Identifying Potential Natural Antibiotics from Unani Formulas through Machine Learning Approaches
Abstract
1. Introduction
2. Results
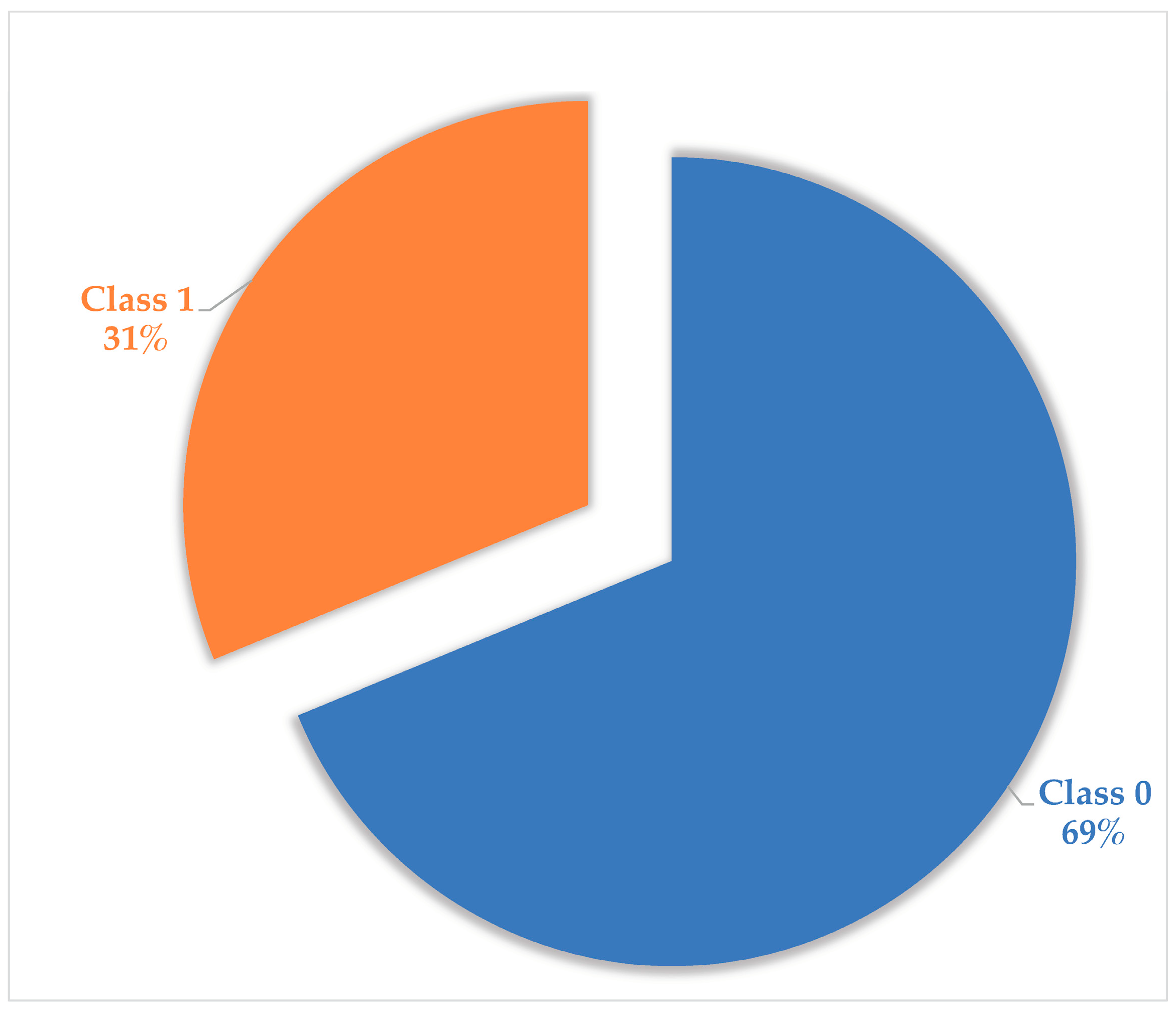
2.1. Pre-Processing of Unani Data
2.2. Machine Learning Model
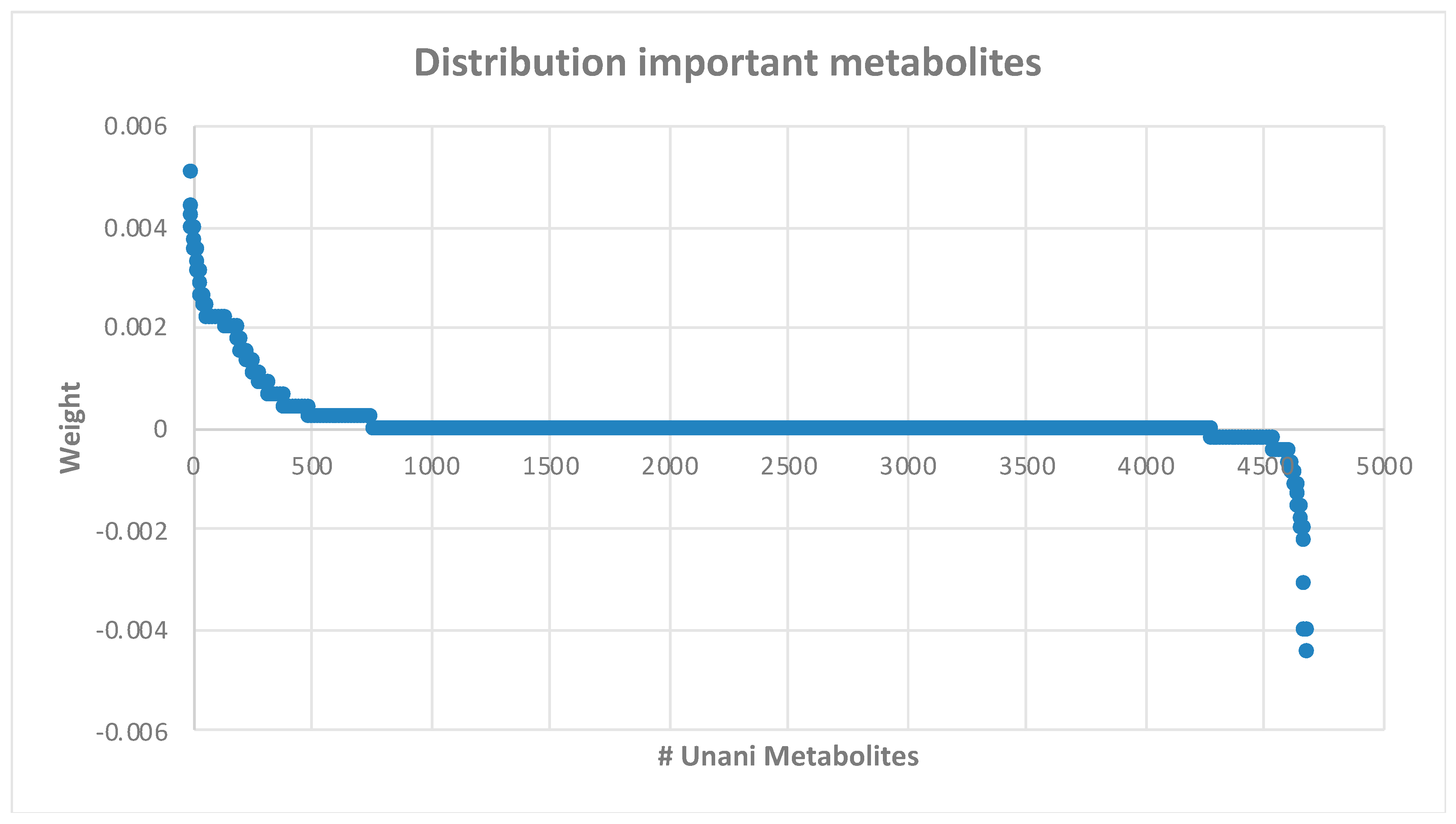
2.3. Ranking and Selecting Features
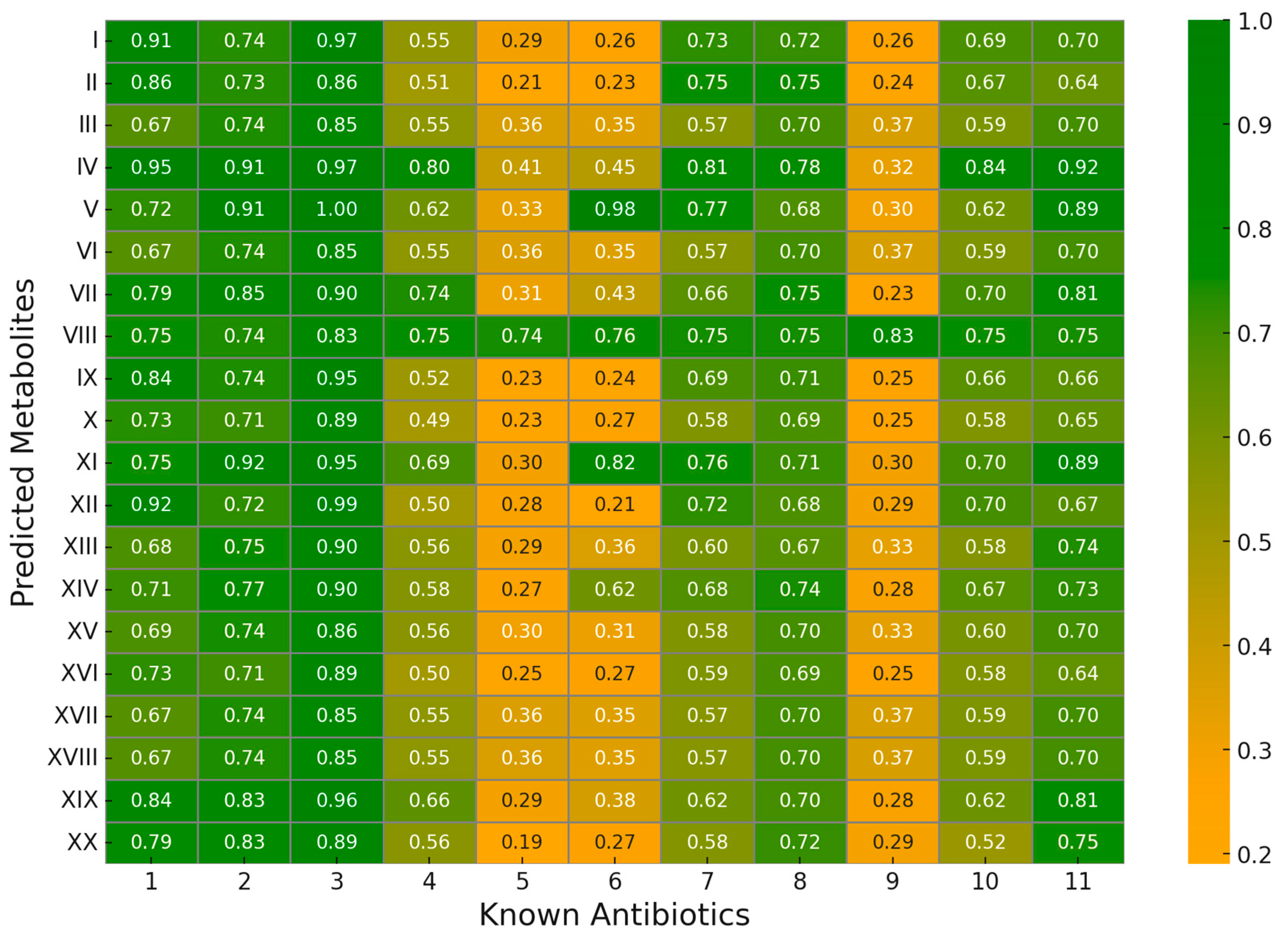
2.4. Validation of Predicted Results
2.4.1. Literature Validation
2.4.2. Structural Similarity
3. Materials and Methods
3.1. Collecting Data
3.2. Data Pre-Processing
3.3. Machine Learning Modeling
3.4. Ranking and Selecting Feature
3.5. Validation
- A and B are the two vectors being compared, and alpha and beta are the weights assigned to the false positives and false negatives, respectively.
- i is the index of the feature being compared.
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. 2014. Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 30 October 2021).
- Cassini, A.; Diaz Högberg, L.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar , M.E.; Devleesschauwe, B.; Cecchini, M.; et al. Attributable Deaths and Disability-Adjusted Life-Years Caused by Infections with Antibiotic-Resistant Bacteria in the EU and the European Economic Area in 2015: A Population-Level Modelling Analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 180, 688–702. [Google Scholar] [CrossRef] [PubMed]
- Corsello, S.M.; Bittker, J.A.; Liu, Z.; Gould, J.; McCarren, P.; Hirschman, J.E.; Johnston, S.E.; Vrcic, A.; Wong, B.; Khan, M.; et al. The Drug Repurposing Hub: A Next-Generation Drug Library and Information Resource. Nat. Med. 2017, 23, 405–408. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15—Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef] [PubMed]
