First Detection of High-Level Aminoglycoside-Resistant Klebsiella pneumoniae and Enterobacter cloacae Isolates Due to 16S rRNA Methyltransferases with and Without blaNDM in Uruguay
Abstract
1. Introduction
2. Results
2.1. Isolates and Antibiotic Susceptibility Testing
2.2. Characterization of rmtB-Carrying Isolates
2.3. Characterization of the rmtC-Carrying Isolate
2.4. Characterization of the rmtD-Carrying Isolate
3. Discussion
4. Materials and Methods
4.1. Strains
4.2. 16S rRNA-Methyltransferase and Carbapenemase Detection
4.3. Conjugation Assays
4.4. Whole Genome Sequencing
4.5. In Silico Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. WHO Bacterial Priority Pathogens List 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2024; pp. 1–72. [Google Scholar]
- CDC. COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022; US Department of Health and Human Services: Atlanta, GA, USA, 2022; Volume 399, pp. 629–655.
- Coque, T.M.; Cantón, R.; Pérez-Cobas, A.E.; Fernández-de-Bobadilla, M.D.; Baquero, F. Antimicrobial Resistance in the Global Health Network: Known Unknowns and Challenges for Efficient Responses in the 21st Century. Microorganisms 2023, 11, 1050. [Google Scholar] [CrossRef] [PubMed]
- García-Betancur, J.C.; Appel, T.M.; Esparza, G.; Gales, A.C.; Levy-Hara, G.; Cornistein, W.; Vega, S.; Nuñez, D.; Cuellar, L.; Bavestrello, L.; et al. Update on the epidemiology of carbapenemases in Latin America and the Caribbean. Expert Rev. Anti-Infect. Ther. 2021, 19, 197–213. [Google Scholar] [CrossRef]
- Garcia-Fulgueiras, V.; Zapata, Y.; Papa-Ezdra, R.; Ávila, P.; Caiata, L.; Seija, V.; Rodriguez, A.E.R.; Magallanes, C.; Villalba, C.M.; Vignoli, R. First characterization of K. pneumoniae ST11 clinical isolates harboring blaKPC-3 in Latin America. Rev. Argent. Microbiol. 2020, 52, 211–216. [Google Scholar] [CrossRef]
- Garcia-Fulgueiras, V.; Magallanes, C.; Reyes, V.; Cayota, C.; Galiana, A.; Vieytes, M.; Vignoli, R.; Márquez, C. In Vivo High Plasticity of Multi-Drug Resistant ST258 Klebsiella pneumoniae. Microb. Drug Resist. 2021, 27, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Bail, L.; Ito, C.A.S.; Arend, L.N.V.S.; Pilonetto, M.; da Silva Nogueira, K.; Tuon, F.F. Distribution of genes encoding 16S rRNA methyltransferase in plazomicin-nonsusceptible carbapenemase-producing Enterobacterales in Brazil. Diagn. Microbiol. Infect. Dis. 2021, 99, 115239. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Wachino, J.I.; Arakawa, Y. Aminoglycoside Resistance: The Emergence of Acquired 16S Ribosomal RNA Methyltransferases. Infect. Dis. Clin. N. Am. 2016, 30, 523. [Google Scholar] [CrossRef]
