.A.; Rahman, S.; Ballah, F.M.; Ievy, S.; Siddique, M.P.; Rahman, M.; Kafi, M.A.; Rahman, M.T. Detection of blaTEM, blaCTX-M, blaCMY, and blaSHV Genes among Extended-Spectrum Beta-Lactamase-Producing Escherichia coli Isolated from Migratory Birds Travelling to Bangladesh. Microb. Ecol. 2022, 83, 942–950. [Google Scholar] [CrossRef]
- Zhang, Z.; Guo, H.; Li, X.; Li, W.; Yang, G.; Ni, W.; Zhan, M.; Lu, L.; Zhang, Z.; Li, X.; et al. Genetic Diversity and Characteristics of blaNDM-Positive Plasmids in Escherichia coli. Front. Microbiol. 2021, 12, 729952. [Google Scholar] [CrossRef]
- Elsayed, A.G.A.; Badr, D.F.; El Kheir, N.Y.A.; Zaki, M.E.S.; Mossad, A.E.M.; Mahmoud, E.M.F. Prevalence of extended-spectrum beta-lactamase and molecular detection of blaTEM, blaSHV, and blaCTX-M genotypes among gram-negative Bacilli isolates from hospital acquired infections in pediatrics, one institutional study. Ital. J. Pediatr. 2024, 50, 31. [Google Scholar] [CrossRef]
- Al-Tahish, G.A.A.; Al-Yosaffi, E.A.; Othman, A.M.; Al-Shamahy, H.A.; Al-Haddad, A.M.; Al-Moyed, K.A.; Al-Shawkany, A.-A.-R.M. Prevalence of blatem, blashv, and blactx-m genes among esbl-producing Escherichia coli isolated from the blood samples of icus patients of university hospitals in Sana’a city, Yemen. Univ. J. Pharm. Res. 2024, 8, 6. [Google Scholar] [CrossRef]
- Hamwi, A.M.; Salem-Sokhn, E. High frequency and molecular characterization of ESBL-producing Enterobacteriaceae isolated from wound infections in North Lebanon. Expert. Rev. Anti-Infect. Ther. 2023, 21, 901–909. [Google Scholar] [CrossRef]
- Telling, K.; Brauer, A.; Laht, M.; Kalmus, P.; Toompere, K.; Kisand, V.; Maimets, M.; Remm, M.; Tenson, T.; Lutsar, I. Characteristics of Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae and Contact to Animals in Estonia. Microorganisms 2020, 8, 1130. [Google Scholar] [CrossRef]
- Tadesse, S.; Mulu, W.; Genet, C.; Kibret, M.; Belete, M.A. Emergence of High Prevalence of Extended-Spectrum Beta-Lactamase and Carbapenemase-Producing Enterobacteriaceae Species among Patients in Northwestern Ethiopia Region. Biomed. Res. Int. 2022, 2022, 5727638. [Google Scholar] [CrossRef]
- Vargas-Gutierrez, P.; Silva-Sanchez, J.; Uribe-Salas, F.J.; Lopez-Jasso, F.; Juarez-Perez, E.Y.; del Rocio Gonzalez-Martinez, M.; Camacho, H.B. Molecular Characterization of Extended Spectrum β-Lactamase-Producing Escherichia coli: Insights into the O25b-ST131 Clone in Mexican Urinary Tract Infections. Jundishapur J. Microbiol. 2024, 17. [Google Scholar] [CrossRef]
