Brave New World of Artificial Intelligence: Its Use in Antimicrobial Stewardship—A Systematic Review
Abstract
1. Introduction
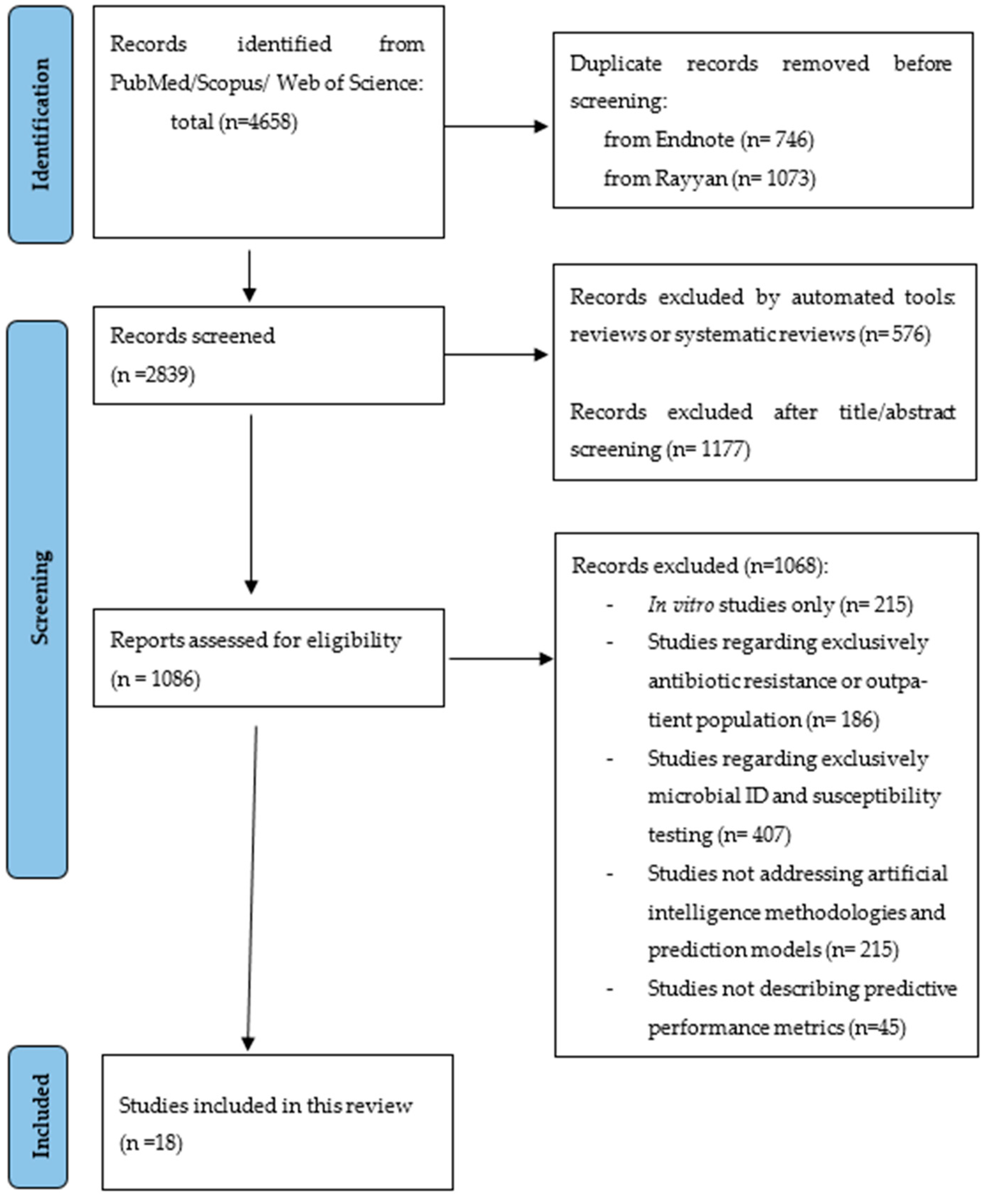
2. Results
2.1. Characteristics of the Included Studies
2.2. Risk of Bias/Quality Assessment
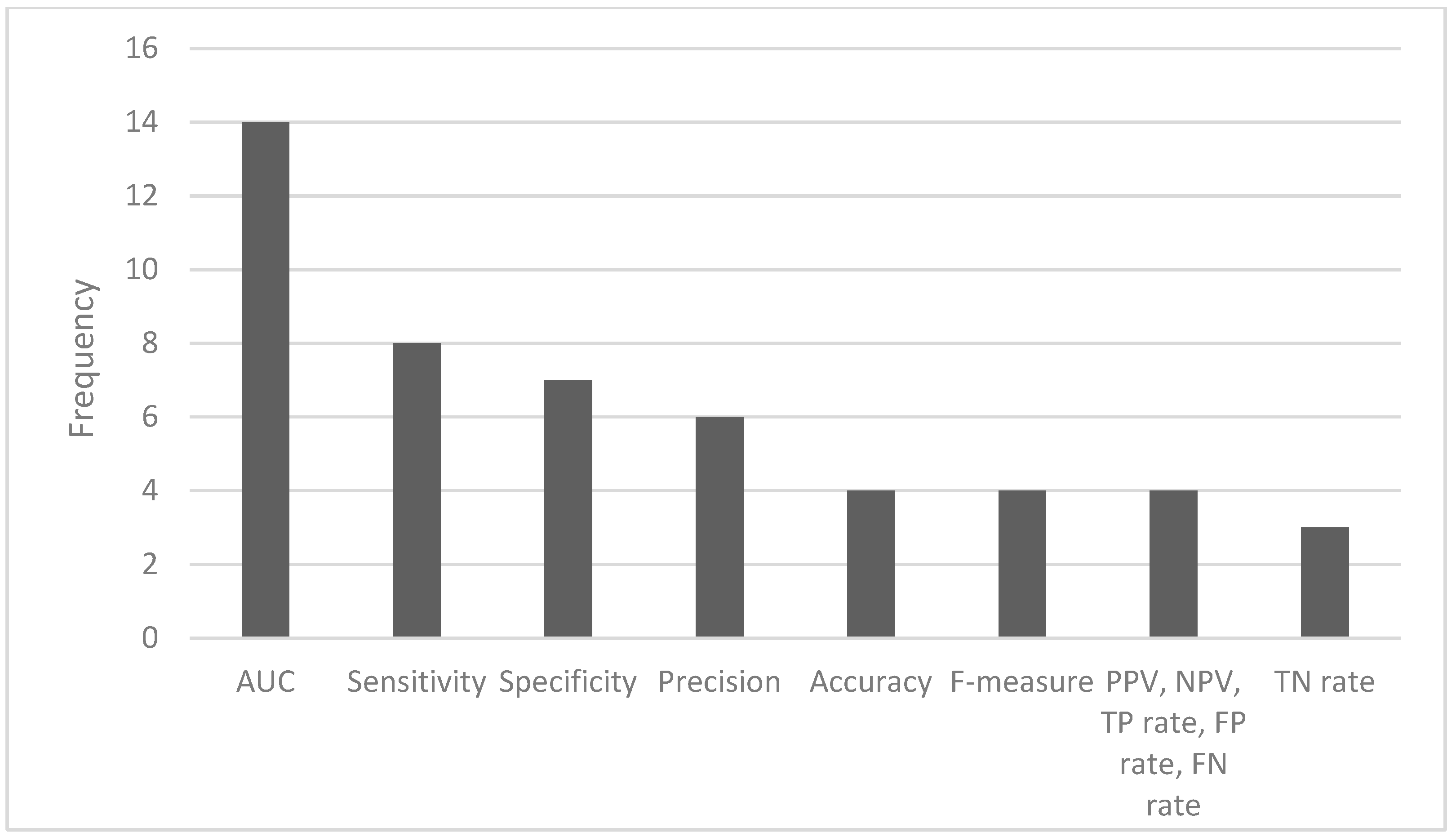
2.3. Predictive Performance of Artificial Intelligence Algorithms
3. Discussion
3.1. Main Findings
3.2. AI and Antimicrobial Stewardship
3.3. Limitations of the Studies Included
3.4. Limitations of the Review
4. Materials and Methods
4.1. Data Source and Search Strategy
4.2. Eligibility Criteria
4.3. Data Extraction and Synthesis
4.4. Risk of Bias (ROB) Assessment
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tripartite and UNEP Support OHHLEP’s Definition of ‘One Health’. Available online: https://www.who.int/news/item/01-12-2021-tripartite-and-unep-support-ohhlep-s-definition-of-one-health (accessed on 20 September 2023).