- Serio, A.W.; Keepers, T.; Andrews, L.; Krause, K.M. Aminoglycoside Revival: Review of a Historically Important Class of Antimicrobials Undergoing Rejuvenation. EcoSal Plus 2018, 8, 10–1128. [Google Scholar] [CrossRef]
- Costello, S.E.; Deshpande, L.M.; Davis, A.P.; Mendes, R.E.; Castanheira, M. Aminoglycoside-modifying enzyme and 16S ribosomal RNA methyltransferase genes among a global collection of Gram-negative isolates. J. Glob. Antimicrob. Resist. 2019, 16, 278–285. [Google Scholar] [CrossRef]
- Sellera, F.P.; Fuentes-Castillo, D.; Furlan, J.P.R. One Health Spread of 16S Ribosomal RNA Methyltransferase-Harboring Gram-Negative Bacterial Genomes: An Overview of the Americas. Pathogens 2023, 12, 1164. [Google Scholar] [CrossRef]
- Delgado-Blas, J.F.; Ovejero, C.M.; David, S.; Serna, C.; Pulido-Vadillo, M.; Montero, N.; Aanensen, D.M.; Abadia-Patiño, L.; Gonzalez-Zorn, B. Global scenario of the RmtE pan-aminoglycoside-resistance mechanism: Emergence of the rmtE4 gene in South America associated with a hospital-related IncL plasmid. Microb. Genom. 2023, 9, 000946. [Google Scholar] [CrossRef]
- Bado, I.; Papa-Ezdra, R.; Delgado-Blas, J.F.; Gaudio, M.; Gutiérrez, C.; Cordeiro, N.F.; García-Fulgueiras, V.; Araujo Pirez, L.; Seija, V.; Medina, J.C.; et al. Molecular Characterization of Carbapenem-Resistant Acinetobacter baumannii in the Intensive Care Unit of Uruguay’s University Hospital Identifies the First rmtC Gene in the Species. Microb. Drug Resist. 2018, 24, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Coppola, N.; Cordeiro, N.F.; Trenchi, G.; Esposito, F.; Fuga, B.; Fuentes-Castillo, D.; Lincopan, N.; Iriarte, A.; Bado, I.; Vignoli, R. Imported One-Day-Old Chicks as Trojan Horses for Multidrug-Resistant Priority Pathogens Harboring mcr-9, rmtG, and Extended-Spectrum β-Lactamase Genes. Appl. Environ. Microbiol. 2022, 88, e01675-21. [Google Scholar] [CrossRef] [PubMed]
- Bhavnani, S.M.; Ambrose, P.G.; Craig, W.A.; Dudley, M.N.; Jones, R.N. SENTRY Antimicrobial Surveillance Program. Outcomes evaluation of patients with ESBL- and non–ESBL-producing Escherichia coli and Klebsiella species as defined by CLSI reference methods: Report from the SENTRY Antimicrobial Surveillance Program. Diagn. Microbiol. Infect. Dis. 2006, 54, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, S.J.; Harmer, C.J.; Hall, R.M. Evolution and typing of IncC plasmids contributing to antibiotic resistance in Gram-negative bacteria. Plasmid 2018, 99, 40–55. [Google Scholar] [CrossRef]