- Szymankiewicz, M.; Stefaniuk, E.; Baraniak, A.; Nowikiewicz, T. Clinical and Molecular Findings of Infections Caused by Extended-Spectrum β-Lactamase-Producing Enterobacterales in Patients with Solid Tumors: A Single-Center Study. Microb. Drug Resist. 2021, 27, 1470–1481. [Google Scholar] [CrossRef]
- Huang, J.; Lv, C.; Li, M.; Rahman, T.; Chang, Y.-F.; Guo, X.; Song, Z.; Zhao, Y.; Li, Q.; Ni, P.; et al. Carbapenem-resistant Escherichia coli exhibit diverse spatiotemporal epidemiological characteristics across the globe. Commun. Biol. 2024, 7, 51. [Google Scholar] [CrossRef]
- Huang, L.; Hu, H.; Xu, C.; Zhou, M.; Li, Y.; Li, Y.; Wu, S.; Dong, N. Characterization of NDM-5-Producing Escherichia coli Strains Isolated from Pediatric Patients with Bloodstream Infections in a Chinese Hospital. Genes 2023, 14, 520. [Google Scholar] [CrossRef]
- Flament-Simon, S.-C.; García, V.; Duprilot, M.; Mayer, N.; Alonso, M.P.; García-Meniño, I.; Blanco, J.E.; Blanco, M.; Nicolas-Chanoine, M.-H.; Blanco, J. High Prevalence of ST131 Subclades C2-H30Rx and C1-M27 Among Extended-Spectrum β-Lactamase-Producing Escherichia coli Causing Human Extraintestinal Infections in Patients From Two Hospitals of Spain and France During 2015. Front. Cell Infect Microbiol. 2020, 10, 125. [Google Scholar] [CrossRef]
- Miranda-Estrada, L.I.; Ruíz-Rosas, M.; Molina-López, J.; Parra-Rojas, I.; González-Villalobos, E.; Castro-Alarcón, N. Relationship between virulence factors, resistance to antibiotics and phylogenetic groups of uropathogenic Escherichia coli in two locations in Mexico. Enfermedades Infecc. Y Microbiol. Clin. 2017, 35, 426–433. [Google Scholar] [CrossRef]
- Total Number of Households by Province, Zambia. Zambia Statistics Agency. 2022. Available online: https://www.zamstats.gov.zm/total-number-of-household-by-province-zambia-2022/ (accessed on 24 July 2024).
- 2022 Census—Zambia Statistics Agency. Available online: https://www.zamstats.gov.zm/2022-census/ (accessed on 9 September 2024).
- M100 Ed34|Performance Standards for Antimicrobial Susceptibility Testing, 34th Edition. Available online: https://clsi.org/standards/products/microbiology/documents/m100/ (accessed on 10 September 2024).
