Potent Antimicrobial Azoles: Synthesis, In Vitro and In Silico Study
Abstract
1. Introduction
2. Results and Discussion
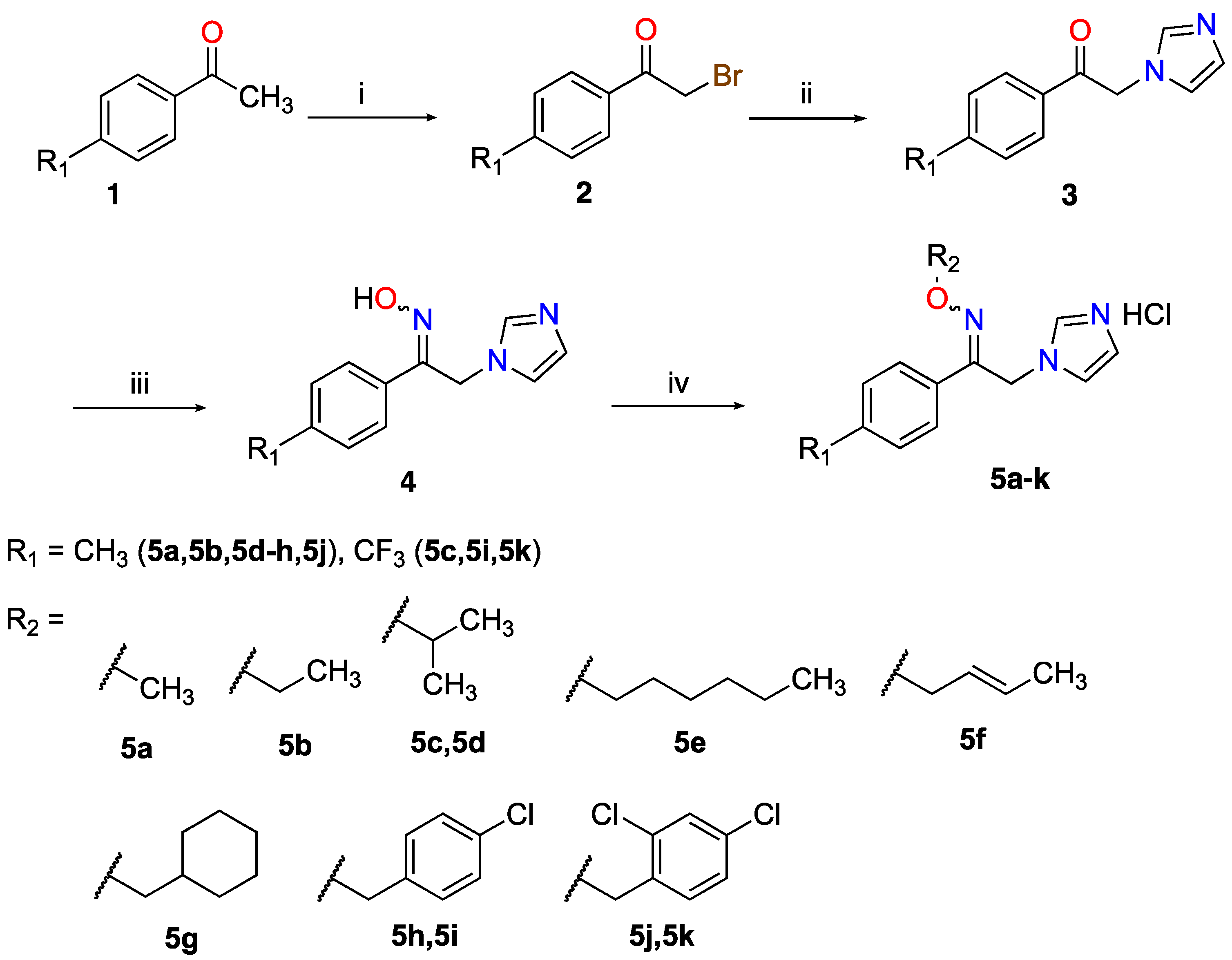
2.1. Chemistry
2.2. Antimicrobial Activity
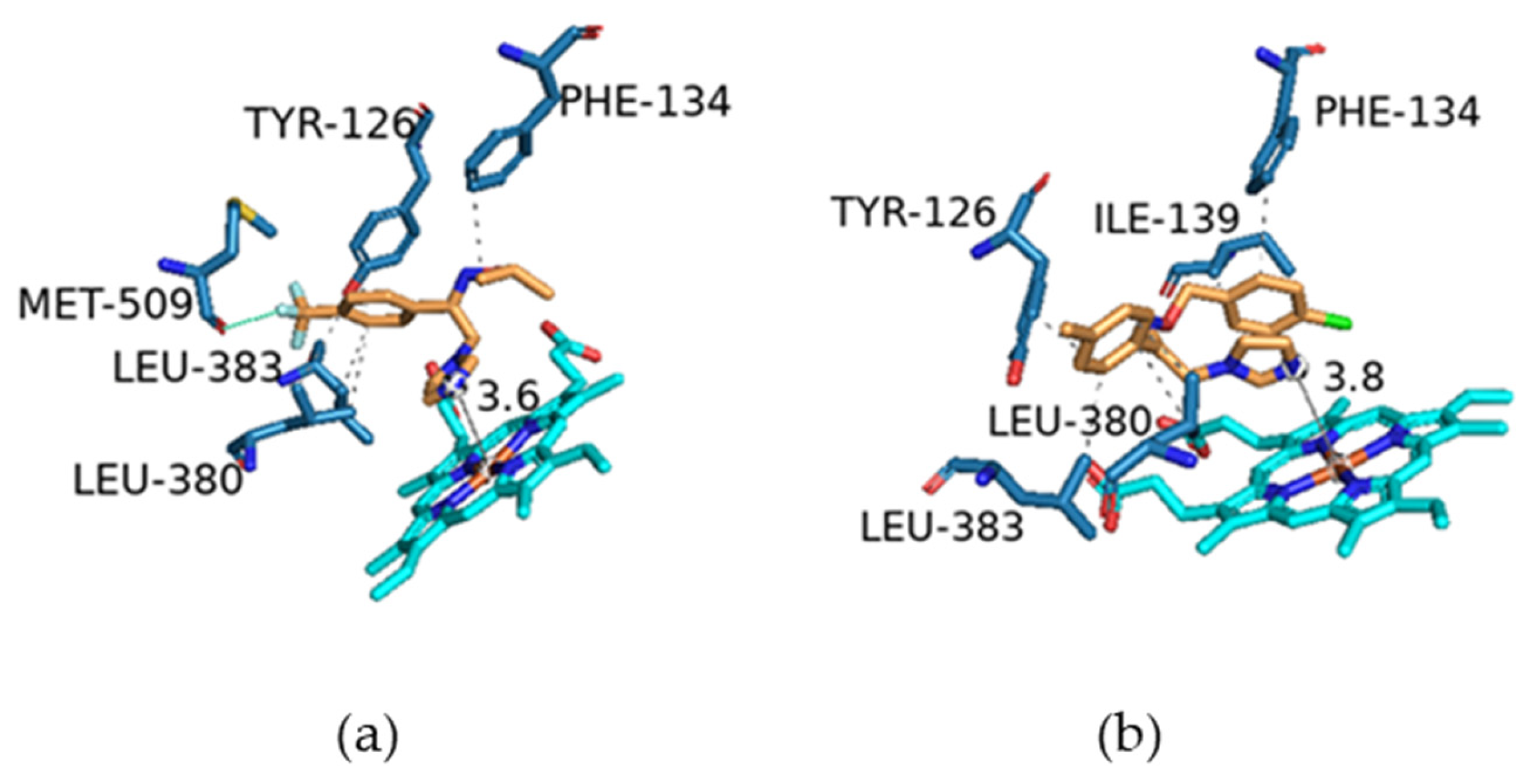
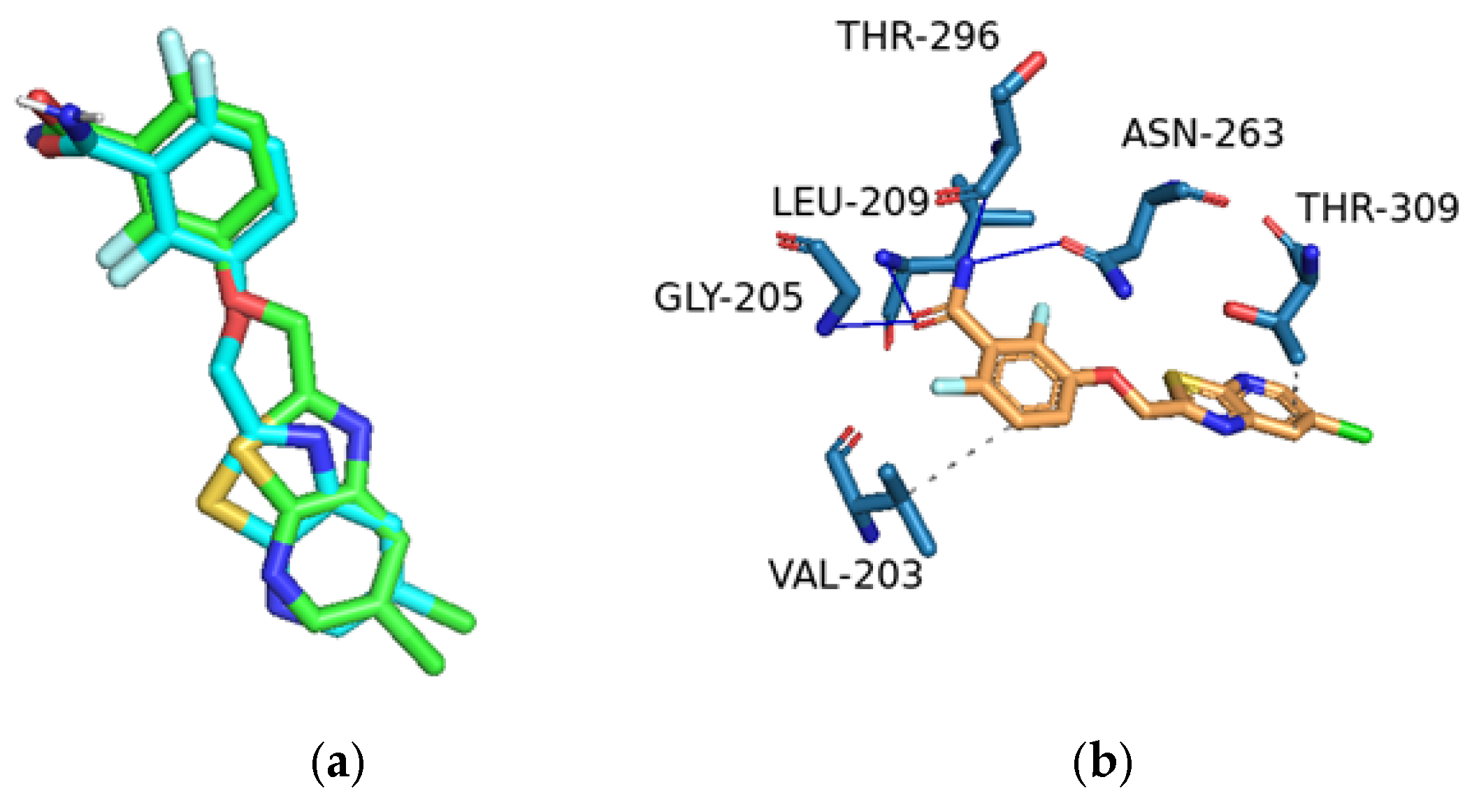
2.3. Molecular Modelling
3. Materials and Methods
3.1. Chemistry
Preparation of the Azole Compounds
- 2-Bromo-1-(4-trifluormethylphenyl)/1-(4-methylphenyl)ethanone (2)
- 1-(4-trifluoromethylphenyl)/1-(4-methylphenyl)-2-(1H-imidazol-1-yl)ethanone (3)
- 1-(4-Trifluoromethylphenyl)/1-(4-methylphenyl)-2-(1H-imidazol-1-yl)ethanone oxime (4)
- 1-(4-Trifluoromethylphenyl)/1-(4-methylphenyl)-2-(1H-imidazol-1-yl)ethanone oxime ethers (5a–k)
- 1-(4-Methylphenyl)-2-(1H-imidazol-1-yl)ethanone O-methyloxime hydrochloride (5a)
- 1-(4-Methylphenyl)-2-(1H-imidazol-1-yl)ethanone O-ethyloxime hydrochloride (5b)
- 1-(4-Trifluoromethylphenyl)-2-(1H-imidazol-1-yl)ethanone O-isopropyloxime hydrochloride (5c)
- 1-(4-Methylphenyl)-2-(1H-imidazol-1-yl)ethanone O-isopropyloxime hydrochloride (5d)
- 1-(4-Methylphenyl)-2-(1H-imidazol-1-yl)ethanone O-hexyloxime hydrochloride (5e)
- 1-(4-Methylphenyl)-2-(1H-imidazol-1-yl)ethanone O-crotyloxime hydrochloride (5f)
- 1-(4-Methylphenyl)-2-(1H-imidazol-1-yl)ethanone O-cyclohexylmethyloxime hydrochloride (5g)
- 1-(4-Methylphenyl)-2-(1H-imidazol-1-yl)ethanone O-(4-chlorophenyl)methyloxime hydrochloride (5h)
- 1-(4-Trifluoromethylphenyl)-2-(1H-imidazol-1-yl)ethanone O-(4-chlorophenyl)methyloxime hydrochloride (5i)
- 1-(4-Methylphenyl)-2-(1H-imidazol-1-yl)ethanone O-(2,4-dichlorophenyl)methyloxime hydrochloride (5j)