- Tijet, N.; Andres, P.; Chung, C.; Lucero, C.; Low, D.E.; Galas, M.; Corso, A.; Petroni, A.; Melano, R.G. rmtD2, a new allele of a 16S rRNA methylase gene, has been present in Enterobacteriaceae isolates from Argentina for more than a decade. Antimicrob. Agents Chemother. 2011, 55, 904–909. [Google Scholar] [CrossRef]
- Liebert, C.A.; Hall, R.M.; Summers, A.O. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. MMBR 1999, 63, 507–522. [Google Scholar] [CrossRef]
- Porto, A.; Ayala, J.; Gutkind, G.; Di Conza, J. A novel OXA-10-like beta-lactamase is present in different Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 2010, 66, 228–229. [Google Scholar] [CrossRef]
- Taylor, E.; Bal, A.M.; Balakrishnan, I.; Brown, N.M.; Burns, P.; Clark, M.; Diggle, M.; Donaldson, H.; Eltringham, I.; Folb, J.; et al. A prospective surveillance study to determine the prevalence of 16S rRNA methyltransferase-producing Gram-negative bacteria in the UK. J. Antimicrob. Chemother. 2021, 76, 2428–2436. [Google Scholar] [CrossRef]
- Arca-Suárez, J.; Rodiño-Janeiro, B.K.; Pérez, A.; Guijarro-Sánchez, P.; Vázquez-Ucha, J.C.; Cruz, F.; Gómez-Garrido, J.; Alioto, T.S.; Álvarez-Tejado, M.; Gut, M.; et al. Emergence of 16S rRNA methyltransferases among carbapenemase-producing Enterobacterales in Spain studied by whole-genome sequencing. Int. J. Antimicrob. Agents 2022, 59, 106456. [Google Scholar] [CrossRef]
- Thomas, G.R.; Corso, A.; Pasterán, F.; Shal, J.; Sosa, A.; Pillonetto, M.; de Souza Peral, R.T.; Hormazábal, J.C.; Araya, P.; Saavedra, S.Y.; et al. Increased Detection of Carbapenemase-Producing Enterobacterales Bacteria in Latin America and the Caribbean during the COVID-19 Pandemic. Emerg. Infect. Dis. 2022, 28, E1–E8. [Google Scholar] [CrossRef]
- Tada, T.; Miyoshi-Akiyama, T.; Dahal, R.K.; Mishra, S.K.; Ohara, H.; Shimada, K.; Kirikae, T.; Pokhrel, B.M. Dissemination of multidrug-resistant Klebsiella pneumoniae clinical isolates with various combinations of carbapenemases (NDM-1 and OXA-72) and 16S rRNA methylases (ArmA, RmtC and RmtF) in Nepal. Int. J. Antimicrob. Agents 2013, 42, 372–374. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, K.L.; Escudero, J.A.; Hidalgo, L.; Gonzalez-Zorn, B. 16S rRNA Methyltransferase RmtC in Salmonella enterica Serovar Virchow. Emerg. Infect. Dis. 2010, 16, 712. [Google Scholar] [CrossRef] [PubMed]
