Anti-Microbial Activities of Mussel-Derived Recombinant Proteins against Gram-Negative Bacteria
Abstract
1. Introduction
2. Results
2.1. Recombinant MAP-Fused Functional Peptides (MAP-FPs)
2.2. Determination of Minimal Inhibitory Concentrations (MICs) of MAP-FPs
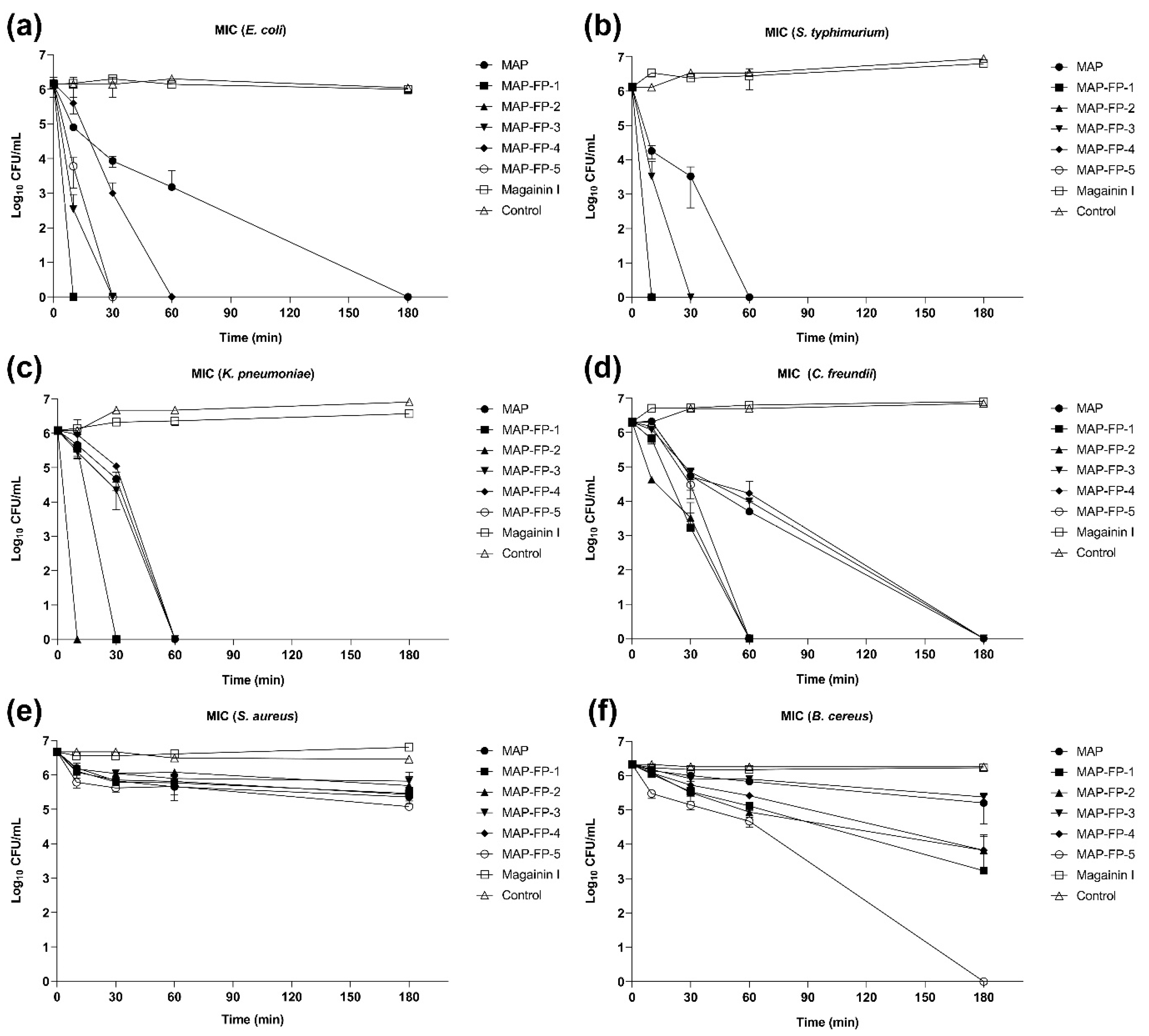
2.3. Time-Dependent Microbicidal Activity of MAP-FPs
2.4. Cytotoxicity of the MAP-FPs
2.5. Thermal Stability of the MAP-FPs
3. Discussion
4. Materials and Methods
4.1. Preparation of Recombinant Proteins
4.2. Microorganisms
4.3. Minimal Inhibitory Concentrations (MICs) Determinations
4.4. Time-Kill Kinetics (TKK) Assay
4.5. Cytotoxicity
4.6. Thermal Stability Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniam, B.; Prateek, R.S.; Saraf, M.; Kar, P.; Singh, S.P.; Thakur, V.K.; Singh, A.; Gupta, R.K. Antibacterial and Antiviral Functional Materials: Chemistry and Biological Activity toward Tackling COVID-19-like Pandemics. ACS Pharmacol. Transl. Sci. 2020, 4, 8–54. [Google Scholar] [CrossRef] [PubMed]
- Huemer, M.; Mairpady, S.S.; Brugger, S.D.; Zinkernagel, A.S. Antibiotic resistance and persistence-Implications for human health and treatment perspectives. EMBO Rep. 2020, 21, e51034. [Google Scholar] [CrossRef] [PubMed]
- Filippone, E.J.; Kraft, W.K.; Farber, J.L. The Nephrotoxicity of Vancomycin. Clin. Pharmacol. Ther. 2017, 102, 459–469. [Google Scholar] [CrossRef]
- Bulet, P.; Hetru, C.; Dimarcq, J.L.; Hoffmann, D. Antimicrobial peptides in insects; structure and function. Dev. Comp. Immunol. 1999, 23, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Steinstraesser, L.; Kraneburg, U.; Jacobsen, F.; Al-Benna, S. Host defense peptides and their antimicrobial-immunomodulatory duality. Immunobiology 2011, 216, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Nijnik, A.; Philpott, D.J. Modulating immunity as a therapy for bacterial infections. Nat. Rev. Microbiol. 2012, 10, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Pasupuleti, M.; Schmidtchen, A.; Malmsten, M. Antimicrobial peptides: Key components of the innate immune system. Crit. Rev. Biotechnol. 2012, 32, 143–171. [Google Scholar] [CrossRef]
- Aisenbrey, C.; Marquette, A.; Bechinger, B. The Mechanisms of Action of Cationic Antimicrobial Peptides Refined by Novel Concepts from Biophysical Investigations. Adv. Exp. Med. Biol. 2019, 1117, 33–64. [Google Scholar]