- 1-(4-Trifluoromethylphenyl)-2-(1H-imidazol-1-yl)ethanone O-(2,4-dichlorophenyl)methyloxime hydrochloride (5k)
3.2. Antimicrobial Assays
3.3. Molecular Modelling Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qadri, H.; Shah, A.H.; Alkhanani, M.; Almilaibary, A.; Mir, M.A. Immunotherapies against human bacterial and fungal infectious diseases: A review. Front. Med. 2023, 10, 1135541. [Google Scholar] [CrossRef] [PubMed]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A. Antimicrobial resistance: A growing serious threat for global public health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef] [PubMed]
- Rusu, A.; Moga, I.M.; Uncu, L.; Hancu, G. The role of five-membered heterocycles in the molecular structure of antibacterial drugs used in therapy. Pharmaceutics 2023, 15, 2554. [Google Scholar] [CrossRef] [PubMed]
- Pobitra, B.; Hazarika, S.; Chettri, A.; Sharma, D.; Deka, S.; Venugopala, K.N.; Shinu, P.; Al-Shar’i, N.A.; Bardaweel, S.K.; Deb, P.K. Heterocyclic compounds as antimicrobial agents. In Viral, Parasitic, Bacterial, and Fungal Infections; Academic Press: New York, NY, USA, 2023; pp. 781–804. [Google Scholar]
- Mishra, A.C.; Upadhyay, J.; Dixit, P.P.; Baheti, K.; Thore, S.N. Synthesis, antimicrobial evaluation, and molecular docking studies of Mannich base analogs derived from 2, 3-dihydro-1, 3, 4-oxadiazole-2 (3 H)-thione scaffold. Chem. Pap. 2024, 78, 6627–6647. [Google Scholar] [CrossRef]
- Goel, T.; Jain, N.; Bansode, D. Review on Pyrazole Hybrids as Anti-microbial Agents. Lett. Org. Chem. 2024, 21, 320–332. [Google Scholar] [CrossRef]