- Arcari, G.; Carattoli, A. Global spread and evolutionary convergence of multidrug-resistant and hypervirulent Klebsiella pneumoniae high-risk clones. Pathog. Glob. Health 2023, 117, 328. [Google Scholar] [CrossRef]
- Villa, L.; Feudi, C.; Fortini, D.; Brisse, S.; Passet, V.; Bonura, C.; Endimiani, A.; Mammina, C.; Ocampo, A.M.; Jimenez, J.N.; et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb. Genom. 2017, 3, e000110. [Google Scholar] [CrossRef] [PubMed]
- Cejas, D.; Elena, A.; Nuñez, D.G.; Platero, P.S.; De Paulis, A.; Magariños, F.; Alfonso, C.; Berger, M.A.; Fernández-Canigia, L.; Gutkind, G.; et al. Changing epidemiology of KPC-producing Klebsiella pneumoniae in Argentina: Emergence of hypermucoviscous ST25 and high-risk clone ST307. J. Glob. Antimicrob. Resist. 2019, 18, 238–242. [Google Scholar] [CrossRef]
- Krul, D.; Rodrigues, L.S.; Siqueira, A.C.; Mesa, D.; dos Santos, M.; Vasconcelos, T.M.; Spalanzani, R.N.; Cardoso, R.; Ricieri, M.C.; Motta, F.d.A.; et al. High-risk clones of carbapenem resistant Klebsiella pneumoniae recovered from pediatric patients in Southern Brazil. Braz. J. Microbiol. 2024, 55, 1437–1443. [Google Scholar] [CrossRef]
- Ocampo, A.M.; Chen, L.; Cienfuegos, A.V.; Roncancio, G.; Chavda, K.D.; Kreiswirth, B.N.; Jiménez, J.N. A Two-Year Surveillance in Five Colombian Tertiary Care Hospitals Reveals High Frequency of Non-CG258 Clones of Carbapenem-Resistant Klebsiella pneumoniae with Distinct Clinical Characteristics. Antimicrob. Agents Chemother. 2015, 60, 332–342. [Google Scholar] [CrossRef]
- González-Espinosa, F.; Di Pilato, V.; Magariños, F.; Di Conza, J.; Rossolini, G.M.; Gutkind, G.; Radice, M.; Cejas, D. Genomic characterization of carbapenemase-producing Klebsiella pneumoniae ST307 revealed multiple introductions in Buenos Aires, Argentina. J. Glob. Antimicrob. Resist. 2024, 37, 176–178. [Google Scholar] [CrossRef]
- Faccone, D.; Gomez, S.A.; de Mendieta, J.M.; Sanz, M.B.; Echegorry, M.; Albornoz, E.; Lucero, C.; Ceriana, P.; Menocal, A.; Martino, F.; et al. Emergence of Hyper-Epidemic Clones of Enterobacterales Clinical Isolates Co-Producing KPC and Metallo-Beta-Lactamases during the COVID-19 Pandemic. Pathogens 2023, 12, 479. [Google Scholar] [CrossRef]
- Allel, K.; Peters, A.; Conejeros, J.; Martínez, J.R.W.; Spencer-Sandino, M.; Riquelme-Neira, R.; Rivas, L.; Rojas, P.; Chea, C.O.; García, P.; et al. Antibiotic Consumption During the Coronavirus Disease 2019 Pandemic and Emergence of Carbapenemase-Producing Klebsiella pneumoniae Lineages Among Inpatients in a Chilean Hospital: A Time-Series Study and Phylogenomic Analysis. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2023, 77, S20. [Google Scholar] [CrossRef]