- Torrent, M.; Navarro, S.; Moussaoui, M.; Nogués, M.V.; Boix, E. Eosinophil cationic protein high-affinity binding to bacteria-wall lipopolysaccharides and peptidoglycans. Biochemistry 2008, 47, 3544–3555. [Google Scholar] [CrossRef] [PubMed]
- Bin, H.A.; Jiang, X.; Bergen, P.J.; Zhu, Y. Antimicrobial Peptides: An Update on Classifications and Databases. Int. J. Mol. Sci. 2021, 22, 11691. [Google Scholar]
- Tally, F.P.; DeBruin, M.F. Development of daptomycin for gram-positive infections. J. Antimicrob. Chemother. 2000, 46, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.N. Microbiological features of vancomycin in the 21st century: Minimum inhibitory concentration creep, bactericidal/static activity, and applied breakpoints to predict clinical outcomes or detect resistant strains. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2006, 42 (Suppl. S1), S13–S24. [Google Scholar] [CrossRef]
- Rai, A.; Ferrão, R.; Palma, P.; Patricio, T.; Parreira, P.; Anes, E.; Tonda-Turo, C.; Martins, M.C.L.; Alves, N.; Ferreira, L. Antimicrobial peptide-based materials: Opportunities and challenges. J. Mater. Chem. B 2022, 10, 2384–2429. [Google Scholar] [CrossRef]
- Rodríguez, A.A.; Otero-González, A.; Ghattas, M.; Ständker, L. Discovery, Optimization, and Clinical Application of Natural Antimicrobial Peptides. Biomedicines 2021, 9, 1381. [Google Scholar] [CrossRef]
- Choi, Y.S.; Kang, D.G.; Lim, S.; Yang, Y.J.; Kim, C.S.; Cha, H.J. Recombinant mussel adhesive protein fp-5 (MAP fp-5) as a bulk bioadhesive and surface coating material. Biofouling 2011, 27, 729–737. [Google Scholar] [CrossRef]
- Kaushik, N.K.; Kaushik, N.; Pardeshi, S.; Sharma, J.G.; Lee, S.H.; Choi, E.H. Biomedical and Clinical Importance of Mussel-Inspired Polymers and Materials. Mar. Drugs 2015, 13, 6792–6817. [Google Scholar] [CrossRef]
- Choi, B.H.; Cheong, H.; Jo, Y.K.; Bahn, S.Y.; Seo, J.H.; Cha, H.J. Highly purified mussel adhesive protein to secure biosafety for in vivo applications. Microb. Cell Factories 2014, 13, 52. [Google Scholar] [CrossRef]
- Cha, H.J.; Hwang, D.S.; Lim, S.; White, J.D.; Matos-Perez, C.A.; Wilker, J.J. Bulk adhesive strength of recombinant hybrid mussel adhesive protein. Biofouling 2009, 25, 99–107. [Google Scholar] [CrossRef]