- David, L.; Brata, A.M.; Mogosan, C.; Pop, C.; Czako, Z.; Muresan, L.; Ismaiel, A.; Dumitrascu, D.I.; Leucuta, D.C.; Stanculete, M.F.; et al. Artificial Intelligence and Antibiotic Discovery. Antibiotics 2021, 10, 1376. [Google Scholar] [CrossRef]
- Gao, P.; Nasution, A.K.; Yang, S.; Chen, Z.; Ono, N.; Kanaya, S.; Altaf-Ul-Amin, M.D. On finding natural antibiotics based on TCM formulae. Methods 2023, 214, 35–45. [Google Scholar] [CrossRef]
- Nasution, A.K.; Ono, N.; Kanaya, S.; Ul-Amin, M.A. Investigating Potential Natural Antibiotics Plants Based on Unani Formula Using Supervised Network Analysis and Machine Learning Approach. In Proceedings of the 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Istanbul, Turkey, 3–5 December 2023; IEEE: New York, NY, USA, 2023; pp. 3111–3117. [Google Scholar]
- Nasution, A.K.; Wijaya, S.H.; Gao, P.; Islam, R.M.; Huang, M.; Ono, N.; Kanaya, S.; Altaf-Ul-Amin, M. Prediction of Potential Natural Antibiotics Plants Based on Jamu Formula Using Random Forest Classifier. Antibiotics 2022, 11, 1199. [Google Scholar] [CrossRef]
- Ekor, M. The Growing Use of Herbal Medicines: Issues Relating to Adverse Reactions and Challenges in Monitoring Safety. Front. Pharmacol. 2014, 4, 177. [Google Scholar] [CrossRef]
- Wijaya, S.H.; Tanaka, Y.; Altaf-Ul-Amin, M.; Hirai Morita, A.; Mochamad Afendi, F.; Batubara, I.; Ono, N.; Darusman, L.K.; Kanaya, S. Utilization of KNApSAcK Family Databases for Developing Herbal Medicine Systems. J. Comput. Aided Chem. 2016, 17, 1–7. [Google Scholar] [CrossRef]
