Aeromonas spp. in Freshwater Bodies: Antimicrobial Resistance and Biofilm Assembly
Abstract
1. Introduction
2. Results
2.1. Characterization of Aeromonas Isolates
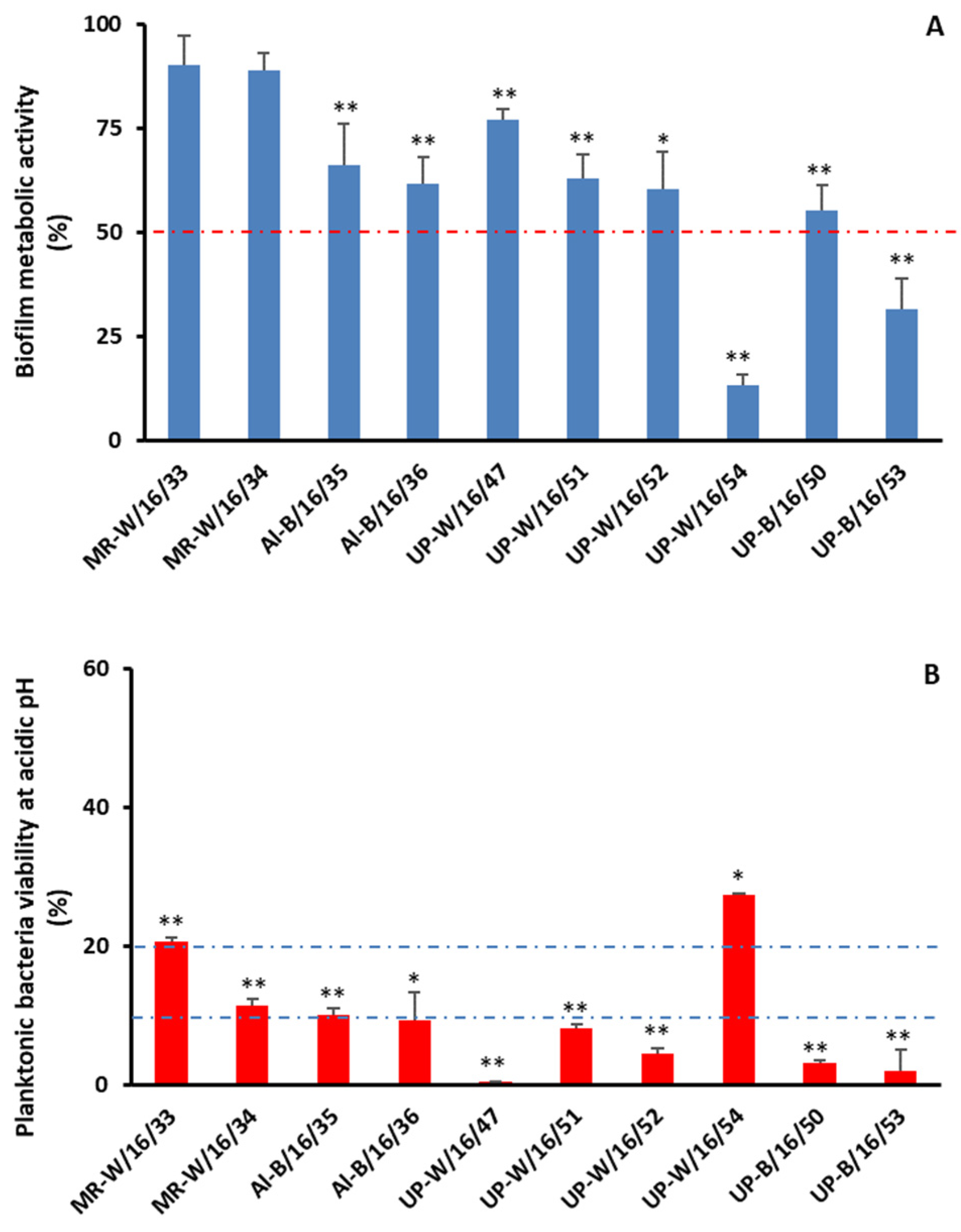
2.2. Biofilm Assembly
2.3. Chlorination
3. Discussion
4. Materials and Methods
4.1. Sampling Collection
4.2. Microorganism Isolation and Identification
4.3. Antimicrobial Susceptibility Testing
4.4. Biofilm Assay
4.5. Mobility Assays and Hemolytic Activity
4.6. Chlorination
4.6.1. Biofilms
4.6.2. Planktonic Bacteria
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fernández-Bravo, A.; Figueras, M.J. An Update on the Genus Aeromonas: Taxonomy, Epidemiology, and Pathogenicity. Microorganisms 2020, 8, 129. [Google Scholar] [CrossRef]
- van der Wielen, P.W.J.J.; Bakker, G.; Atsma, A.; Lut, M.; Roeselersd, G.; de Graaf, B. survey of indicator parameters to monitor regrowth in unchlorinated drinking water. Environ. Sci. Water Res. Technol. 2016, 2, 683–692. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef]
- Pessoa, R.B.G.; de Oliveira, W.F.; Correia, M.T.S.; Fontes, A.; Coelho, L.C.B.B. Aeromonas and Human Health Disorders: Clinical Approaches. Front. Microbiol. 2022, 13, 868890. [Google Scholar] [CrossRef] [PubMed]
- Conte, D.; Palmeiro, J.K.; Bavaroski, A.A.; Rodrigues, L.S.; Cardozo, D.; Tomaz, A.P.; Camargo, J.O.; Dalla-Costa, L.M. Antimicrobial resistance in Aeromonas species isolated from aquatic environments in Brazil. J. Appl. Microbiol. 2021, 131, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Nhinh, D.T.; Le, D.V.; Van, K.V.; Huong Giang, N.T.; Dang, L.T.; Hoai, T.D. Prevalence, Virulence Gene Distribution and Alarming the Multidrug Resistance of Aeromonas hydrophila Associated with Disease Outbreaks in Freshwater Aquaculture. Antibiotics 2021, 10, 532. [Google Scholar] [CrossRef] [PubMed]
- Skwor, T.; Stringer, S.; Haggerty, J.; Johnson, J.; Duhr, S.; Johnson, M.; Seckinger, M.; Stemme, M. Prevalence of potentially pathogenic antibiotic-resistant Aeromonas spp. in treated urban wastewater effluents versus recipient riverine populations: A 3-year comparative study. Appl. Environ. Microbiol. 2020, 86, e02053-19. [Google Scholar] [CrossRef] [PubMed]
- Carusi, J.; Kabuki, D.Y.; de Seixas Pereira, P.M.; Cabral, L. Aeromonas spp. in drinking water and food: Occurrence, virulence potential and antimicrobial resistance. Food Res. Int. 2024, 175, 113710. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.L. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 2009, 157, 2893–2902. [Google Scholar] [CrossRef] [PubMed]