- Hwang, D.S.; Gim, Y.; Yoo, H.J.; Cha, H.J. Practical recombinant hybrid mussel bioadhesive fp-151. Biomaterials 2007, 28, 3560–3568. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.S.; Sim, S.B.; Cha, H.J. Cell adhesion biomaterial based on mussel adhesive protein fused with RGD peptide. Biomaterials 2007, 28, 4039–4046. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.J.; Cheong, H.; Choi, E.S.; Yun, S.H.; Choi, B.H.; Park, K.S.; Kim, I.S.; Park, D.H.; Cha, H.J. Accelerated skin wound healing using electrospun nanofibrous mats blended with mussel adhesive protein and polycaprolactone. J. Biomed. Mater. Res. Part A 2017, 105, 218–225. [Google Scholar] [CrossRef]
- Rothstein, D.M.; Spacciapoli, P.; Tran, L.T.; Xu, T.; Roberts, F.D.; Dalla, S.M.; Buxton, D.K.; Oppenheim, F.G.; Friden, P. Anticandida activity is retained in P-113, a 12-amino-acid fragment of histatin 5. Antimicrob. Agents Chemother. 2001, 45, 1367–1373. [Google Scholar] [CrossRef] [PubMed]
- Dharmar, M.; Shanmugam, G.; Menakha, S.; Jagatheesh, K.; Sangeetha, M.; Thangavel, M.; Rajendran, V.; Krishnan, M.; Rengasamy, B.; Namasivayam, E. Molecular insights of newly identified potential peptide inhibitors of hypoxia inducible factor 1α causing breast cancer. J. Mol. Struct. 2019, 1177, 558–563. [Google Scholar]
- Lee, J.H.; Engler, J.A.; Collawn, J.F.; Moore, B.A. Receptor mediated uptake of peptides that bind the human transferrin receptor. Eur. J. Biochem. 2001, 268, 2004–2012. [Google Scholar] [CrossRef] [PubMed]
- Bruhn, K.W.; Spellberg, B. Transferrin-mediated iron sequestration as a novel therapy for bacterial and fungal infections. Curr. Opin. Microbiol. 2015, 27, 57–61. [Google Scholar] [CrossRef]
- Czechowicz, P.; Jaśkiewicz, M.; Neubauer, D.; Gościniak, G.; Kamysz, W. Anticandidal Activity of Omiganan and Its Retro Analog Alone and in Combination with Fluconazole. Probiotics Antimicrob. Proteins 2021, 13, 1173–1182. [Google Scholar] [CrossRef]
- Melo, M.N.; Castanho, M.A. Omiganan interaction with bacterial membranes and cell wall models. Assigning a biological role to saturation. Biochim. Biophys. Acta 2007, 1768, 1277–1290. [Google Scholar] [CrossRef]
- Obeid, M. Anticancer activity of targeted proapoptotic peptides and chemotherapy is highly improved by targeted cell surface calreticulin-inducer peptides. Mol. Cancer Ther. 2009, 8, 2693–2707. [Google Scholar] [CrossRef]