- Hossain, S.F.; Wijaya, S.H.; Huang, M.; Batubara, I.; Kanaya, S.; Farhad, M.A.-U.-A. Prediction of Plant-Disease Relations Based on Unani Formulas by Network Analysis. In Proceedings of the 2018 IEEE 18th International Conference on Bioinformatics and Bioengineering (BIBE), Taichung, Taiwan, 29–31 October 2018; IEEE: New York, NY, USA, 2018; pp. 348–351. [Google Scholar]
- Choudhury, P.R.; Talukdar, A.D.; Nath, D.; Saha, P.; Nath, R. Traditional Folk Medicine and Drug Discovery: Prospects and Outcome. Adv. Pharm. Biotechnol. Recent Prog. Future Appl. 2020, 3, 3–13. [Google Scholar]
- Wijaya, S.H.; Nasution, A.K.; Batubara, I.; Gao, P.; Huang, M.; Ono, N.; Kanaya, S.; Altaf-Ul-Amin, M. Deep Learning Approach for Predicting the Therapeutic Usages of Unani Formulas towards Finding Essential Compounds. Life 2023, 13, 439. [Google Scholar] [CrossRef] [PubMed]
- Itrat, M. Methods of health promotion and disease prevention in Unani medicine. J. Educ. Health Promot. 2020, 9, 168. [Google Scholar] [CrossRef] [PubMed]
- Husain, A.; Sofi, G.; Tajuddin, T.; Dang, R.; Kumar, N. Unani System of Medicine—Introduction and Challenges. Med. J. Islam. World Acad. Sci. 2010, 18, 27–30. [Google Scholar]
- Makbul, S.A.A.; Jahan, N.; Kalam, M.A. Bio-active Compounds from Unani Medicinal Plants and Their Application in Urolithiasis. In Natural Bio-Active Compounds; Swamy, M., Akhtar, M., Eds.; Springer: Singapore, 2019. [Google Scholar]
- SY, I. Unani System of Medicine; Department of AYUSH, Ministry of Health and Family Welfare, Government of India: New Delhi, India, 2013.