- Ansari, M.I.; Schiwon, K.; Malik, A.; Grohmann, E. Biofilm formation by environmental bacteria. In Environmental Protection Strategies for Sustainable Development; Malik, A., Grohmann, E., Eds.; Springer: Rotterdam, The Netherlands, 2012; pp. 341–377. [Google Scholar]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, Y.; Ichijo, T.; Uchii, K.; Nasu, M. Changes in bacterial diversity and community structure in drinking water distribution system revealed by high throughput sequencing. J. Microorg. Control 2023, 28, 27–34. [Google Scholar] [CrossRef]
- Lu, Y.W.; Liang, X.X.; Wang, C.Y.; Chen, D.; Liu, H. Synergistic nanowire-assisted electroporation and chlorination for inactivation of chlorine-resistant bacteria in drinking water systems via inducing cell pores for chlorine permeation. Water Res. 2023, 229, 119399. [Google Scholar] [CrossRef]
- Luo, L.W.; Wu, Y.H.; Chen, G.Q.; Wang, H.B.; Wang, Y.H.; Tong, X.; Bai, Y.; Xu, Y.Q.; Zhang, Z.W.; Ikuno, N.; et al. Chlorine-resistant bacteria (CRB) in the reverse osmosis system for wastewater reclamation: Isolation, identification and membrane fouling mechanisms. Water Res. 2022, 209, 117966. [Google Scholar] [CrossRef]
- Cho, M.; Kim, J.; Kim, J.Y.; Yoon, J.; Kim, J.H. Mechanisms of Escherichia coli inactivation by several disinfectants. Water Res. 2010, 44, 3410–3418. [Google Scholar] [CrossRef] [PubMed]
- Raposo, A.; Mansilha, C.; Veber, A.; Melo, A.; Rodrigues, J.; Matias, R.; Rebelo, H.; Grossinho, J.; Cano, M.; Almeida, C.; et al. Occurrence of polycyclic aromatic hydrocarbons, microplastics and biofilms in Alqueva surface water at touristic spots. Sci. Total Environ. 2022, 850, 157983. [Google Scholar] [CrossRef] [PubMed]
- Sousa, M.; Morgado, P.; Rodrigues, J.; Matias, R.; Nogueira, I.; Jordao, L. Caracterização da população bacteriana em barragens na bacia hidrografica do Sado. Bol. Epidemiol. Obs. 2019, 11, 44–48. [Google Scholar]
- Stepanovic, S.; Vukovic, D.; Hola, V.; di Bonaventura, G.; Djukic, S.; Cirkovic, I.; Ruzicka, F. Quantification of Biofilm in Microtiter Plates: Overview of Testing Conditions and Practical Recommendations for Assessment of Biofilm Production by Staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Kirov, S.M.; Tassell, B.C.; Semmler, A.B.; O’Donovan, L.A.; Rabaan, A.A.; Shaw, J.G. Lateral flagella and swarming motility in Aeromonas species. J. Bacteriol. 2002, 184, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Decreto-Lei n° 152/2017, de 7 de Dezembro. Available online: https://diariodarepublica.pt/dr/detalhe/decreto-lei/152-2017-114315242 (accessed on 17 January 2024).