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Rev. Antimicrob. Resist. Arch. Pharm. Pract. 2016, 7, 110. [Google Scholar]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell. Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef]
- González-Zorn, B.; Escudero, J.A. Ecology of Antimicrobial Resistance: Humans, Animals, Food and Environment. Int. Microbiol. 2012, 15, 101–109. [Google Scholar] [CrossRef]
- Rice, L.B. Antimicrobial Stewardship and Antimicrobial Resistance. Med. Clin. N. Am. 2018, 102, 805–818. [Google Scholar] [CrossRef]
- McGowan, J.E.; Finland, M. Usage of Antibiotics in a General Hospital: Effect of Requiring Justification. J. Infect. Dis. 1974, 130, 165–168. [Google Scholar] [CrossRef]
- Lanckohr, C.; Bracht, H. Antimicrobial Stewardship. Curr. Opin. Crit. Care 2022, 28, 551–556. [Google Scholar] [CrossRef]
- Donà, D.; Barbieri, E.; Daverio, M.; Lundin, R.; Giaquinto, C.; Zaoutis, T.; Sharland, M. Implementation and Impact of Pediatric Antimicrobial Stewardship Programs: A Systematic Scoping Review. Antimicrob. Resist. Infect. Control 2020, 9, 3. [Google Scholar] [CrossRef]
- Contejean, A.; Abbara, S.; Chentouh, R.; Alviset, S.; Grignano, E.; Gastli, N.; Casetta, A.; Willems, L.; Canouï, E.; Charlier, C.; et al. Antimicrobial Stewardship in High-Risk Febrile Neutropenia Patients. Antimicrob. Resist. Infect. Control 2022, 11, 52. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Alhumaid, S.; Mutair, A.A.; Garout, M.; Abulhamayel, Y.; Halwani, M.A.; Alestad, J.H.; Bshabshe, A.A.; Sulaiman, T.; AlFonaisan, M.K.; et al. Application of Artificial Intelligence in Combating High Antimicrobial Resistance Rates. Antibiotics 2022, 11, 784. [Google Scholar] [CrossRef] [PubMed]
- Brotherton, A.L. Metrics of Antimicrobial Stewardship Programs. Med. Clin. N. Am. 2018, 102, 965–976. [Google Scholar] [CrossRef]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6, 6.2.10. [Google Scholar] [CrossRef]
- Peiffer-Smadja, N.; Rawson, T.M.; Ahmad, R.; Buchard, A.; Georgiou, P.; Lescure, F.-X.; Birgand, G.; Holmes, A.H. Machine Learning for Clinical Decision Support in Infectious Diseases: A Narrative Review of Current Applications. Clin. Microbiol. Infect. 2020, 26, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Weis, C.V.; Jutzeler, C.R.; Borgwardt, K. Machine Learning for Microbial Identification and Antimicrobial Susceptibility Testing on MALDI-TOF Mass Spectra: A Systematic Review. Clin. Microbiol. Infect. 2020, 26, 1310–1317. [Google Scholar] [CrossRef]
- McCoy, A.; Das, R. Reducing Patient Mortality, Length of Stay and Readmissions through Machine Learning-Based Sepsis Prediction in the Emergency Department, Intensive Care Unit and Hospital Floor Units. BMJ Open Qual. 2017, 6, e000158. [Google Scholar] [CrossRef]
- Calvert, J.S.; Price, D.A.; Chettipally, U.K.; Barton, C.W.; Feldman, M.D.; Hoffman, J.L.; Jay, M.; Das, R. A Computational Approach to Early Sepsis Detection. Comput. Biol. Med. 2016, 74, 69–73. [Google Scholar] [CrossRef]
- Saybani, M.R.; Shamshirband, S.; Golzari Hormozi, S.; Wah, T.Y.; Aghabozorgi, S.; Pourhoseingholi, M.A.; Olariu, T. Diagnosing Tuberculosis With a Novel Support Vector Machine-Based Artificial Immune Recognition System. Iran. Red. Crescent Med. J. 2015, 17. [Google Scholar] [CrossRef]
- Maiellaro, P.; Cozzolongo, R.; Marino, P. Artificial Neural Networks for the Prediction of Response to Interferon Plus Ribavirin Treatment in Patients with Chronic Hepatitis C. Curr. Pharm. Des. 2004, 10, 2101–2109. [Google Scholar] [CrossRef]
- Li, S.; Tang, B.; He, H. An Imbalanced Learning Based MDR-TB Early Warning System. J. Med. Syst. 2016, 40, 164. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, M.; Kabanza, F.; Nault, V.; Valiquette, L. Evaluation of a Machine Learning Capability for a Clinical Decision Support System to Enhance Antimicrobial Stewardship Programs. Artif. Intell. Med. 2016, 68, 29–36. [Google Scholar] [CrossRef] [PubMed]
- De Vries, S.; Ten Doesschate, T.; Totté, J.E.E.; Heutz, J.W.; Loeffen, Y.G.T.; Oosterheert, J.J.; Thierens, D.; Boel, E. A Semi-Supervised Decision Support System to Facilitate Antibiotic Stewardship for Urinary Tract Infections. Comput. Biol. Med. 2022, 146, 105621. [Google Scholar] [CrossRef]
- Corbin, C.K.; Sung, L.; Chattopadhyay, A.; Noshad, M.; Chang, A.; Deresinksi, S.; Baiocchi, M.; Chen, J.H. Personalized Antibiograms for Machine Learning Driven Antibiotic Selection. Commun. Med. 2022, 2, 38. [Google Scholar] [CrossRef]