- Yadav, V.; Krishnan, A.; Vohora, D. A systematic review on Piper longum L.: Bridging traditional knowledge and pharmacological evidence for future translational research. J. Ethnopharmacol. 2020, 247, 112255. [Google Scholar] [CrossRef] [PubMed]
- Gawai, K.R.; Kodam, K.M.; Kuchekar, B.S.; Chabukswar, A.R.; Jagdale, S.C.; Lokhande, P.D. Antibacterial Activity of Extracts of Piper longum. J. Pharmacol. Toxicol. 2007, 2, 574–579. [Google Scholar]
- Moein, M.R.; Zomorodian, K.; Pakshir, K.; Yavari, F.; Motamedi, M.; Zarshenas, M.M. Trachyspermum ammi (L.) Sprague: Chemical composition of essential oil and antimicrobial activities of respective fractions. J. Evid. Based Complement. Altern. Med. 2015, 20, 50–56. [Google Scholar] [CrossRef]
- Celedon, J.M.; Bohlmann, J. Genomics-based discovery of plant genes for synthetic biology of terpenoid fragrances: A case study in sandalwood oil biosynthesis. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 47–67. [Google Scholar]
- Zhang, L.-L.; Zhang, L.-F.; Hu, Q.-P.; Hao, D.-L.; Xu, J.-G. Chemical composition, antibacterial activity of Cyperus rotundus rhizomes essential oil against Staphylococcus aureus via membrane disruption and apoptosis pathway. Food Control 2017, 80, 290–296. [Google Scholar] [CrossRef]
- Badet, C. Antibacterial activity of grape (Vitis vinifera, Vitis rotundifolia) seeds. In Nuts and Seeds in Health and Disease Prevention; Elsevier: Amsterdam, The Netherlands, 2011; pp. 545–552. [Google Scholar]
- Radulescu, C.; Buruleanu, L.C.; Nicolescu, C.M.; Olteanu, R.L.; Bumbac, M.; Holban, G.C.; Simal-Gandara, J. Phytochemical profiles, antioxidant and antibacterial activities of grape (Vitis vinifera L.) seeds and skin from organic and conventional vineyards. Plants 2020, 9, 1470. [Google Scholar] [CrossRef]
- El Mihyaoui, A.; da Silva, J.C.G.; Charfi, S.; Candela Castillo, M.E.; Lamarti, A.; Arnao, M.B. Chamomile (Matricaria chamomilla L.): A review of ethnomedicinal use, phytochemistry and pharmacological uses. Life 2022, 12, 479. [Google Scholar] [CrossRef]
- Malu, S.P.; Obochi, G.O.; Tawo, E.N.; Nyong, B.E. Antibacterial activity and medicinal properties of ginger (Zingiber officinale). Glob. J. Pure Appl. Sci. 2009, 15, 3–4. [Google Scholar] [CrossRef][Green Version]
- Srivastava, N.; Nandi, I.; Ibeyaima, A.; Gupta, S.; Sarethy, I.P. Microbial Diversity of a Himalayan Forest and Characterization of Rare Actinomycetes for Antimicrobial Compounds. 3 Biotech 2019, 9, 27. [Google Scholar] [CrossRef]
- Patra, A.K. An Overview of Antimicrobial Properties of Different Classes of Phytochemicals. In Dietary Phytochemicals and Microbes; Patra, A.K., Ed.; Springer: Dordrecht, The Netherlands, 2012; pp. 1–32. [Google Scholar]
- Allemailem, K. Antimicrobial Potential of Naturally Occurring Bioactive Secondary Metabolites. J. Pharm. Bioallied Sci. 2021, 13, 155. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Massih, R.M.; Debs, E.; Othman, L.; Attieh, J.; Cabrerizo, F.M. Glucosinolates, a Natural Chemical Arsenal: More to Tell than the Myrosinase Story. Front. Microbiol. 2023, 14, 1130208. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.E.; Bergaentzlé, M.; Bindler, F.; Marchioni, E.; Lintz, A.; Ennahar, S. In Vitro Efficacies of Various Isothiocyanates from Cruciferous Vegetables as Antimicrobial Agents against Foodborne Pathogens and Spoilage Bacteria. Food Control 2013, 30, 318–324. [Google Scholar] [CrossRef]