- Hiruma, J.; Nojyo, H.; Harada, K.; Kano, R. Development of treatment strategies by comparing the minimum inhibitory concentrations and minimum fungicidal concentrations of azole drugs in dermatophytes. J. Dermatol. 2024. [Google Scholar] [CrossRef]
- Houst, J.; Spizek, J.; Havlicek, V. Antifungal Drugs. Metabolites 2020, 10, 106. [Google Scholar] [CrossRef]
- Rani, N.; Singh, R.; Kumar, P. Imidazole and Derivatives Drugs Synthesis: A Review. Curr. Org. Synt. 2023, 20, 630–662. [Google Scholar] [CrossRef]
- Kosmalski, T.; Kupczyk, D.; Baumgart, S.; Paprocka, R.; Studzińska, R. A review of biologically active oxime ethers. Molecules 2023, 28, 5041. [Google Scholar] [CrossRef]
- Monk, B.C.; Sagatova, A.A.; Hosseini, P.; Ruma, Y.N.; Wilson, R.K.; Keniya, M.V. Fungal Lanosterol 14α-demethylase: A target for next-generation antifungal design. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140206. [Google Scholar] [CrossRef]
- Monk, B.C.; Keniya, M.V. Roles for structural biology in the discovery of drugs and agrochemicals targeting sterol 14α-demethylases. J. Fungi 2021, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, A.; Mohammadnejadi, E.; Karami, P.; Razzaghi-Asl, N. Current status and structure activity relationship of privileged azoles as antifungal agents (2016–2020). Int. J. Antimicrob. Agent. 2022, 59, 106518. [Google Scholar] [CrossRef] [PubMed]
- Maksimov, A.Y.; Balandina, S.Y.; Topanov, P.A.; Mashevskaya, I.V.; Chaudhary, S. Organic antifungal drugs and targets of their action. Curr. Top. Med. Chem. 2021, 21, 705–736. [Google Scholar] [CrossRef] [PubMed]
- Sagatova, A.A.; Keniya, M.V.; Wilson, R.K.; Sabherwal, M.; Tyndall, J.D.; Monk, B.C. Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14α-demethylase. Sci. Rep. 2016, 6, 26213. [Google Scholar] [CrossRef]