- Feretzakis, G.; Loupelis, E.; Sakagianni, A.; Kalles, D.; Martsoukou, M.; Lada, M.; Skarmoutsou, N.; Christopoulos, C.; Valakis, K.; Velentza, A.; et al. Using Machine Learning Techniques to Aid Empirical Antibiotic Therapy Decisions in the Intensive Care Unit of a General Hospital in Greece. Antibiotics 2020, 9, 50. [Google Scholar] [CrossRef]
- Shi, Z.-Y.; Hon, J.-S.; Cheng, C.-Y.; Chiang, H.-T.; Huang, H.-M. Applying Machine Learning Techniques to the Audit of Antimicrobial Prophylaxis. Appl. Sci. 2022, 12, 2586. [Google Scholar] [CrossRef]
- Stracy, M.; Snitser, O.; Yelin, I.; Amer, Y.; Parizade, M.; Katz, R.; Rimler, G.; Wolf, T.; Herzel, E.; Koren, G.; et al. Minimizing Treatment-Induced Emergence of Antibiotic Resistance in Bacterial Infections. Science 2022, 375, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Bolton, W.J.; Rawson, T.M.; Hernandez, B.; Wilson, R.; Antcliffe, D.; Georgiou, P.; Holmes, A.H. Machine Learning and Synthetic Outcome Estimation for Individualised Antimicrobial Cessation. Front. Digit. Health 2022, 4, 997219. [Google Scholar] [CrossRef]
- Feretzakis, G.; Sakagianni, A.; Loupelis, E.; Kalles, D.; Skarmoutsou, N.; Martsoukou, M.; Christopoulos, C.; Lada, M.; Petropoulou, S.; Velentza, A.; et al. Machine Learning for Antibiotic Resistance Prediction: A Prototype Using Off-the-Shelf Techniques and Entry-Level Data to Guide Empiric Antimicrobial Therapy. Healthc. Inform. Res. 2021, 27, 214–221. [Google Scholar] [CrossRef]
- Bystritsky, R.J.; Beltran, A.; Young, A.T.; Wong, A.; Hu, X.; Doernberg, S.B. Machine Learning for the Prediction of Antimicrobial Stewardship Intervention in Hospitalized Patients Receiving Broad-Spectrum Agents. Infect. Control Hosp. Epidemiol. 2020, 41, 1022–1027. [Google Scholar] [CrossRef]
- Eickelberg, G.; Sanchez-Pinto, L.N.; Luo, Y. Predictive Modeling of Bacterial Infections and Antibiotic Therapy Needs in Critically Ill Adults. J. Biomed. Inform. 2020, 109, 103540. [Google Scholar] [CrossRef]
- Chowdhury, A.S.; Lofgren, E.T.; Moehring, R.W.; Broschat, S.L. Identifying Predictors of Antimicrobial Exposure in Hospitalized Patients Using a Machine Learning Approach. J. Appl. Microbiol. 2020, 128, 688–696. [Google Scholar] [CrossRef]
- Moehring, R.W.; Phelan, M.; Lofgren, E.; Nelson, A.; Dodds Ashley, E.; Anderson, D.J.; Goldstein, B.A. Development of a Machine Learning Model Using Electronic Health Record Data to Identify Antibiotic Use Among Hospitalized Patients. JAMA Netw. Open 2021, 4, e213460. [Google Scholar] [CrossRef]
- Goodman, K.E.; Heil, E.L.; Claeys, K.C.; Banoub, M.; Bork, J.T. Real-World Antimicrobial Stewardship Experience in a Large Academic Medical Center: Using Statistical and Machine Learning Approaches to Identify Intervention “Hotspots” in an Antibiotic Audit and Feedback Program. Open Forum Infect. Dis. 2022, 9, ofac289. [Google Scholar] [CrossRef]
- Mancini, A.; Vito, L.; Marcelli, E.; Piangerelli, M.; De Leone, R.; Pucciarelli, S.; Merelli, E. Machine Learning Models Predicting Multidrug Resistant Urinary Tract Infections Using “DsaaS”. BMC Bioinform. 2020, 21, 347. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.G.; Aung, A.-H.; Lian, W.; Lye, D.C.; Ooi, C.-K.; Chow, A. Risk Prediction Models to Guide Antibiotic Prescribing: A Study on Adult Patients with Uncomplicated Upper Respiratory Tract Infections in an Emergency Department. Antimicrob. Resist. Infect. Control 2020, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Kanjilal, S.; Oberst, M.; Boominathan, S.; Zhou, H.; Hooper, D.C.; Sontag, D. A Decision Algorithm to Promote Outpatient Antimicrobial Stewardship for Uncomplicated Urinary Tract Infection. Sci. Transl. Med. 2020, 12, eaay5067. [Google Scholar] [CrossRef]
- Oonsivilai, M.; Mo, Y.; Luangasanatip, N.; Lubell, Y.; Miliya, T.; Tan, P.; Loeuk, L.; Turner, P.; Cooper, B.S. Using Machine Learning to Guide Targeted and Locally-Tailored Empiric Antibiotic Prescribing in a Children’s Hospital in Cambodia. Wellcome Open Res. 2018, 3, 131. [Google Scholar] [CrossRef] [PubMed]
- Artificial Intelligence to Guide Antibiotic Choice in Recurrent Uti: Is It the Right Way for Improving Antimicrobial Stewardship? UROLUTS. Available online: https://uroluts.uroweb.org/webcast/artificial-intelligence-to-guide-antibiotic-choice-in-recurrent-uti-is-it-the-right-way-for-improving-antimicrobial-stewardship/ (accessed on 22 December 2023).