- Al-Anany, A.M.; Fatima, R.; Nair, G.; Mayol, J.T.; Hynes, A.P. Temperate phage-antibiotic synergy across antibiotic classes reveals new mechanism for preventing lysogeny. mBio 2024, 15, e0050424. [Google Scholar] [CrossRef] [PubMed]
- Qin, K.; Shi, X.; Yang, K.; Xu, Q.; Wang, F.; Chen, S.; Xu, T.; Liu, J.; Wen, W.; Chen, R.; et al. Phage-antibiotic synergy suppresses resistance emergence of Klebsiella pneumoniae by altering the evolutionary fitness. mBio 2024, e0139324. [Google Scholar] [CrossRef] [PubMed]
- Georjon, H.; Bernheim, A. The highly diverse antiphage defence systems of bacteria. Nat. Rev. Microbiol. 2023, 21, 686–700. [Google Scholar] [CrossRef]
- Araújo, L.; Papa-Ezdra, R.; Ávila, P.; Iribarnegaray, V.; Bado, I.; Telechea, H.; Garcia-Fulgueiras, V.; Vignoli, R. Great Plasticity in a Great Pathogen: Capsular Types, Virulence Factors and Biofilm Formation in ESBL-Producing Klebsiell pneumoniae from Pediatric Infections in Uruguay. Antibiotics 2024, 13, 170. [Google Scholar] [CrossRef]
- Romina, P.E.; Lucía, A.; Leticia, C.; Federica, F.; Pablo, Á.; Verónica, S.; Antonio, G.; Inés, B.; Rafael, V. In vitro effectiveness of ceftazidime-avibactam in combination with aztreonam on carbapenemase-producing Enterobacterales. J. Glob. Antimicrob. Resist. 2023, 35, 62–66. [Google Scholar] [CrossRef]
- Bado, I.; Cordeiro, N.F.; Robino, L.; García-Fulgueiras, V.; Seija, V.; Bazet, C.; Gutkind, G.; Ayala, J.A.; Vignoli, R. Detection of class 1 and 2 integrons, extended-spectrum β-lactamases and qnr alleles in enterobacterial isolates from the digestive tract of Intensive Care Unit inpatients. Int. J. Antimicrob. Agents 2010, 36, 453–458. [Google Scholar] [CrossRef]
- Bado, I.; Gutiérrez, C.; García-Fulgueiras, V.; Cordeiro, N.F.; Pirez, L.A.; Seija, V.; Bazet, C.; Rieppi, G.; Vignoli, R. CTX-M-15 in combination with aac(6′)-Ib-cr is the most prevalent mechanism of resistance both in Escherichia coli and Klebsiella pneumoniae, including K. pneumoniae ST258, in an ICU in Uruguay. J. Glob. Antimicrob. Resist. 2016, 6, 5–9. [Google Scholar] [CrossRef]
- Hidalgo, L.; Hopkins, K.L.; Gutierrez, B.; Ovejero, C.M.; Shukla, S.; Douthwaite, S.; Prasad, K.N.; Woodford, N.; Gonzalez-Zorn, B. Association of the novel aminoglycoside resistance determinant RmtF with NDM carbapenemase in Enterobacteriaceae isolated in India and the UK. J. Antimicrob. Chemother. 2013, 68, 1543–1550. [Google Scholar] [CrossRef]