- Shafiei, M.; Peyton, L.; Hashemzadeh, M.; Foroumadi, A. History of the development of antifungal azoles: A review on structures, SAR, and mechanism of action. Bioorg. Chem. 2020, 104, 104240. [Google Scholar] [CrossRef]
- Sobel, J.D.; Nyirjesy, P. Oteseconazole: An advance in treatment of recurrent vulvovaginal candidiasis. Future Microbiol. 2021, 16, 1453–1461. [Google Scholar] [CrossRef]
- Jegasothy, B.V.; Pakes, G.E. Oxiconazole nitrate: Pharmacology, efficacy, and safety of a new imidazole antifungal agent. Clin. Ther. 1991, 13, 126–141. [Google Scholar]
- Alsterholm, M.; Karami, N.; Faergemann, J. Antimicrobial activity of topical skin pharmaceuticals—An in vitro study. Acta Derm. Venereol. 2010, 90, 239–245. [Google Scholar] [CrossRef]
- Nenoff, P.; Koch, D.; Kruger, C.; Drechsel, C.; Mayser, P. New insights on the antibacterial efficacy of miconazole in vitro. Mycoses 2017, 60, 552–557. [Google Scholar] [CrossRef]
- Kaul, G.; Akhir, A.; Shukla, M.; Shafi, H.; Akunuri, R.; Pawar, G.; Ghouse, M.; Srinivas, N.; Chopra, S. Oxiconazole Potentiates Gentamicin against Gentamicin-Resistant Staphylococcus aureus In Vitro and In Vivo. Microbiol. Spectr. 2023, 11, e0503122. [Google Scholar] [CrossRef]
- Azevedo, M.M.; Teixeira-Santos, R.; Silva, A.P.; Cruz, L.; Ricardo, E.; Pina-Vaz, C.; Rodrigues, A.G. The effect of antibacterial and non-antibacterial compounds alone or associated with antifugals upon fungi. Front. Microbiol. 2015, 6, 669. [Google Scholar] [CrossRef] [PubMed]