- Romeo, L.; Iori, R.; Rollin, P.; Bramanti, P.; Mazzon, E. Isothiocyanates: An Overview of Their Antimicrobial Activity against Human Infections. Molecules 2018, 23, 624. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; Kumar, P.; Marwaha, R.; Narasimhan, B. Synthesis, Antimicrobial Evaluation and QSAR Studies of 2-Chlorobenzoic Acid Derivatives. Drug Res. 2013, 64, 208–213. [Google Scholar] [CrossRef]
- Alibi, S.; Crespo, D.; Navas, J. Plant-Derived Small Molecules with Antibacterial Activity. Antibiotics 2021, 10, 231. [Google Scholar] [CrossRef]
- Maisch, N.A.; Bereswill, S.; Heimesaat, M.M. Antibacterial Effects of Vanilla Ingredients Provide Novel Treatment Options for Infections with Multidrug-Resistant Bacteria—A Recent Literature Review. Eur. J. Microbiol. Immunol. 2022, 12, 53–62. [Google Scholar] [CrossRef]
- Casillas-Vargas, G.; Ocasio-Malavé, C.; Medina, S.; Morales-Guzmán, C.; García Del Valle, R.; Carballeira, N.M.; Sanabria-Ríos, D.J. Antibacterial Fatty Acids: An Update of Possible Mechanisms of Action and Implications in the Development of the Next-Generation of Antibacterial Agents. Prog. Lipid Res. 2021, 82, 101093. [Google Scholar] [CrossRef]
- Pratama, O.A.; Tunjung, W.A.S.; Sutikno, S.; Daryono, B.S. Bioactive Compound Profile of Melon Leaf Extract (Cucumis melo L. ‘Hikapel’) Infected by Downy Mildew. Biodiversitas J. Biol. Divers. 2019, 20. [Google Scholar] [CrossRef]
- Zuegg, J.; Muldoon, C.; Adamson, G.; McKeveney, D.; Le Thanh, G.; Premraj, R.; Becker, B.; Cheng, M.; Elliott, A.G.; Huang, J.X.; et al. Carbohydrate Scaffolds as Glycosyltransferase Inhibitors with In Vivo Antibacterial Activity. Nat. Commun. 2015, 6, 7719. [Google Scholar] [CrossRef] [PubMed]
- Aryal, B.; Niraula, P.; Khadayat, K.; Adhikari, B.; Khatri Chhetri, D.; Sapkota, B.K.; Bhattarai, B.R.; Aryal, N.; Parajuli, N. Antidiabetic, Antimicrobial, and Molecular Profiling of Selected Medicinal Plants. Evid. Based Complement. Altern. Med. 2021, 2021, 5510099. [Google Scholar] [CrossRef] [PubMed]
- Nićiforović, N.; Abramovič, H. Sinapic Acid and Its Derivatives: Natural Sources and Bioactivity. Compr. Rev. Food Sci. Food Saf. 2014, 13, 34–51. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Stewart, J.D.; Ioannou, I.; Allais, F. Sinapic Acid and Sinapate Esters in Brassica: Innate Accumulation, Biosynthesis, Accessibility via Chemical Synthesis or Recovery from Biomass, and Biological Activities. Front. Chem. 2021, 9, 664602. [Google Scholar] [CrossRef] [PubMed]
- Bhatwalkar, S.B.; Mondal, R.; Krishna, S.B.N.; Adam, J.K.; Govender, P.; Anupam, R. Antibacterial Properties of Organo-Sulfur Compounds of Garlic (Allium sativum). Front. Microbiol. 2021, 12, 613077. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, G.G.F.; Locatelli, J.; Freitas, P.C.; Silva, G.L. Antibacterial Activity of Plant Extracts and Phytochemicals on Antibiotic-Resistant Bacteria. Braz. J. Microbiol. 2000, 31, 247–256. [Google Scholar] [CrossRef]
- Su, Y.; Peng, B.; Li, H.; Cheng, Z.; Zhang, T.; Zhu, J.; Li, D.; Li, M.; Ye, J.; Du, C.; et al. Pyruvate Cycle Increases Aminoglycoside Efficacy and Provides Respiratory Energy in Bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, E1578–E1587. [Google Scholar] [CrossRef]
- Ye, J.; Su, Y.; Lin, X.; Lai, S.; Li, W.; Ali, F.; Zheng, J.; Peng, B. Alanine Enhances Aminoglycosides-Induced ROS Production as Revealed by Proteomic Analysis. Front. Microbiol. 2018, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, Z.; Yang, T.; Jiang, M.; Wang, J.; Cheng, Z.; Yang, M.; Zhu, J.; Zhang, T.; Li, H.; et al. Glutamine Promotes Antibiotic Uptake to Kill Multidrug-Resistant Uropathogenic Bacteria. Sci. Transl. Med. 2021, 13, eabj0716. [Google Scholar] [CrossRef]