- Papa-Ezdra, R.; Caiata, L.; Palacio, R.; Outeda, M.; Cabezas, L.; Bálsamo, A.; Vignoli, R.; Bado, I.; Seija, V. Prevalence and molecular characterisation of carbapenemase-producing Enterobacterales in an outbreak-free setting in a single hospital in Uruguay. J. Glob. Antimicrob. Resist. 2021, 24, 58–62. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcìa-Fernandez, A.; Larsen, M.; Lund, O.; Voldby Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 1–8. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 1–9. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Carver, T.; Berriman, M.; Tivey, A.; Patel, C.; Böhme, U.; Barrell, B.G.; Parkhill, J.; Rajandream, M.-A. Artemis and ACT: Viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 2008, 24, 2672–2676. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
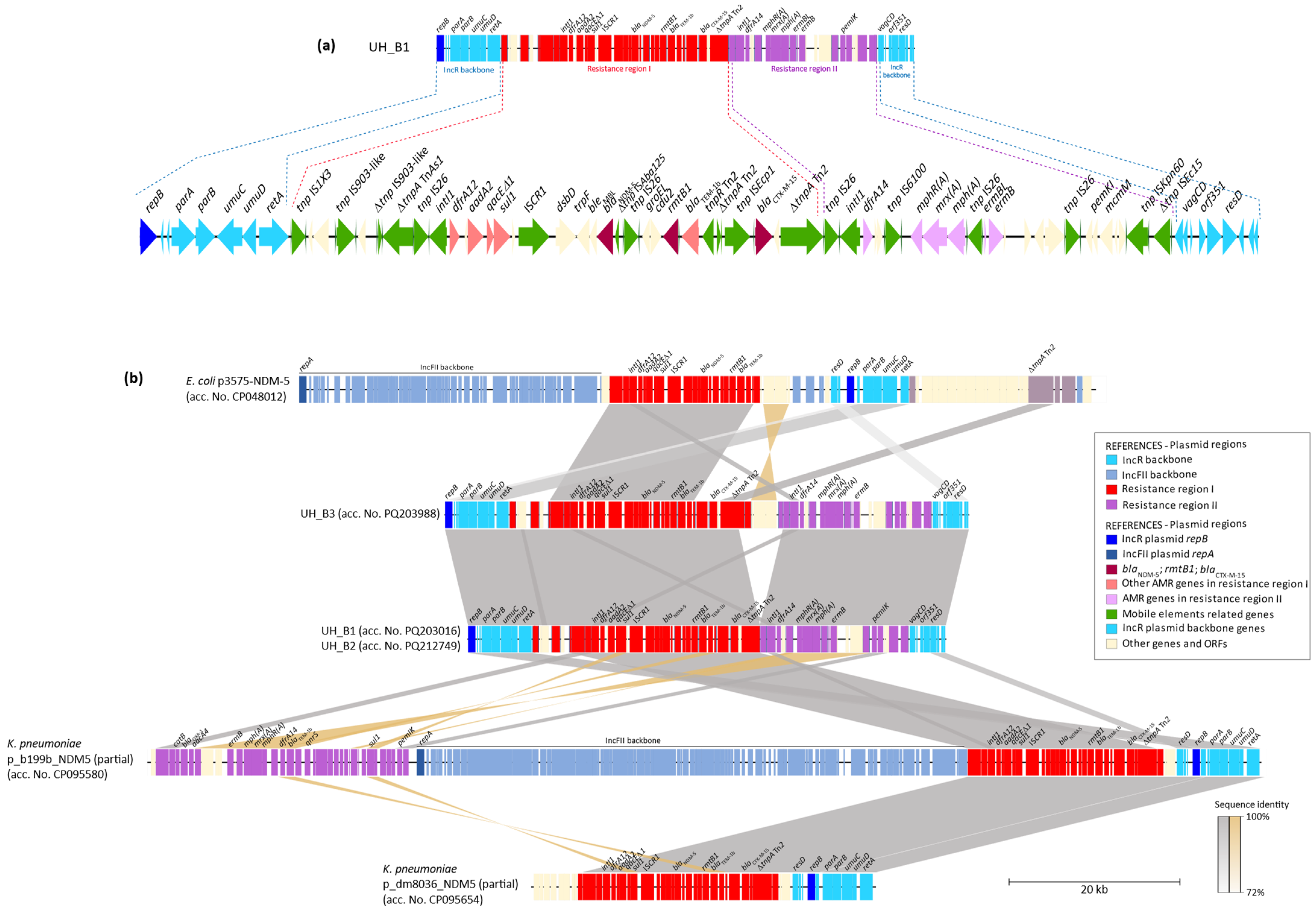
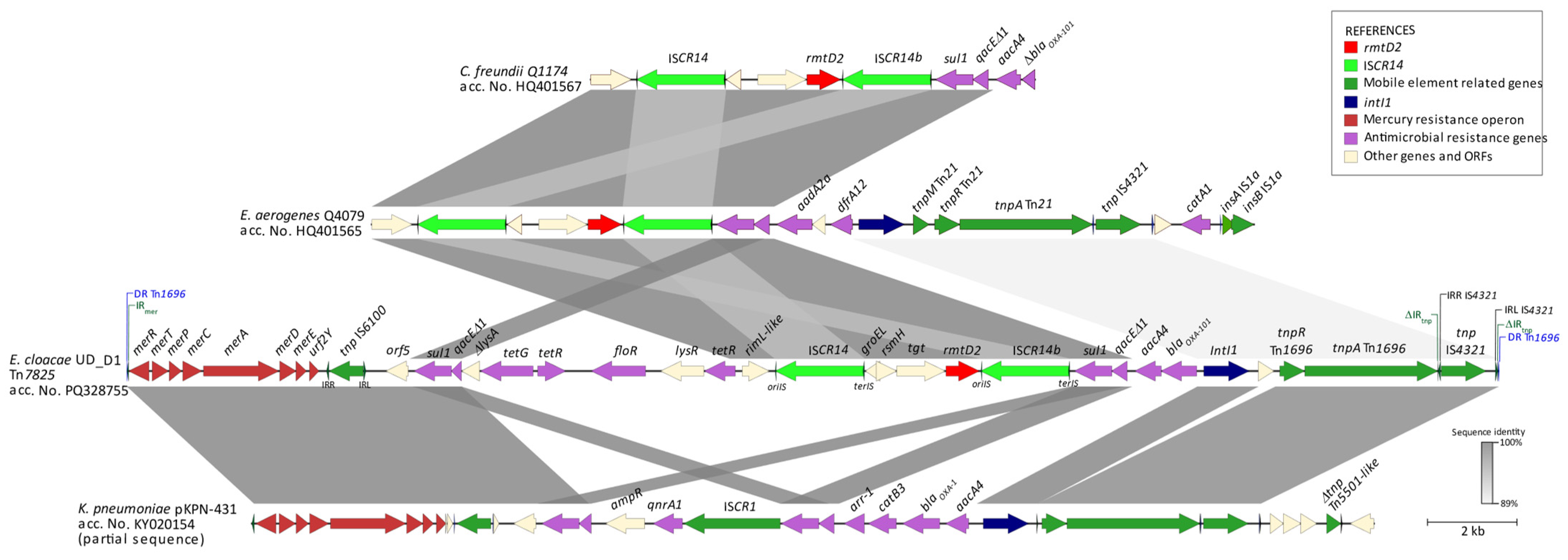
| Strain | Isolation Date and Hospital | Sample | Species and Sequence Type | Rmt/CARB | MIC (mg/L) | Incompatibility Group and Plasmid Size (bp) | Antibiotic Resistance Genes | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AK | GM | PTZ | CTX | CAZ | CZA | FEP | ATM | IPM | MEM | ETP | CIP | SXT | COL | TIG | |||||||
| UH_B1 | 08/2022 H1 | Blood | K. pneumoniae ST307 | RmtB/NDM-5 | >32 | >8 | >64 | >32 | >32 | >256 | 16 | >16 | 4 | 2 | >2 | >2 | >2 | ≤1 | ≤0.5 | IncR (55,925 bp) | dfrA12, aadA2, sul1, blaNDM-5, rmtB, blaTEM-1B, blaCTX-M-15, dfrA14, mph(A), ermB, qacEΔ1 |
| IncFIB(K)/IncFII (217,500 bp) | sul2, aph(3″)-Ib, aph(6)-Id, blaTEM-1B | ||||||||||||||||||||
| UH_B2 | 08/2022 H2 | Urine | K. pneumoniae ST307 | RmtB/NDM-5 | >32 | >8 | >64 | >32 | >32 | >256 | 16 | >16 | 2 | 2 | >2 | >2 | >2 | ≤1 | ≤0.5 | IncR (55,916 bp) | dfrA12, aadA2, sul1, blaNDM-5, rmtB, blaTEM-1B, blaCTX-M-15, dfrA14, mph(A), ermB, qacEΔ1 |
| IncFIB(K)/IncFII (217,500 bp) | sul2, aph(3″)-Ib, aph(6)-Id, blaTEM-1B | ||||||||||||||||||||
| IncM1 (68,728 bp) | blaCTX-M-8, blaTEM-1B, qnrE1 | ||||||||||||||||||||
| IncFIB(K)/IncFII (129,906 bp) | qacEΔ1, mph(A), catB3, sul1, aac(6′)-Ib-cr, aadA2, aac(3)-IIa, blaTEM-1B, blaSHV-28, blaOXA-1, aac(6′)-Ib-cr, dfrA12 | ||||||||||||||||||||
| UH_B3 | 09/2023 H1 | Abscess | K. pneumoniae ST307 | RmtB/NDM-5 | >32 | >8 | >64 | >32 | >32 | >256 | >16 | >16 | 4 | 8 | >2 | >2 | >2 | ≤1 | ≤0.5 | IncR (61,310 bp) | dfrA12, aadA2, sul1, blaNDM-5, rmtB, blaTEM-1B, blaCTX-M-15, dfrA14, mph(A), ermB, qacEΔ1 |