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Leggett, R.M.; Ramirez-Gonzalez, R.H.; Clavijo, B.J.; Waite, D.; Davey, R.P. Sequencing quality assessment tools to enable data-driven informatics for high throughput genomics. Front. Genet. 2013, 4, 288. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-sites: Rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2016, 2, e000056. [Google Scholar] [CrossRef]
- Tavaré, S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect. Math. Life Sci. 1986, 17, 57–86. [Google Scholar]
- Letunic, I.; Bork, P. Intesractive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]

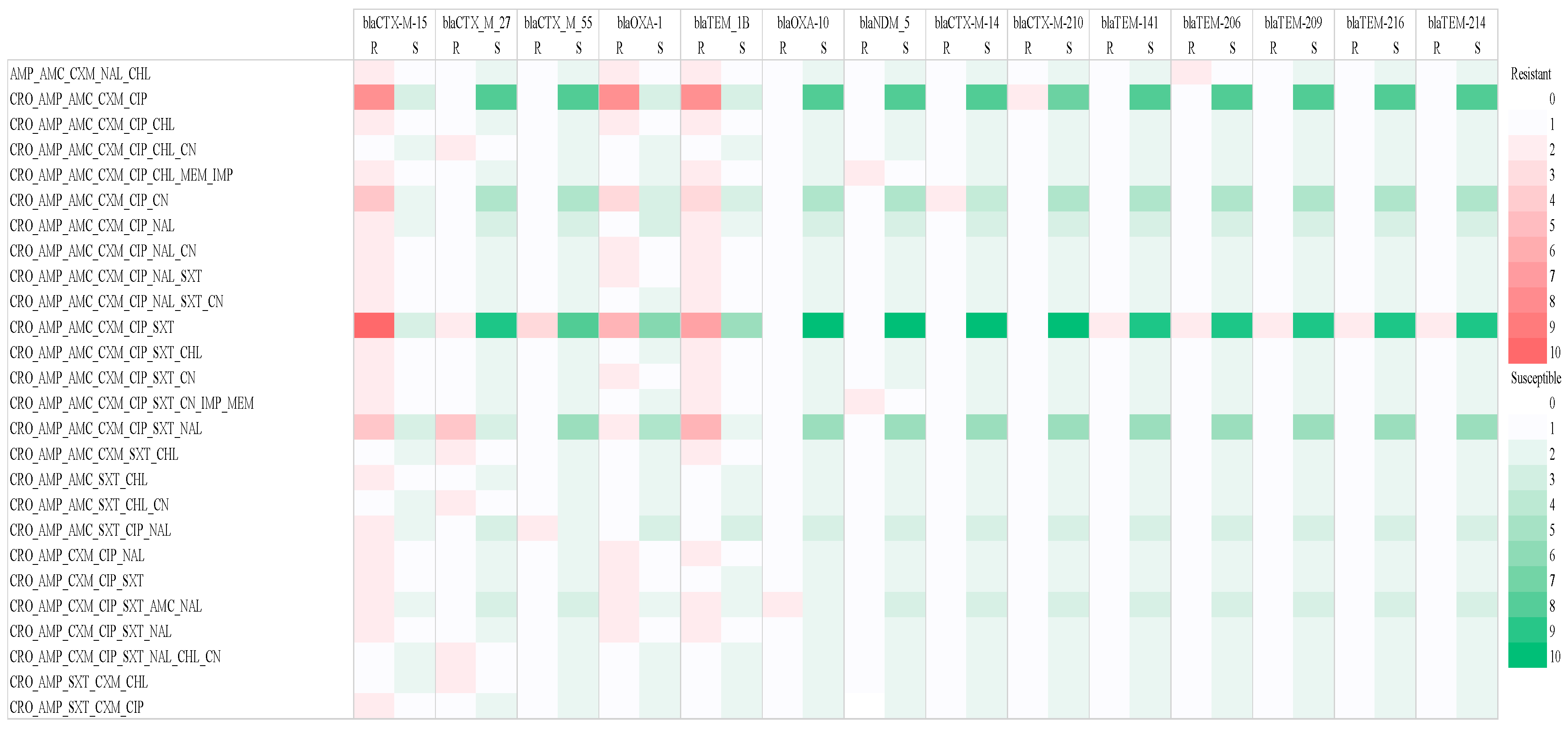



| Resistant Phenotype | Phylogroup | |
|---|---|---|
| Environment | Clinical | |
| AMP_AMC_CXM_NAL_CHL | B2 | |
| CRO_AMP_AMC_CXM_CIP | B2 | A, B2, F |
| CRO_AMP_AMC_CXM_CIP_CHL | B2 | |
| CRO_AMP_AMC_CXM_CIP_CHL_CN | A | |
| CRO_AMP_AMC_CXM_CIP_CHL_MEM_IMP | A | |
| CRO_AMP_AMC_CXM_CIP_CN | B2 | A, B2 |
| CRO_AMP_AMC_CXM_CIP_NAL | A, B1 | |
| CRO_AMP_AMC_CXM_CIP_NAL_SXT | C | |
| CRO_AMP_AMC_CXM_CIP_NAL_SXT_CN | A | |
| CRO_AMP_AMC_CXM_CIP_SXT | A, B2, C, D, G | |
| CRO_AMP_AMC_CXM_CIP_SXT_CHL | B2 | |
| CRO_AMP_AMC_CXM_CIP_SXT_CN | B2 | |
| CRO_AMP_AMC_CXM_CIP_SXT_CN_IMP_MEM | C | |
| CRO_AMP_AMC_CXM_CIP_SXT_NAL | B2 | A, B2 |
| CRO_AMP_AMC_CXM_SXT_CHL | B1 | |
| CRO_AMP_AMC_SXT_CHL | B1 | |
| CRO_AMP_AMC_SXT_CHL_CN | B1 | |
| CRO_AMP_AMC_SXT_CIP_NAL | A, C | |
| CRO_AMP_CXM_CIP_NAL | B2 | |
| CRO_AMP_CXM_CIP_SXT | B2 | |
| CRO_AMP_CXM_CIP_SXT_AMC_NAL | B1, B2 | |
| CRO_AMP_CXM_CIP_SXT_NAL | B2 | |
| CRO_AMP_CXM_CIP_SXT_NAL_CHL_CN | B2 | |
| CRO_AMP_SXT_CXM_CHL | B2 | |
| CRO_AMP_SXT_CXM_CIP | B2 | |
| Antibiotic | Clinical Sources | Environmental Sources | |||||
|---|---|---|---|---|---|---|---|
| Resistant | Intermediate | Susceptible | Resistant | Intermediate | Susceptible | p-Value | |
| Tetracyclines | 4 (3.39%) | 2 (1.69%) | 28 (23.72%) | 2 (1.69%) | 0 (0%) | 47 (39.83%) | 0.07 |
| Chloramphenicol | 0 (0%) | 1 (0.84%) | 33 (27.96%) | 1 (0.84%) | 5 (4.24%) | 43 (36.44%) | 0.33 |
| Aminoglycosides | 15 (12.71%) | 8 (6.78%) | 11 (9.32%) | 14 (11.86%) | 11 (9.32%) | 22 (18.64%) | 0.71 |
| Trimethoprim | 25 (21.18%) | 0 (0%) | 9 (7.62%) | 20 (16.94%) | 2 (1.69%) | 20 (16.94%) | 0.24 |
| Fluoroquinolones | 0 (0%) | 1 (0.84%) | 33 (27.96%) | 0 (0%) | 0 (0%) | 48 (40.67%) | 0.32 |