- Tang, R.; Luo, R.; Tang, S.; Song, H.; Chen, X. Machine Learning in Predicting Antimicrobial Resistance: A Systematic Review and Meta-Analysis. Int. J. Antimicrob. Agents 2022, 60, 106684. [Google Scholar] [CrossRef]
- Greenhalgh, T.; Wherton, J.; Papoutsi, C.; Lynch, J.; Hughes, G.; A’Court, C.; Hinder, S.; Fahy, N.; Procter, R.; Shaw, S. Beyond Adoption: A New Framework for Theorizing and Evaluating Nonadoption, Abandonment, and Challenges to the Scale-Up, Spread, and Sustainability of Health and Care Technologies. J. Med. Internet Res. 2017, 19, e367. [Google Scholar] [CrossRef]
- Liyanage, H.; Liaw, S.-T.; Jonnagaddala, J.; Schreiber, R.; Kuziemsky, C.; Terry, A.L.; De Lusignan, S. Artificial Intelligence in Primary Health Care: Perceptions, Issues, and Challenges: Primary Health Care Informatics Working Group Contribution to the Yearbook of Medical Informatics 2019. Yearb. Med. Inform. 2019, 28, 041–046. [Google Scholar] [CrossRef] [PubMed]
- Beil, M.; Proft, I.; Van Heerden, D.; Sviri, S.; Van Heerden, P.V. Ethical Considerations about Artificial Intelligence for Prognostication in Intensive Care. Intensive Care Med. Exp. 2019, 7, 70. [Google Scholar] [CrossRef]
- WHO Regulatory Considerations on Artificial Intelligence for Health; World Health Organization: Geneva, Switzerland, 2023; ISBN 978-92-4-007887-1.
- Gianfrancesco, M.A.; Tamang, S.; Yazdany, J.; Schmajuk, G. Potential Biases in Machine Learning Algorithms Using Electronic Health Record Data. JAMA Intern. Med. 2018, 178, 1544. [Google Scholar] [CrossRef]
- Abdullah, Y.I.; Schuman, J.S.; Shabsigh, R.; Caplan, A.; Al-Aswad, L.A. Ethics of Artificial Intelligence in Medicine and Ophthalmology. Asia-Pac. J. Ophthalmol. 2021, 10, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Cochrane Handbook for Systematic Reviews of Interventions, 2nd ed.; Higgins, J., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2019. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. Syst. Rev. 2021, 10, 89. [Google Scholar] [CrossRef] [PubMed]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A Web and Mobile App for Systematic Reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef] [PubMed]
- Study Quality Assessment Tools|NHLBI, NIH. Available online: https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools (accessed on 30 November 2023).
- Wolff, R.F.; Moons, K.G.M.; Riley, R.D.; Whiting, P.F.; Westwood, M.; Collins, G.S.; Reitsma, J.B.; Kleijnen, J.; Mallett, S.; for the PROBAST Group. PROBAST: A Tool to Assess the Risk of Bias and Applicability of Prediction Model Studies. Ann. Intern. Med. 2019, 170, 51. [Google Scholar] [CrossRef]
- Moons, K.G.M.; Wolff, R.F.; Riley, R.D.; Whiting, P.F.; Westwood, M.; Collins, G.S.; Reitsma, J.B.; Kleijnen, J.; Mallett, S. PROBAST: A Tool to Assess Risk of Bias and Applicability of Prediction Model Studies: Explanation and Elaboration. Ann. Intern. Med. 2019, 170, W1. [Google Scholar] [CrossRef]

| Study | Year of Publication | Country | No. Centers | Study Time Frame | Target Population | No. Patients | Infection Site | No. Features | Objective | Algorithm | Performance Measurement | Main Results |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [20] | 2016 | Canada | 1 | February to November 2012 | Patients monitored by APSS who received at least one prescription of piperacillin–tazobactam at the Centre Hospitalier Universitaire de Sherbrooke | 421 hospitalizations | Not specified | Not specified | To evaluate the ability of the algorithm to discover rules for identifying inappropriate prescriptions of piperacillin-tazobactam | Supervised learning module of APSS, temporal induction of classification models algorithm | PPV, sensitivity, accuracy, precision | The combined system achieved an overall PPV (precision) of identifying confirmed inappropriate prescriptions of 74% (95% CI, 68–79), with sensitivity (recall) of 96% (95% CI, 92–98), and accuracy of 79% (95% CI, 74–83). |
| [21] | 2022 | Netherlands | 1 | January 2017–December 2018 | Inpatients of the UMC Utrecht | 906 cultures from 810 patients | UTI | 36 | To report on the design and evaluation of a CDSS to predict UTI before the urine culture results are available | CDSS using the RESSEL method; supervised models implemented in the Scikit-learn package: LR, SVM, RF, XGB and k-NN | Accuracy, sensitivity, specificity, PPV, NPV, AUC, Nneg, Npos | The predictive performance of the best-performing semi-supervised model (RF enhanced with RESSEL) had an accuracy of 76.77 (±0.97), sensitivity of 81.28 (±1.16), specificity of 70.75 (±1.85), and AUC of 80.02 (±1.00). |