- Bajusz, D.; Rácz, A.; Héberger, K. Why Is Tanimoto Index an Appropriate Choice for Fingerprint-Based Similarity Calculations? J. Cheminform. 2015, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Scalfani, V.F.; Patel, V.D.; Fernandez, A.M. Visualizing Chemical Space Networks with RDKit and NetworkX. J. Cheminform. 2022, 14, 87. [Google Scholar] [CrossRef]
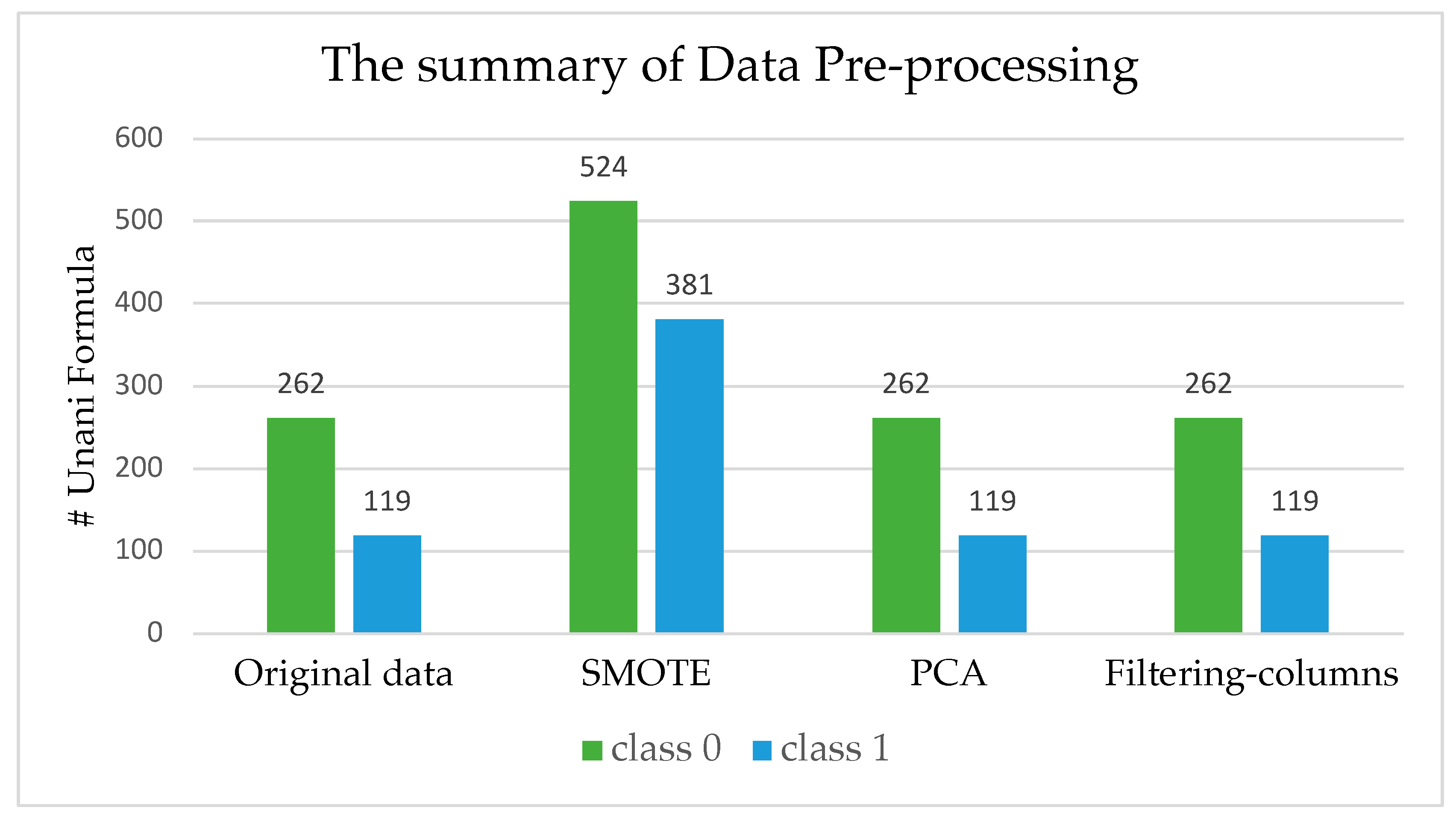

| Dataset | Number of Unani Formula | Number of Metabolite Features |
|---|---|---|
| Original data | 381 | 4688 |
| SMOTE | 905 | 4688 |
| PCA | 381 | 21 |
| Filtering columns | 381 | 501 |
| Machine Learning | Original Data | SMOTE | Feature Selection | PCA |
|---|---|---|---|---|
| AdaBoost | 0.674596 | 0.742910 | 0.677193 | 0.681545 |
| Bagging | 0.697323 | 0.818048 | 0.698200 | 0.687685 |
| BernouliNB | 0.626361 | 0.614733 | 0.658715 | 0.615117 |
| Decision Tree | 0.666712 | 0.659669 | 0.680656 | 0.67979 |
| Extra Trees | 0.630793 | 0.828361 | 0.648348 | 0.632604 |
| Gradient Boosting | 0.685042 | 0.806262 | 0.685919 | 0.681556 |
| K-Nearest Neighbors | 0.688540 | 0.706446 | 0.688551 | 0.689417 |
| Linier Discriminant Analysis | 0.601003 | 0.750645 | 0.640351 | 0.664912 |
| Logistic Regression | 0.648257 | 0.764273 | 0.688551 | 0.662303 |
| Multi-Layer Perceptron | 0.694691 | 0.838200 | 0.694680 | 0.690305 |
| Random Forest | 0.687697 | 0.825046 | 0.691194 | 0.688596 |
| Support Vector Machine | 0.697323 | 0.776427 | 0.698200 | 0.691171 |
| Measurement | Value | Derivation |
|---|---|---|
| Sensitivity | 0.9182 | TPR = TP/(TP + FN) |
| Specificity | 0.7436 | SPC = TN/(FP + TN) |
| Precision | 0.7710 | PPV = TP/(TP + FP) |
| Negative Predictive Value | 0.9063 | NPV = TN/(TN + FN) |
| False Positive Rate | 0.2564 | FPR = FP/(FP + TN) |
| False Discovery Rate | 0.2290 | FDR = FP/(FP + TP) |
| False Negative Rate | 0.0818 | FNR = FN/(FN + TP) |
| Accuracy | 0.8382 | ACC = (TP + TN)/(P + N) |
| F1-Score | 0.8382 | F1 = 2TP/(2TP + FP + FN) |
| Matthews Correlation Coefficient | 0.6695 | TP × TN − FP×FN/sqrt((TP + FP) × (TP + FN) × (TN + FP) × (TN + FN)) |
| Id | Metabolite | Formula | Molecular Mass (g/mol) |
|---|---|---|---|
| (I) | 2-hydroxyethyl hexadecanoate | C18H36O3 | 300.48 |
| (II) | N-[(+)-12-hydroxy-7 isojasmonyl]isoleucine | C18H29NO5 | 323.43 |
| (III) | gluconapin | C11H19NO9S2 | 373.40 |
| (IV) | 3-phenylpropionitrile | C9H9N | 131.18 |
| (V) | flamenol | C7H8O3 | 140.14 |
| (VI) | glucobrassicanapin(1−) | C16H19N2O9S2 | 446.46 |
| (VII) | 2-chlorobenzoic acid | C7H5ClO2 | 156.57 |
| (VIII) | 8-nitroguanosine 3′,5′-cyclic monophosphate | C10H11N6O9P | 390.21 |
| (IX) | 1,3-dioctanoylglycerol | C19H36O5 | 344.49 |
| (X) | PG(18:2(9Z,12Z)/16:0) | C40H75O10P | 758.99 |
| (XI) | 3-methoxybenzaldehyde | C8H8O2 | 138.15 |
| (XII) | Methyl stearate | C19H38O2 | 298.50 |
| (XIII) | 1-naphthyl β-D-glucoside | C16H18O6 | 306.32 |
| (XIV) | sinapine | C16H24NO5+ | 310.36 |
| (XV) | 3-butenyldesulfoglucosinolate | C12H23NO7S | 341.39 |
| (XVI) | 1-oleoyl-2-hexadecanoyl-sn-glycero-3-phospho-(1′-sn-glycerol-3′-phosphate) | C18H34O2 | 282.46 |
| (XVII) | glucobrassicanapin | C12H21NO9S2 | 399.43 |
| (XVIII) | gluconapin(1−) | C11H18NO9S2 | 373.40 |
| (XIX) | 5-methoxyindole-3-acetic acid | C11H11NO3 | 205.21 |
| (XX) | α-allenylagmatine | C8H16N4 | 168.24 |
| Metabolite Name | Properties/Pharmacological Activities | Drug Target Protein | References |