| Lincosamide Streptogramins’ B | 3 (2.54%) | 12 (10.17%) | 19 (16.1%) | 2 (1.69%) | 13 (11.01%) | 33 (27.69%) | 0.33 |
| Sulphonamide | 18 (15.25%) | 2 (1.69%) | 14 (11.86%) | 17 (14.4%) | 12 (10.17%) | 10 (8.47%) | 0.07 |
| Disinfectants | 1 (0.84%) | 1 (0.84%) | 32 (27.12%) | 1 (0.84%) | 0 (0%) | 48 (40.67%) | 0.41 |
| AMR Genes | Overall n (%) | Environmental n (%) | Clinical n (%) |
|---|---|---|---|
| Trimethoprim | |||
| dfrA12 | 8 (13.8) | 1 (12.5) | 7 (87.5) |
| dfrA14 | 10 (17.2) | 0 (0.0) | 10 (100.0) |
| dfrA1 | 1 (1.7) | 0 (0.0) | 1 (100.0) |
| dfrA17 | 29 (50.0) | 6 (20.7) | 23 (79.3) |
| dfrA27 | 1 (1.7) | 1 (100.0) | 0 (0.0) |
| dfrB4 | 1 (1.7) | 0 (0.0) | 1 (100.0) |
| Sulphonamides | |||
| sul1 | 37 (63.7) | 7 (18.9) | 30 (81.1) |
| sul2 | 42 (72.4) | 7 (16.7) | 35 (83.3) |
| sul3 | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Tetracycline | |||
| tet (A) | 37 (63.7) | 6 (16.2) | 31 (83.2) |
| tet (B) | 17 (29.3) | 1 (5.9) | 16 (94.1) |
| tet (M) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Acquired quinolone resistance | |||
| qnrB1 | 3 (5.1) | 0 (0.0) | 3 (100) |
| qnrS1 | 3 (5.1) | 2 (66.7) | 1 (33.3) |
| qnrB6 | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| aac (6′)-ib-cr5 | 19 (32.7) | 3 (15.8) | 16 (84.2) |
| MLSB | |||
| mdf(A) | 58 (100.0) | 9 (17.6) | 42 (82.4) |
| erm(B) | 5 (8.6) | 0 (0.0) | 5 (100.0) |
| mph(A) | 38 (65.8) | 6 (16.2) | 31 (83.8) |
| Phenicols | |||
| catA1 | 8 (15.4) | 1 (12.5) | 7 (87.5) |
| catB3 | 8 (15.4) | 0 (0.0) | 8 (100.0) |
| floR | 1 (1.9) | 0 (0.0) | 1 (100.0) |
| cmlA1 | 1 (1.9) | 0 (0.0) | 1 (100.0) |
| Disinfectants | |||
| qacE | 43 (82.7) | 8 (19.0) | 34 (81.0) |
| sitABCD | 40 (76.9) | 7 (17.9) | 32 (82.1) |
| Beta-Lactam | Urine | Pus | Blood | Water | Meat | Equipment | Vegetables |
|---|---|---|---|---|---|---|---|
| blaCTM-X-14 | 1 (1.7%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| blaCTM-X-15 | 20 (34.5%) | 6 (10.4%) | 1 (1.7%) | 2 (3.5%) | 1 (1.7%) | 1 (1.7%) | 3 (5.2%) |
| blaCTM-X-55 | 1 (1.7%) | 1 (1.7%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| blaCTM-X-27 | 8 (13.8%) | 1 (1.7%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (1.7%) | 0 (0.0%) |
| blaOXA-1 | 15 (25.9%) | 1 (1.7%) | 0 (0.0%) | 1 (1.7%) | 0 (0.0%) | 1 (1.7%) | 1 (1.7%) |