| [22] | 2022 | USA | 5 | Stanford hospitals: January 2009–December 2019; Boston hospitals: 2007–2016 | Patients who presented to Stanford emergency departments, Massachusetts General Hospital, and Brigham and Women’s Hospital in Boston | Stanford: N = 8342 infections from 6920 adult patients. Boston: N = 15,806 uncomplicated urinary tract infections from 13,862 unique female patients. Our dataset is split by time into training, validation, and test sets containing Ntrain = 5804 patient infections from 2009 to 2017, Nval = 1218 patient infections from 2018, and Ntest = 1320 patient infections from 2019. | Stanford: unspecified infection; Boston: UTI | Boston: The total number of features used in this portion of the analysis was 788. Stanford: In total, the sparse feature matrix contained 43,220 columns. | To investigate the utility of ML-based clinical decision support for antibiotic prescribing stewardship. | LR, RF, gradient boosted tree, lasso, ridge | AUROC, prevalence, average precision, antibiogram coverage rate | Stanford dataset: personalized antibiograms reallocate clinician antibiotic selections with a coverage rate of 85.9%, similar to clinician performance (84.3% p = 0.11). The best model class for selection of vancomycin+meropenem was gradient boosted tree, with average precision of 0.99 [0.99, 0.99] and AUROC of 0.73 [0.65, 0.81]. Boston dataset: personalized antibiograms coverage rate of 90.4%, a significant improvement over clinicians (88.1% p < 0.0001). The best model class for the selection of levofloxacin was LASSO, with an average precision of 0.96 [0.95, 0.96] and AUROC of 0.64 [0.60, 0.67]. |
| [23] | 2020 | Greece | 1 | January 2017–December 2018 | ICU patients in a public tertiary hospital | 345 | Invasive, respiratory, urinary, mucocutaneous, and wound infections | 23,067 (binary, numerical, and categorical in total) | To compare the performance of eight ML algorithms to assess antibiotic susceptibility predictions | ML toolkit: WEKA—Data Mining Software in Java Workbench; LIBLINEAR LR and linear SVM; SVMs; SMO; instance-based learning (k-NN); J48; RF; RIPPER; MLP | TP rate, FP rate, precision, recall, F-measure, mmc, AUROC, precision-recall plot | The best performances were obtained with the RIPPER algorithm (F-measure of 0.678) and the MLP classifier (AUROC of 0.726). |
| [24] | 2022 | Taiwan | 25 | May 2013 to May 2014 | Patients with healthcare-associated infections receiving at least one antimicrobial drug | 7377 | Healthcare-associated infection (bloodstream, urinary, pneumonia and surgical site infection). | 26 | To develop accurate and efficient ML models for auditing appropriate surgical antimicrobial prophylaxis | Supervised ML classifiers (Auto-WEKA (Bayesian optimisation method), MLP (artificial neural network), decision tree, SimpleLogistic (LogitBoost e CART algorithm), bagging, SMOTE and AdaBoost) | TP rate, TN rate, FP, FN, AUC, precision, specificity, sensitivity, weighted average for the multiclass model, execution time | The ML technique with the best performance metrics was the MLP, with a sensitivity of 0.967, specificity of 0.992, precision of 0.967, and AUC of 0.992. |
| [25] | 2022 | Israel | 1 | June 2007 to January 2019 | Patients with UTI and wound infections from Maccabi Healthcare Services (MHS) with at least one record of a positive wound infection culture | 140,349 UTI and 7365 wound infections. | UTI and wound | Not specified | To understand and predict the personal risk of treatment-induced gain of resistance | ML | Personal predicted risk | Choosing the antibiotic treatment with the minimal ML-predicted risk of emergence of resistance reduces the overall risk of emergence of resistance by 70% for UTIs and 74% for wound infections compared to the risk for physician-prescribed treatments. |
| [26] | 2022 | USA | 1 | 2008 to 2019 | Patients who received intravenous antibiotic treatment for a duration between 1 and 21 days during an ICU stay, at Beth Israel Deaconess Medical Centre, Boston | 18,988 (22,845 unique stays) | Respiratory (pneumonia) and UTI | 43 | To estimate patients’ ICU LOS and mortality outcomes for any given day under the alternative scenarios of if they were to stop vs. continue antibiotic treatment | AI-based CDSS: recurrent neural network autoencoder and a synthetic control-based approach. It uses a bidirectional LSTM autoencoder; PyTorch was used to create a bidirectional LSTM RNN | Patients’ ICU LOS (days, mean delta, root mean squared error), mortality outcomes, to stop vs. continue ATB treatment (mean days reduction); day(s), mean delta (days, p-value), MAPE, MAE, RMSE, AUROC | The model reliably estimates patient outcomes under the contrasting scenarios of stopping or continuing ATB treatment: impact days where the potential effect of the unobserved scenario was assessed showed that stopping ATB therapy earlier had a statistically significant shorter LOS (mean reduction 2.71 days, p-value < 0.01). No impact on mortality was observed. |