| IncFIB(K)/IncFII (212,501 bp) | dfrA14, mph(A), blaCTX-M-15, blaTEM-1B, aph(6)-Id, aph(3″)-Ib, sul2 | ||||||||||||||||||||
| UH_C1 | 11/2017 H3 | Urine | K. pneumoniae ST258 | RmtC/NDM-1 | >32 | >8 | >64 | >32 | >32 | >256 | >16 | >16 | >8 | 16 | >2 | >2 | ≤2 | ≤1 | ≤0.5 | IncC (138,998 bp) | blaCMY-6, aac(6′)-Ib, sul1, rmtC, blaNDM-1 |
| ColRNAI (15,271 bp) | aac(6′)-Ib | ||||||||||||||||||||
| IncFIB(K)/IncFII (137,364 bp) | mph(A) | ||||||||||||||||||||
| UH_D1 | 12/2010 H1 | Rectal colonization | E. cloacae ST88 | RmtD/- | >32 | >8 | >64 | >32 | 32 | ≤0.5 | ≤2 | >16 | ≤0.5 | ≤1 | 2 | >2 | >2 | ≤1 | ≤0.5 | IncFIB(K)/IncFII (149,562 bp) | sul1, tet(G), floR, rmtD2, sul1, blaOXA-101 |
| IncFII (130,409 bp) | blaTEM-258, aac(3)-IIa, blaTEM-1a | ||||||||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papa-Ezdra, R.; Cordeiro, N.F.; Ferreira, F.; García-Fulgueiras, V.; Araújo, L.; Mota, M.I.; Outeda, M.; Seija, V.; Vignoli, R.; Bado, I. First Detection of High-Level Aminoglycoside-Resistant Klebsiella pneumoniae and Enterobacter cloacae Isolates Due to 16S rRNA Methyltransferases with and Without blaNDM in Uruguay. Antibiotics 2024, 13, 1029. https://doi.org/10.3390/antibiotics13111029
Papa-Ezdra R, Cordeiro NF, Ferreira F, García-Fulgueiras V, Araújo L, Mota MI, Outeda M, Seija V, Vignoli R, Bado I. First Detection of High-Level Aminoglycoside-Resistant Klebsiella pneumoniae and Enterobacter cloacae Isolates Due to 16S rRNA Methyltransferases with and Without blaNDM in Uruguay. Antibiotics. 2024; 13(11):1029. https://doi.org/10.3390/antibiotics13111029
Chicago/Turabian StylePapa-Ezdra, Romina, Nicolás F. Cordeiro, Federica Ferreira, Virginia García-Fulgueiras, Lucía Araújo, María Inés Mota, Matilde Outeda, Verónica Seija, Rafael Vignoli, and Inés Bado. 2024. "First Detection of High-Level Aminoglycoside-Resistant Klebsiella pneumoniae and Enterobacter cloacae Isolates Due to 16S rRNA Methyltransferases with and Without blaNDM in Uruguay" Antibiotics 13, no. 11: 1029. https://doi.org/10.3390/antibiotics13111029
APA StylePapa-Ezdra, R., Cordeiro, N. F., Ferreira, F., García-Fulgueiras, V., Araújo, L., Mota, M. I., Outeda, M., Seija, V., Vignoli, R., & Bado, I. (2024). First Detection of High-Level Aminoglycoside-Resistant Klebsiella pneumoniae and Enterobacter cloacae Isolates Due to 16S rRNA Methyltransferases with and Without blaNDM in Uruguay. Antibiotics, 13(11), 1029. https://doi.org/10.3390/antibiotics13111029