- Lin, L.; Chi, J.; Yan, Y.; Luo, R.; Feng, X.; Zheng, Y.; Xian, D.; Li, X.; Quan, G.; Liu, D.; et al. Membrane-disruptive peptides/peptidomimetics-based therapeutics: Promising systems to combat bacteria and cancer in the drug-resistant era. Acta Pharm. Sin. B 2021, 11, 2609–2644. [Google Scholar] [CrossRef] [PubMed]
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.; Black, D.S.; Walsh, W.R.; Kumar, N. A New Era of Antibiotics: The Clinical Potential of Antimicrobial Peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef]
- Matsuzaki, K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta 2009, 1788, 1687–1692. [Google Scholar] [CrossRef]
- Sykes, R.B.; Bonner, D.P. Aztreonam: The first monobactam. Am. J. Med. 1985, 78, 2–10. [Google Scholar] [CrossRef]
- Yotsuji, A.; Mitsuyama, J.; Hori, R.; Yasuda, T.; Saikawa, I.; Inoue, M.; Mitsuhashi, S. Mechanism of action of cephalosporins and resistance caused by decreased affinity for penicillin-binding proteins in Bacteroides fragilis. Antimicrob. Agents Chemother. 1988, 32, 1848–1853. [Google Scholar] [CrossRef] [PubMed]
- Domingues, M.M.; Inácio, R.G.; Raimundo, J.M.; Martins, M.; Castanho, M.A.; Santos, N.C. Biophysical characterization of polymyxin B interaction with LPS aggregates and membrane model systems. Biopolymers 2012, 98, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.P.; Messersmith, P.B.; Israelachvili, J.N.; Waite, J.H. Mussel-Inspired Adhesives and Coatings. Annu. Rev. Mater. Res. 2011, 41, 99–132. [Google Scholar] [CrossRef] [PubMed]
- Tagliaferri, T.L.; Jansen, M.; Horz, H.P. Fighting Pathogenic Bacteria on Two Fronts: Phages and Antibiotics as Combined Strategy. Front. Cell. Infect. Microbiol. 2019, 9, 22. [Google Scholar] [CrossRef]
- Römling, U.; Balsalobre, C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 2012, 272, 541–561. [Google Scholar] [CrossRef]
- Salwiczek, M.; Qu, Y.; Gardiner, J.; Strugnell, R.A.; Lithgow, T.; McLean, K.M.; Thissen, H. Emerging rules for effective antimicrobial coatings. Trends Biotechnol. 2014, 32, 82–90. [Google Scholar] [CrossRef]

| Peptide | Sequence | Molecular Weight | Hydrophobic Residues (%) | Net Charge at pH 7.0 |
|---|---|---|---|---|
| MAP-FP-1 | AKRHHGYKRKFH | 1564.82 | 42 | 5.27 |
| MAP-FP-2 | LKKLAKLALAF | 1215.59 | 27 | 3.00 |
| MAP-FP-3 | THRPPMWSPVWP | 1490.75 | 17 | 1.09 |
| MAP-FP-4 | ILRWPWWPWRRK | 1780.16 | 33 | 4.00 |
| MAP-FP-5 | KLAKLAKKLAKLAK | 1524.01 | 43 | 6.00 |
| Magainin I | GIGKFLHSAGKFGKAFVGEIMKS | 2409.88 | 30 | 3.09 |