| [27] | 2021 | Greece | 1 | January to December 2018 | Patients admitted to the internal medicine wards of a public hospital | 499 patients (11,496 instances) | Not specified | 6 (attributes of sex, age, sample type, Gram stain, 44 antimicrobial substances, and the antibiotic susceptibility results) | To assess the effectiveness of AutoML-trained models to predict AMR | AutoML techniques using Microsoft Azure AutoML; SMOTE; algorithms: StackEnsemble, VotingEnsemble, MaxAbsScaler, LightGBM, SparseNormalizer, XGBoostClassifier | AUROC, AUCW, APSW, F1W, and ACC | The stack ensemble technique achieved the best results in the original and balanced dataset, with an AUCW metric of 0.822 and 0.850, respectively. |
| [28] | 2020 | EUA | 1 | December 2015 to August 2017 | Patients hospitalized who received at least one antimicrobial from a list of those routinely tracked by the ASP at University of California, San Francisco Medical Centre | 9651 | Bloodstream, UTI, etc. | More than 200 | To predict whether antibiotic therapy required stewardship intervention on any given day compared to the criterion standard of note left by the antimicrobial stewardship team in the patient’s chart | LR and boosted tree models | AUROC, Brier score, sensitivity, specificity, PPV, and NPV | Logistic regression and boosted tree models had AUROCs of 0.73 (95% CI, 0.69–0.77) and 0.75 (95% CI, 0.72–0.79) (p = 0.07), respectively. |
| [29] | 2020 | Israel | 1 | 2001 to 2012 | ICU adult patients are patients suspected of having a community-acquired bacterial infection | 10,290 patients (12,232 ICU encounters) | Non-specified bacterial infection | Not specified | To identify ICU patients with low risk of bacterial infection as candidates for earlier EAT discontinuation | ML algorithms, including ridge regression, RF, SVC, XG Boost, K- NN, and MLP | AUROC, NPV, F1, precision, recall, high sensitivity threshold, TN, FP, FN, TP | Using structured longitudinal data collected up to 24, 48, and 72 h after starting EAT, the best models identified patients at low risk of bacterial infections with AUROCs up to 0.8 and negative predictive values > 93%. The T = 24 h RF model was the best performing model within this timepoint: AUC of 0.774, F1 of 0.424, NPV of 0.944, precision of 0.277, recall of 0.905, high sensitivity threshold of 0.258. |
| [30] | 2019 | USA | 27 | October 2015 to September 2017 | Patients from the Duke Antimicrobial Stewardship Outreach Network (DASON) (Duke University School of Medicine) | 382,943 | Not specified | More than 100 features, including demographic data, length of stay, comorbidity, etc. | To identify patient- and facility-level predictors of antimicrobial usage in hospitalized patients using an ML approach, which can be used to inform a risk adjustment model to facilitate assessment of antimicrobial utilization | SVR and CB models | Root-mean-square error values | Both the SVR and CB models show better predictive accuracy than the null LM and null NB-GLM models (null statistical models) for all SAAR (external comparator) groups. CB performed better than SVR, according to the RMSE values (5.51 vs. 7.17 for all antibiotics, respectively). |
| [31] | 2021 | USA | 3 | October 2015 to September 2017 | Adult and pediatric inpatient from Duke University Health System | 170,294 | Not specified | 204 | To evaluate whether variables derived from the electronic health records accurately identify inpatient antimicrobial use | A 2-stage RF ML modeling | AUROC and absolute error | Models accurately identified antimicrobial exposure in the testing dataset: the majority of AUCs were above 0.8, with a mean AUC of 0.85. |