- Bertran, X.; Rubio, M.; Gómez, L.; Llovet, T.; Muñoz, C.; Navarro, F.; Miro, E. Taxonomic Identification of Different Species of the Genus Aeromonas by Whole-Genome Sequencing and Use of Their Species-Specific β-Lactamases as Phylogenetic Markers. Antibiotics 2021, 10, 354. [Google Scholar] [CrossRef]
- Shin, H.B.; Yoon, J.; Lee, Y.; Kim, M.S.; Lee, K. Comparison of MALDI-TOF MS, housekeeping gene sequencing, and 16S rRNA gene sequencing for identification of Aeromonas clinical isolates. Yonsei Med. J. 2015, 56, 550–555. [Google Scholar] [CrossRef]
- Soler, L.; Yáñez, M.A.; Chacon, M.R.; Aguilera-Arreola, M.G.; Catalán, V.; Figueras, M.J.; Martínez-Murcia, A.J. Phylogenetic analysis of the genus Aeromonas based on two housekeeping genes. Int. J. Syst. Evol. Microbiol. 2004, 54, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Dhanapala, P.M.; Kalupahana, R.S.; Kalupahana, A.W.; Wijesekera, D.P.H.; Kottawatta, S.A.; Jayasekera, N.K.; Silva-Fletcher, A.; Jagoda, S.S.S.S. Characterization and Antimicrobial Resistance of Environmental and Clinical Aeromonas Species Isolated from Fresh Water Ornamental Fish and Associated Farming Environment in Sri Lanka. Microorganisms 2021, 9, 2106. [Google Scholar] [CrossRef] [PubMed]
- Sadique, A.; Neogi, S.B.; Bashar, T.; Sultana, M.; Johura, F.-T.; Islam, S.; Hasan, N.A.; Huq, A.; Colwell, R.R.; Alam, M. Dynamics, Diversity, and Virulence of Aeromonas spp. in Homestead Pond Water in Coastal Bangladesh. Front. Public Health 2021, 9, 692166. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). AWaRe Classification of Antibiotics for Evaluation and Monitoring of Use. 2023. Available online: https://www.who.int/publications/i/item/WHO-MHP-HPS-EML-2023.04 (accessed on 12 January 2024).
- Heuzenroeder, M.W.; Wong, C.Y. Distribution of two hemolytic toxin genes in clinical and environmental isolates of Aeromonas spp.: Correlation with virulence in a suckling mouse model. FEMS Microbiol. Lett. 1999, 174, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Lau, T.V.; Puah, S.M.; Tan, J.M.A.; Merino, S.; Puthucheary, S.D.; Chua, K.H. Flagellar motility mediates biofilm formation in Aeromonas dhakensis. Microb. Pathog. 2023, 177, 106059. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The Microbial “Protective Clothing” in Extreme Environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef] [PubMed]
- Watnick, P.; Kolter, R. Biofilm, city of microbes. J. Bacteriol. 2000, 182, 2675–2679. [Google Scholar] [CrossRef] [PubMed]
- Karem, K.L.; Foster, J.W.; Bej, A.K. Adaptive acid tolerance response (ATR) in Aeromonas hydrophila. Microbiology 1994, 140, 1731–1736. [Google Scholar] [CrossRef]
- Nojoumi, S.A.; Smith, D.G.; Rowbury, R.J. Tolerance to acid in pH 5.0-grown organisms of potentially pathogenic gram-negative bacteria. Lett. Appl. Microbiol. 1995, 21, 359–363. [Google Scholar] [CrossRef]
- Scoaris, D.O.; Colacite, J.; Nakamura, C.V.; Ueda-Nakamura, T.; de Abreu Filho, B.A.; Dias Filho, B.P. Virulence and antibiotic susceptibility of Aeromonas spp. isolated from drinking water. Antonie Van Leeuwenhoek 2008, 93, 111–122. [Google Scholar] [CrossRef]
- Wadström, T.; Ljungh, A. Aeromonas and Plesiomonas as food- and waterborne pathogens. Int. J. Food Microbiol. 1991, 12, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, M.; Rodrigues, J.; Reis, L.; Nogueira, I.; Carvalho, P.; Brandão, J.; Duarte, A.; Jordao, L. Pathogens in ornamental waters: A pilot study. Int. J. Environ. Res. Public Health 2016, 13, 216. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2022. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 31 January 2022).