- Sari, S.; Koçak, E.; Kart, D.; Özdemir, Z.; Acar, M.F.; Sayoğlu, B.; Karakurt, A.; Dalkara, S. Azole derivatives with naphthalene showing potent antifungal effects against planktonic and biofilm forms of Candida spp.: An in vitro and in silico study. Int. Microbiol. 2021, 24, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Sari, S.; Sabuncuoğlu, S.; Koçak Aslan, E.; Avci, A.; Kart, D.; Özdemir, Z.; Acar, M.F.; Sayoğlu, B.; Alagöz, M.A.; Karakurt, A.; et al. Azoles containing naphthalene with activity against Gram-positive bacteria: In vitro studies and in silico predictions for flavohemoglobin inhibition. J. Biomol. Struct. Dyn. 2022, 40, 10220–10229. [Google Scholar] [CrossRef] [PubMed]
- Chunquan, S.; Zhang, W.; Ji, H.; Zhang, M.; Song, Y.; Xu, H.; Zhu, J.; Miao, Z.; Jiang, Q.; Yao, J.; et al. Structure-based optimization of azole antifungal agents by CoMFA, CoMSIA, and molecular docking. J. Med. Chem. 2006, 49, 2512–2525. [Google Scholar]
- Carradori, S.; Ammazzalorso, A.; De Filippis, B.; Şahin, A.F.; Akdemir, A.; Orekhova, A.; Bonincontro, G.; Simonetti, G. Azole-Based Compounds That Are Active against Candida Biofilm: In Vitro, In Vivo and In Silico Studies. Antibiotics 2022, 11, 1375. [Google Scholar] [CrossRef]
- Karakurt, A.; Bozbey, İ.; Uslu, H.; Sari, S.; Özdemir, Z.; Şalva, E. Synthesis and cytotoxicity studies on new pyrazolecontaining oxime ester derivatives. Trop. J. Pharm. Res. 2019, 18, 1315–1322. [Google Scholar] [CrossRef]
- Rykaczewski, K.A.; Wearing, E.R.; Blackmun, D.E.; Schindler, C.S. Reactivity of oximes for diverse methodologies and synthetic applications. Nat. Synth. 2022, 1, 24–36. [Google Scholar] [CrossRef]
- CLSI Document M07-A10; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—10th ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015.