| Peptide | Gram-Negative | Gram-Positive | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | S. typhimurium | K. pneumoniae | C. freundii | S. aureus | B. cereus | |||||||
| MIC (µM) | p Value | MIC (µM) | p Value | MIC (µM) | p Value | MIC (µM) | p Value | MIC (µM) | p Value | MIC (µM) | p Value | |
| MAP | 8 | <0.0001 | 8 | <0.0001 | n. d. | - | n. d. | - | n. d. | - | n. d. | - |
| MAP-FP-1 | 8 | <0.0001 | 8 | <0.0001 | 8 | <0.0001 | 8 | 0.0001 | n. d. | - | n. d. | - |
| MAP-FP-2 | 4 | <0.0001 | 4 | <0.0001 | 8 | 0.0001 | 8 | 0.0011 | n. d. | - | n. d. | - |
| MAP-FP-3 | 8 | <0.0001 | 8 | <0.0001 | 8 | 0.0091 | n. d. | - | n. d. | - | n. d. | - |
| MAP-FP-4 | 4 | <0.0001 | 4 | <0.0001 | 8 | <0.0001 | 8 | <0.0001 | n. d. | - | n. d. | - |
| MAP-FP-5 | 4 | <0.0001 | 4 | 0.0001 | 8 | <0.0001 | 8 | <0.0001 | n. d. | - | n. d. | - |
| Magainin | 4 | <0.0001 | 4 | <0.0001 | 4 | <0.0001 | 8 | <0.0001 | n. d. | - | n. d. | - |
| Peptide | CC50 (µM) | |
|---|---|---|
| Vero | HEK293T | |
| MAP | >12 | >12 |
| MAP-FP-1 | 8.3 | >12 |
| MAP-FP-2 | >12 | >12 |
| MAP-FP-3 | >12 | >12 |
| MAP-FP-4 | 5.8 | >12 |
| MAP-FP-5 | 7.3 | >12 |
| Day of Storage | MAP | MAP-FP-2 | ||
|---|---|---|---|---|
| Recovery ± RSD | MIC (µM) | Recovery ± RSD | MIC (µM) | |
| 0 | 100.00 ± 0.78 | 8 | 100.00 ± 3.61 | 4 |
| 14 | 100.02 ± 0.19 | 8 | 90.89 ± 1.44 | 4 |
| 28 | 96.36 ± 0.30 | 8 | 91.33 ± 2.60 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.Y.; Oh, Y.B.; Park, J.S.; Min, Y.-H.; Park, M.C. Anti-Microbial Activities of Mussel-Derived Recombinant Proteins against Gram-Negative Bacteria. Antibiotics 2024, 13, 239. https://doi.org/10.3390/antibiotics13030239
Kim DY, Oh YB, Park JS, Min Y-H, Park MC. Anti-Microbial Activities of Mussel-Derived Recombinant Proteins against Gram-Negative Bacteria. Antibiotics. 2024; 13(3):239. https://doi.org/10.3390/antibiotics13030239
Chicago/Turabian StyleKim, Dong Yun, You Bin Oh, Je Seon Park, Yu-Hong Min, and Min Chul Park. 2024. "Anti-Microbial Activities of Mussel-Derived Recombinant Proteins against Gram-Negative Bacteria" Antibiotics 13, no. 3: 239. https://doi.org/10.3390/antibiotics13030239
APA StyleKim, D. Y., Oh, Y. B., Park, J. S., Min, Y.-H., & Park, M. C. (2024). Anti-Microbial Activities of Mussel-Derived Recombinant Proteins against Gram-Negative Bacteria. Antibiotics, 13(3), 239. https://doi.org/10.3390/antibiotics13030239