- Bandeira, M.; Carvalho, P.A.; Duarte, A.; Jordao, L. Exploring dangerous connections between Klebsiella pneumoniae biofilms and healthcare-associated infections. Pathogens 2014, 19, 720–731. [Google Scholar] [CrossRef]
- Gavín, R.; Merino, S.; Altarriba, M.; Canals, R.; Shaw, J.G.; Tomás, J.M. Lateral flagella are required for increased cell adherence, invasion and biofilm formation by Aeromonas spp. FEMS Microbiol. Lett. 2003, 224, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Morgado, P.I.; Jose, S.; Wanke, R.M.; Antunes, A.M.; Cardoso, A.S.; Jordao, L. Integration of cellular and molecular endpoints to assess the toxicity of polycyclic aromatic hydrocarbons in HepG2 cell line. Environ. Toxicol. Chem. 2017, 36, 3404–3414. [Google Scholar] [CrossRef] [PubMed]
| Source | ID | Vitek-MS | 16S rRNA |
|---|---|---|---|
| Water | Mz-W/21/18 | A. salmonicida/bestiarium | A. salmonicida |
| (Natural) | Mz-W/21/60 | A. sobria | A. hydrophila |
| Mz-W/21/58 | A. sobria | A. veronii | |
| Mo-W/21/09 | A. salmonicida/bestiarium | A. salmonicida | |
| Mo-W/21/15 | A. salmonicida/bestiarium | A. salmonicida | |
| Mo-W/21/65 | A. sobria | A. hydrophila/veronii | |
| Mo-W/21/10 | A. media | A. media | |
| Am-W/21/06 | A. salmonicida/bestiarium | A. popoffii | |
| Am-W/21/53 | A. sobria | A. veronii | |
| Am-W/21/05 | A. veronii | A. veronii | |
| Am-W/21/07 | A. veronii | A. veronii | |
| MR-W/16/33 | A. veronii | A. veronii | |
| MR-W/16/34 | A. sobria | A. veronii | |
| Biofilm | Al-B/16/35 | A. sobria | A. veronii |
| (Natural) | Al-B/16/36 | A. sobria | A. veronii |
| Water | UP-W/16/47 | A. sobria | A. veronii |
| (Anthropogenic) | UP-W/16/51 | A. hydrophila/caviae | A. veronii |
| UP-W/16/52 | A. veronii | A. veronii | |
| UP-W/16/54 | A. sobria | A. veronii | |
| Biofilm | UP-B/16/50 | A. sobria | A. veronii |
| (Anthropogenic) | UP-B/16/53 | A. sobria | A. veronii |
| Source | ID | CAZ10 | CIP5 | LEV5 | STX25 | FOX30 | IMP10 | MEM10 | CN30 |
|---|---|---|---|---|---|---|---|---|---|
| Water | Mz-W/21/18 | S | S | S | S | S | I | S | S |
| (Natural) | Mz-W/21/60 | S | S | S | S | S | S | S | S |
| Mz-W/21/58 | S | S | S | S | S | R | S | S | |
| Mo-W/21/09 | R | I | S | S | R | R | S | S | |
| Mo-W/21/15 | S | S | S | S | R | R | S | S | |
| Mo-W/21/65 | S | S | S | S | S | I | S | S | |
| Mo-W/21/10 | S | S | S | S | S | S | S | S | |
| Am-W/21/06 | S | S | S | S | S | S | S | S | |
| Am-W/21/53 | S | S | S | S | S | R | I | S | |
| Am-W/21/05 | I | S | S | S | S | S | S | S | |
| Am-W/21/07 | I | S | S | S | S | S | S | S | |
| MR-W/16/33 | S | S | S | S | S | S | S | S | |
| MR-W/16/34 | S | S | S | S | R | R | S | S | |
| Biofilm | Al-B/16/35 | S | S | S | S | S | I | S | S |
| (Natural) | Al-B/16/36 | S | S | S | S | S | R | S | S |
| Water | UP-W/16/47 | S | S | S | S | S | S | S | R |
| (Anthropogenic) | UP-W/16/51 | S | S | S | S | S | R | I | R |