- CLSI Standard M27; Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeast, 4th ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017.
- CLSI Supplement M60; Performance Standards for Antifungal Susceptibility Testing of Yeasts, 2nd ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020.
- CLSI Document M38-A2; Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi, Approved standard—2nd ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008.
- D’Agostino, I.; Mathew, G.E.; Angelini, P.; Venanzoni, R.; Angeles Flores, G.; Angeli, A.; Carradori, S.; Marinacci, B.; Menghini, L.; Abdelgawad, M.A.; et al. Biological investigation of N-methyl thiosemicarbazones as antimicrobial agents and bacterial carbonic anhydrases inhibitors. J. Enzyme Inhib. Med. Chem. 2022, 37, 986–993. [Google Scholar] [CrossRef]
- Kabier, M.; Gambacorta, N.; Trisciuzzi, D.; Kumar, S.; Nicolotti, O.; Mathew, B. MzDOCK: A free ready-to-use GUI based pipeline for molecular docking simulations. J. Comput. Chem. 2024, 45, 1980. [Google Scholar] [CrossRef]
- Singh, U.C.; Kollman, P.A. An approach to computing electrostatic charges for molecules. J. Comput. Chem. 1984, 5, 129–145. [Google Scholar] [CrossRef]
- Jayan, J.; Lee, J.; Kumar, S.; Manoharan, A.; Narayanan, A.P.; Jauhari, R.; Abdelgawad, M.A.; Ghoneim, M.M.; Ebrahim, H.A.; Zachariah, S.M.; et al. Development of a New Class of Monoamine Oxidase-B Inhibitors by Fine-Tuning the Halogens on the Acylhydrazones. ACS Omega 2023, 8, 47606. [Google Scholar] [CrossRef] [PubMed]
- Bienfait, B.; Ertl, P. JSME: A free molecule editor in JavaScript. J. Cheminform 2013, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996, 17, 490–519. [Google Scholar] [CrossRef]
- Koes, D.R.; Baumgartner, M.P.; Camacho, C.J. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J. Chem. Inf. Model. 2013, 53, 1893–18904. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDockVina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Bell, E.W.; Zhang, Y. DockRMSD: An open-source tool for atom mapping and RMSD calculation of symmetric molecules through graph isomorphism. J. Cheminform. 2019, 11, 40. [Google Scholar] [CrossRef]
- Tan, C.M.; Therien, A.G.; Lu, J.; Lee, S.H.; Caron, A.; Gill, C.J.; Lebeau-Jacob, C.; Benton-Perdomo, L.; Monteiro, J.M.; Pereira, P.M.; et al. Restoring Methicillin-Resistant Staphylococcus aureus Susceptibility to β-Lactam Antibiotics. Sci. Transl. Med. 2012, 4, 126ra35. [Google Scholar] [CrossRef]



| Compounds | MIC (µg mL−1) * | ||
|---|---|---|---|
| Escherichia coli (PeruMycA 3) | Bacillus subtilis (PeruMycA 6) | Staphylococcus aureus (ATCC 6538) | |