|---|---|---|---|
| 2-hydroxyethyl hexadecanoate (2-HEP) | Antibacterial; damages bacterial cell membranes and disrupts bacterial growth. Effective against Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa. | Membrane phospholipids | [29] |
| Gluconapin | By myrosinase, and AITC has been demonstrated to be effective against various bacteria, including Escherichia coli, Salmonella Typhimurium, and Staphylococcus aureus | Membrane phospholipids | [30,31] |
| 3-phenylpropionitrile | Antibacterial; disrupts bacterial membranes by forming pores. Effective against Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa. | Bacterial cell membrane proteins (pore-forming) | [33] |
| Flamenol | Antibacterial properties against various bacteria, including Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa | - | [31] |
| Glucobrassicanapin(1−) | Antibacterial; a glucosinolate broken down into isothiocyanates, which have a broad range of antibacterial activities. | Enzyme involved in sulfur metabolism and membrane protein | [34] |
| 2-chlorobenzoic acid (2-CBA) | Antibacterial; disrupts bacterial cell membranes and inhibits DNA synthesis. Effective against Escherichia coli, Staphylococcus aureus, Bacillus subtilis. | DNA replication proteins, membrane proteins | [35,36] |
| 3-methoxybenzaldehyde | Antibacterial; disrupts bacterial cell membranes. Effective against Gram-positive and Gram-negative bacteria. | Membrane lipids and proteins | [37,38,39] |
| 1-naphthyl β-D-glucoside | Antibacterial; inhibits bacterial glycosyltransferases, necessary for cell wall synthesis. Effective against multi-drug-resistant and non-resistant strains. | Cell wall synthesis | [40,41] |
| Sinapine | Antibacterial; disrupts bacterial cell membranes by interacting with phospholipids. Effective against antibiotic-resistant bacteria (e.g., MRSA). | Membrane phospholipids | [42,43] |
| 3-butenyldesulfoglucosinolate | Antibacterial; derived from garlic, known for antimicrobial activity against bacteria such as Staphylococcus aureus. | Membrane proteins | [44,45] |
| Gluconapin(1−) | Antibacterial; converts into isothiocyanates, which disrupt bacterial cell membranes and inhibit enzymes. Effective against a broad spectrum of bacteria. | Membrane proteins | [31,32] |
| 5-methoxyindole-3-acetic acid | Antibacterial; potential as an antioxidant and in the synthesis of agents. Limited information on antibacterial capabilities. | - | - |
| Id | Antibiotics | Formula | Molecular Mass (g/mol) | Rank |
|---|---|---|---|---|
| 1 | Daptomycin | C72H101N17O26 | 1620.69 | 3 |
| 2 | Moxifloxacin | C21H24FN3O4 | 401.43 | 2 |
| 3 | Rifaximin | C43H51N3O11 | 785.89 | 1 |
| 4 | Ciprofloxacin | C17H18FN3O3 | 331.35 | 8 |
| 5 | Sulfamethoxazole | C10H11N3O3S | 253.28 | 11 |
| 6 | Trimethoprim | C14H18N4O3 | 290.32 | 9 |
| 7 | Amoxicillin | C16H19N3O5S | 365.40 | 6 |
| 8 | Cefdinir | C14H13N5O5S2 | 395.41 | 5 |
| 9 | Metronidazole | C6H9N3O3 | 171.15 | 10 |
| 10 | Cephalexin | C16H17N3O4S | 347.39 | 7 |
| 11 | Levofloxacin | C18H20FN3O4 | 361.37 | 4 |
| Machine Learning | Explanation | Advantage |
|---|---|---|
| AdaBoost | An ensemble learning algorithm that combines the predictions of multiple weak learners to produce a single, strong learner | Can handle imbalanced data |
| Bagging | Ensemble learning algorithm that works by creating multiple bootstrap samples of the training data and training a separate model on each sample | Can improve model with imbalanced data |