| blaTEM-1 | 16 (27.6%) | 2 (3.5%) | 0 (0.0%) | 2 (3.5%) | 0 (0.0%) | 2 (3.5%) | 1 (1.7%) |
| blaTEM | 2 (3.5%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| blaTEM-1 | 16 (27.6%) | 2 (3.5%) | 0 (0.0%) | 2 (3.5%) | 0 (0.0%) | 2 (3.5%) | 1 (1.7%) |
| blaEC | 35 (60.4%) | 12 (20.7%) | 2 (3.5%) | 2 (3.5%) | 1 (1.7%) | 3 (5.2%) | 3 (5.2%) |
| blaNDM-5 | 0 (0.0%) | 1 (1.7%) | 0 (0.0%) | 1 (1.7%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasanga, M.; Gajdács, M.; Muleya, W.; Ikhimiukor, O.O.; Mudenda, S.; Kasanga, M.; Chizimu, J.; Shempela, D.M.; Solochi, B.B.; Mwikisa, M.J.; et al. Genotypic Characterisation and Antimicrobial Resistance of Extended-Spectrum β-Lactamase-Producing Escherichia coli in Humans, Animals, and the Environment from Lusaka, Zambia: Public Health Implications and One Health Surveillance. Antibiotics 2024, 13, 951. https://doi.org/10.3390/antibiotics13100951
Kasanga M, Gajdács M, Muleya W, Ikhimiukor OO, Mudenda S, Kasanga M, Chizimu J, Shempela DM, Solochi BB, Mwikisa MJ, et al. Genotypic Characterisation and Antimicrobial Resistance of Extended-Spectrum β-Lactamase-Producing Escherichia coli in Humans, Animals, and the Environment from Lusaka, Zambia: Public Health Implications and One Health Surveillance. Antibiotics. 2024; 13(10):951. https://doi.org/10.3390/antibiotics13100951
Chicago/Turabian StyleKasanga, Maisa, Márió Gajdács, Walter Muleya, Odion O. Ikhimiukor, Steward Mudenda, Maika Kasanga, Joseph Chizimu, Doreen Mainza Shempela, Benjamin Bisesa Solochi, Mark John Mwikisa, and et al. 2024. "Genotypic Characterisation and Antimicrobial Resistance of Extended-Spectrum β-Lactamase-Producing Escherichia coli in Humans, Animals, and the Environment from Lusaka, Zambia: Public Health Implications and One Health Surveillance" Antibiotics 13, no. 10: 951. https://doi.org/10.3390/antibiotics13100951
APA StyleKasanga, M., Gajdács, M., Muleya, W., Ikhimiukor, O. O., Mudenda, S., Kasanga, M., Chizimu, J., Shempela, D. M., Solochi, B. B., Mwikisa, M. J., Yamba, K., Andam, C. P., Chanda, R., Chanda, D., & Kwenda, G. (2024). Genotypic Characterisation and Antimicrobial Resistance of Extended-Spectrum β-Lactamase-Producing Escherichia coli in Humans, Animals, and the Environment from Lusaka, Zambia: Public Health Implications and One Health Surveillance. Antibiotics, 13(10), 951. https://doi.org/10.3390/antibiotics13100951