| 5a | 4.96 (±1.8) | 4.96 (±1.8) | 7.87 (±3.6) |
| 5b | 1.61 (±1.13) | 2.47 (±0.90) | 9.92 (±3.6) |
| 5c | 1.96 (±0.90) | 1.23 (±1.35) | 15.75 (±7.21) |
| 5d | 1.96 (±0.90) | 3.93 (±1.8) | 31.49 (±14.4) |
| 5e | 4.96 (±1.8) | 3.93 (±1.8) | 2.47 (±0.9) |
| 5f | 1.96 (±0.90) | 2.47 (±0.9) | 3.93 (±1.8) |
| 5g | 3.93 (±1.8) | 4.96 (±1.8) | 7.87 (±3.6) |
| 5h | 1.96 (±0.90) | 1.96 (±0.90) | 3.93 (±1.8) |
| 5i | 31.49 (±14.4) | 7.87 (±3.6) | >200 |
| 5j | 9.92 (±3.6) | 3.93 (±1.8) | 62.99 (±28.8) |
| 5k | 19.84 (±7.21) | 15.75 (±7.21) | 9.92 (±3.6) |
| Ciprofloxacin | 31.49 (±14.4) | 125.99 (±57.7) | 200–>200 |
| Compounds | MIC (µg mL−1) * | ||
|---|---|---|---|
| Candida tropicalis (YEPGA 6184) | Candida albicans (YEPGA 6379) | Candida parapsilosis (YEPGA 6551) | |
| 5a | 62.99 (±28.86) | >200 | 31.49 (±14.43) |
| 5b | 7.87 (±3.6) | >200 | 62.99 (±28.86) |
| 5c | 125.99 (±57.33) | >200 | 39.68 (±14.43) |
| 5d | 79.37 (±28.86) | 158.74 (±57.73) | 62.99 (±28.86) |
| 5e | 31.49 (±14.43) | 79.37 (±28.86) | 31.49 (±14.43) |
| 5f | 158.74 (±57.73) | >200 | 62.99 (±28.86) |
| 5g | 79.37 (±28.86) | 125.99 (±57.73) | 31.49 (±14.43) |
| 5h | 158.74 (±57.73) | >200 | 79.37 (±28.86) |
| 5i | >200 | >200 | 79.37 (±28.86) |
| 5j | 125.99 (±57.33) | 62.99 (±28.86) | 39.68 (±14.43) |
| 5k | >200 | 31.49 (±14.43) | 62.99 (±28.86) |
| Fluconazole | 1.81 (± 0.28) | 1.25 (±0.57) | 3.17 (±1.15) |
| Compounds | MIC (µg mL−1) * | ||
|---|---|---|---|
| Trichophyton mentagrophytes (CCF 4823) | Arthroderma quadrifidum (CCF 5792) | Arthroderma gypseum (CCF 6261) | |
| 5a | 31.49 (±14.43) | 15.74 (±7.21) | 39.68 (±14.43) |
| 5b | 15.74 (±7.21) | 7.87 (±7.21) | 9.92 (±3.6) |
| 5c | 0.491 (±0.22) | 0.619 (±0.22) | 7.87 (±3.6) |
| 5d | 15.74 (±7.21) | 2.48 (±0.90) | 15.74 (±7.21) |
| 5e | 7.87 (±7.21) | 2.48 (±0.90) | 1.968 (±0.90) |
| 5f | 15.74 (±7.21) | 7.87 (±7.21) | 31.49 (±14.43) |
| 5g | 1.239 (±0.45) | 3.937 (±1.8) | 1.968 (±0.90) |
| 5h | 0.619 (±0.22) | 0.983 (±0.45) | 2.48 (±0.90) |
| 5i | 4.98 (±1.8) | 2.48 (±0.90) | 3.937 (±1.8) |
| 5j | 1.239 (±0.45) | 1.968 (±0.90) | 1.968 (±0.90) |
| 5k | 3.937 (±1.8) | 1.23 (±0.45) | 15.74 (±7.21) |
| Griseofulvin | 2.52 (±1.15) | >8 | 3.174 (±1.15) |
| Code | Binding Energy (Kcal) | Type of Interactions | Amino Acids Involved | H-DIST (Å) |
|---|---|---|---|---|
| 5c | −8.1 | Hydrophobic | TYR 126 | 3.65 |
| Hydrophobic | PHE 134 | 3.52 | ||
| Hydrophobic | LEU 380 | 3.94 | ||
| Hydrophobic | LEU 380 | 3.91 | ||
| Hydrophobic | LEU 383 | 3.65 | ||
| Halogen Bond | MET 509 | 3.03 | ||
| 5h | −8.2 | Hydrophobic | TYR 126 | 3.57 |
| Hydrophobic | PHE 134 | 3.70 | ||