| BernoulliNB | Naive Bayes classifier that is specifically designed for binary classification problems | A simple but effective algorithm that is often used for imbalanced classification tasks |
| Decision Tree | Decision trees are a type of machine learning model that learns to classify data by constructing a tree of decision rules | Decision trees are relatively robust to imbalanced data, but they can be prone to overfitting |
| Extra Trees | Ensemble learning algorithm that is similar to random forests, but it uses a different approach to bootstrap sampling | Often used for imbalanced classification tasks because they are less likely to overfit than random forests |
| Gradient Boosting | An ensemble learning algorithm that combines the predictions of multiple weak learners in a sequential manner | A powerful algorithm that can be used for a variety of machine learning tasks, including imbalanced classification |
| K-nearest neighbors | K-nearest neighbors (KNN) is a simple but effective machine learning algorithm that classifies data by finding the K most similar training examples to a new data point and predicting the class of the new data point based on the classes of the K most similar training examples | Easy to implement and small dataset can use for imbalanced classification task |
| Linear Discriminant Analysis | Machine learning algorithm that projects the data onto a lower-dimensional space in a way that maximizes the discrimination between the different classes | Good choice for imbalanced data because it is able to find the most important features for discriminating between the classes |
| Logistic Regression | A machine learning algorithm that is used for binary classification problems | Often used for imbalanced classification task |
| Multilayer Perceptron | Multilayer perceptrons (MLPs) are a type of artificial neural network | Can use for classification task and able to learn complex data, even imbalanced classification task |
| Random Forest | Random forests are an ensemble learning algorithm that combines the predictions of multiple decision trees to produce a single prediction | Ability to handle imbalanced data and their resistance to overfitting |
| Support Vector Machine: | Machine learning that can use as classification and regression | Can handle high-dimensional data and imbalanced data |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasution, A.K.; Alqaaf, M.; Islam, R.M.; Wijaya, S.H.; Ono, N.; Kanaya, S.; Altaf-Ul-Amin, M. Identifying Potential Natural Antibiotics from Unani Formulas through Machine Learning Approaches. Antibiotics 2024, 13, 971. https://doi.org/10.3390/antibiotics13100971
Nasution AK, Alqaaf M, Islam RM, Wijaya SH, Ono N, Kanaya S, Altaf-Ul-Amin M. Identifying Potential Natural Antibiotics from Unani Formulas through Machine Learning Approaches. Antibiotics. 2024; 13(10):971. https://doi.org/10.3390/antibiotics13100971
Chicago/Turabian StyleNasution, Ahmad Kamal, Muhammad Alqaaf, Rumman Mahfujul Islam, Sony Hartono Wijaya, Naoaki Ono, Shigehiko Kanaya, and Md. Altaf-Ul-Amin. 2024. "Identifying Potential Natural Antibiotics from Unani Formulas through Machine Learning Approaches" Antibiotics 13, no. 10: 971. https://doi.org/10.3390/antibiotics13100971
APA StyleNasution, A. K., Alqaaf, M., Islam, R. M., Wijaya, S. H., Ono, N., Kanaya, S., & Altaf-Ul-Amin, M. (2024). Identifying Potential Natural Antibiotics from Unani Formulas through Machine Learning Approaches. Antibiotics, 13(10), 971. https://doi.org/10.3390/antibiotics13100971