| UP-W/16/52 | S | S | S | S | S | I | I | R | |
| UP-W/16/54 | S | S | S | S | S | R | I | R | |
| Biofilm | UP-B/16/50 | S | S | S | S | S | S | I | R |
| (Anthropogenic) | UP-B/16/53 | S | S | S | S | S | R | I | R |
| Bacteria (No. of Isolates) | % (No.) of Isolates Resistant to an Antibiotic a | MARindex b | |||
|---|---|---|---|---|---|
| CAZ10 | FOX30 | IMP10 | CN30 | ||
| A. salmonicida | 33% (1) | 67% (2) | 67% (2) | 0% (0) | 0.250 |
| (n = 3) | |||||
| A. veronii | 0% (0) | 7.2% (1) | 50% (7) | 43% (6) | 0.125 |
| (n = 14) | |||||
| Species (No. of Isolates) | ID | Biofilm a | Swimming b | Swarming c | Hemolysis d |
|---|---|---|---|---|---|
| A. salmonicida (3) | Mz-W/21/18 | NBP | + | - | + |
| Mo-W/21/09 | NBP | + | + | + | |
| Mo-W/21/15 | NBP | - | - | + | |
| A. hydrophila (1) | Mz-W/21/60 | NBP | + | - | + |
| A. hydrophila/veronii (1) | Mo-W/21/65 | NBP | - | - | + |
| A. media (1) | Mo-W/21/10 | NBP | + | - | - |
| A. popoffii (1) | Am-W/21/06 | NBP | + | - | + |
| A. veronii | Mz-W/21/58 | WBP | + | - | + |
| (14) | Am-W/21/53 | NBP | ++ | - | + |
| Am-W/21/05 | NBP | + | - | - | |
| Am-W/21/07 | NBP | + | - | - | |
| MR-W/16/33 | SBP | ++ | - | + | |
| MR-W/16/34 | SBP | ++ | + | - | |
| Al-B/16/35 | SBP | + | - | + | |
| Al-B/16/36 | SBP | + | - | + | |
| UP-W/16/47 | SBP | + | - | - | |
| UP-W/16/51 | SBP | ++ | - | + | |
| UP-W/16/52 | SBP | + | - | + | |
| UP-W/16/54 | SBP | ++ | - | + | |
| UP-B/16/50 | SBP | + | + | - | |
| UP-B/16/53 | SBP | + | + | - |
| Bacteria | Water | |||
|---|---|---|---|---|
| Species | ID | pH | Chlorine (mg/L) | Temperature (°C) |
| A. veronii | MR-W/16/33 | 6.60 | --- | 27.0 |
| MR-W/16/34 | 6.60 | --- | 27.0 | |
| Al-B/16/35 | 7.10 | --- | 33.0 | |
| Al-B/16/36 | 7.10 | --- | 33.0 | |
| UP-W/16/47 | 7.93 | --- | 14.7 | |
| UP-W/16/51 | 8.14 | 0.16 | 17.0 | |
| UP-W/16/52 | 8.14 | 0.10 | 17.0 | |
| UP-W/16/54 | 7.91 | 0.10 | 14.0 | |
| UP-B/16/50 | 8.01 | --- | 17.0 | |
| UP-B/16/53 | 7.91 | --- | 14.0 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nascimento, M.; Rodrigues, J.; Matias, R.; Jordao, L. Aeromonas spp. in Freshwater Bodies: Antimicrobial Resistance and Biofilm Assembly. Antibiotics 2024, 13, 166. https://doi.org/10.3390/antibiotics13020166
Nascimento M, Rodrigues J, Matias R, Jordao L. Aeromonas spp. in Freshwater Bodies: Antimicrobial Resistance and Biofilm Assembly. Antibiotics. 2024; 13(2):166. https://doi.org/10.3390/antibiotics13020166
Chicago/Turabian StyleNascimento, Maria, Joao Rodrigues, Rui Matias, and Luisa Jordao. 2024. "Aeromonas spp. in Freshwater Bodies: Antimicrobial Resistance and Biofilm Assembly" Antibiotics 13, no. 2: 166. https://doi.org/10.3390/antibiotics13020166
APA StyleNascimento, M., Rodrigues, J., Matias, R., & Jordao, L. (2024). Aeromonas spp. in Freshwater Bodies: Antimicrobial Resistance and Biofilm Assembly. Antibiotics, 13(2), 166. https://doi.org/10.3390/antibiotics13020166