| Hydrophobic | ILE 139 | 3.79 | ||
| Hydrophobic | LEU 380 | 3.34 | ||
| Hydrophobic | LEU 380 | 3.84 | ||
| Hydrophobic | LEU 383 | 3.23 | ||
| Fluconazole | −7.0 | Hydrophobic | PHE 134 | 3.71 |
| Hydrophobic | ILE 139 | 3.76 | ||
| Hydrophobic | PHE 140 | 3.98 |
| Code | Binding Energy (Kcal) | Type of Interactions | Amino Acid Residues Involved | H-DIST (Å) |
|---|---|---|---|---|
| 5c | −7.0 | Hydrophobic | ILE 197 | 3.85 |
| Hydrophobic | LEU 200 | 3.52 | ||
| Hydrophobic | LEU 200 | 3.42 | ||
| Hydrophobic | ASN 263 | 3.55 | ||
| Hydrophobic | VAL 297 | 3.42 | ||
| Hydrophobic | ILE 311 | 3.74 | ||
| H-Bond | ASN 263 | 2.67 | ||
| H-Bond | THR 265 | 2.41 | ||
| H-Bond | THR 309 | 2.25 | ||
| Halogen Bond | VAL 207 | 3.72 | ||
| Halogen Bond | LEU 209 | 3.16 | ||
| Halogen Bond | THR 296 | 3.69 | ||
| 5h | −9.5 | Hydrophobic | ASP 199 | 3.91 |
| Hydrophobic | LEU 200 | 3.66 | ||
| Hydrophobic | LEU 200 | 3.48 | ||
| Hydrophobic | ILE 228 | 3.80 | ||
| Hydrophobic | ASN 263 | 3.67 | ||
| Hydrophobic | VAL 297 | 3.39 | ||
| Hydrophobic | THR 309 | 3.40 | ||
| Hydrophobic | ILE 311 | 3.78 | ||
| H-Bond | THR 265 | 2.42 | ||
| H-Bond | THR 309 | 2.27 | ||
| 9PC | −9.7 | Hydrophobic | VAL 203 | 3.99 |
| Hydrophobic | THR 309 | 3.68 | ||
| H-Bond | GLY 205 | 2.74 | ||
| H-Bond | LEU 209 | 1.99 | ||
| H-Bond | ASN 263 | 1.91 | ||
| H-Bond | THR 296 | 3.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Özdemir, Z.; Zenni, Y.N.; Karakurt, A.; Sari, S.; Saraç, S.; Akdağ, M.; Merde, İ.B.; Kart, D.; Venanzoni, R.; Flores, G.A.; et al. Potent Antimicrobial Azoles: Synthesis, In Vitro and In Silico Study. Antibiotics 2024, 13, 1044. https://doi.org/10.3390/antibiotics13111044
Özdemir Z, Zenni YN, Karakurt A, Sari S, Saraç S, Akdağ M, Merde İB, Kart D, Venanzoni R, Flores GA, et al. Potent Antimicrobial Azoles: Synthesis, In Vitro and In Silico Study. Antibiotics. 2024; 13(11):1044. https://doi.org/10.3390/antibiotics13111044
Chicago/Turabian StyleÖzdemir, Zeynep, Yaren Nur Zenni, Arzu Karakurt, Suat Sari, Selma Saraç, Mevlüt Akdağ, İrem Bozbey Merde, Didem Kart, Roberto Venanzoni, Giancarlo Angeles Flores, and et al. 2024. "Potent Antimicrobial Azoles: Synthesis, In Vitro and In Silico Study" Antibiotics 13, no. 11: 1044. https://doi.org/10.3390/antibiotics13111044
APA StyleÖzdemir, Z., Zenni, Y. N., Karakurt, A., Sari, S., Saraç, S., Akdağ, M., Merde, İ. B., Kart, D., Venanzoni, R., Flores, G. A., Angelini, P., Kabier, M., Mathew, B., & Carradori, S. (2024). Potent Antimicrobial Azoles: Synthesis, In Vitro and In Silico Study. Antibiotics, 13(11), 1044. https://doi.org/10.3390/antibiotics13111044