| [32] | 2022 | USA | 1 | July 2017 to December 2019 | Patient with antimicrobial orders from University of Maryland Medical Centre | 17,503 | Sepsis/bacteremia, bone/joint, central nervous system, cardiac/vascular, gastrointestinal genitourinary, respiratory, nonsurgical prophylaxis, skin and soft tissue infection, mycobacterial infection, neutropenia, surgical prophylaxis | 33 | To understand which patient and treatment characteristics are associated with either a higher or lower likelihood of intervention in a PAF program and to develop prediction models to identify antimicrobial orders that may be safely excluded from the review | LR, RF | Sensitivity, specificity, C-statistic, the out-of-bag error rate | The RF model had a C-statistic of 0.76 (95% CI, 0.75–0.77), with a sensitivity and specificity of 78% and 58%, respectively. This model would reduce review caseloads by 49%. |
| [33] | 2018 | Italy | 1 | March 2012 to 2019 | Patients with nosocomial (UTI) from Principe di Piemonte Hospital in Senigallia | 1486 | UTI | 6 (5 predictors + MDR resistance) | To design, develop, and evaluate, with a real antibiotic stewardship dataset, a predictive model useful for predicting MDR UTI onset after patient hospitalization | Catboost, support vector machine, and NN | Accuracy, AUROC, AUC-PRC, F1 score, sensitivity, specificity, MCC. FP, FN, TP, and TN | The ML method catboost had the best predictive results (MCC of 0.909; sensitivity of 0.904; F1 score of 0.809; AUC-PRC of 0.853, AUROC of 0.739; ACC of 0.717). |
| [34] | 2020 | Singapore | 1 | June 2016 to November 2018 | Patients with uncomplicated URTI at the emergency department at Tan Tock Seng Hospital | 715 | Upper respiratory tract infections | 50 (univariate analysis), 8 included in the algorithm | To develop prediction models based on local clinical and laboratory data to guide antibiotic prescribing for adult patients with uncomplicated upper respiratory tract infections | LR models, LASSO, and CART | AUC, sensitivity, specificity, PPV, NPV | The AUC on the validation set for the models was similar: LASSO: 0.70 [95% CI: 0.62–0.77], LR: 0.72 [95% CI: 0.65–0.79], decision tree: 0.67 [95% CI: 0.59–0.74]. |
| [35] | 2020 | USA | 2 | 2007 to 2016 | Patients presenting with uncomplicated UTI at Massachusetts General Hospital and the Brigham and Women’s Hospital in Boston | 10,053 (training dataset); 3629 (test set) | UTI | 8 | To predict antibiotic susceptibility using electronic health record data and build a decision algorithm for recommending the narrowest possible antibiotic to which a specimen is susceptible | LR, decision tree, and RF models | AUROC, FN rates | Decision trees and RF were excluded based on their poor validation set performance and relative lack of interpretability. The LR model provided antibiotic stewardship for a common infectious syndrome by maximizing reductions in broad-spectrum antibiotic use while maintaining optimal treatment outcomes. The algorithm achieved a 67% reduction in the use of second-line antibiotics relative to clinicians and reduced inappropriate antibiotic therapy by 18%, close to the rate of clinicians. |
| [36] | 2019 | Cambodia | 1 | February 2013 to January 2016 | Children with at least one positive blood culture from Angkor Hospital for Children | 195 (training set); 48 (model validation) | Bloodstream | 35 | To predict Gram stains and whether bacterial pathogens could be treated with standard empiric antibiotic regimens | RF, LR, decision trees constructed via recursive partitioning, boosted decision trees using adaptive boosting, linear SVM, polynomial SVM, radial SVM, and k-NN | AUROC | The RF method had the best predictive performance overall: AUC of 0.80 (95% CI 0.66–0.94) for predicting susceptibility to ceftriaxone, 0.74 (0.59–0.89) for susceptibility to ampicillin and gentamicin, 0.85 (0.70–1.00) for susceptibility to neither, and 0.71 (0.57–0.86) for Gram stain result. |
| [37] | 2022 | Italy | 2 | January 2012 to December 2020 | Women affected by recurrent UTI who had undergone antimicrobial treatment for uncomplicated lower UTI | 1043 | Recurrent UTI | Not specified | To define an NN for predicting the clinical and microbiological efficacy of antimicrobial treatment of a large cohort of women affected by recurrent UTIs for use in everyday clinical practice | NN | Sensitivity, specificity, HR | The use of artificial NN in women with recurrent cystitis showed a sensitivity of 87.8% and specificity of 97.3% in predicting the clinical and microbiological efficacy of the prescribed antimicrobial treatment. |
| Study | Demographics | Adult | Paediatric | Clinical | Laboratory/Microbiological | Comorbidities | Type of Infection | ICU |
|---|---|---|---|---|---|---|---|---|
| [20] | Yes | Yes | No | Yes | Yes | No | No | Yes |
| [21] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No |
| [22] | Yes | Yes | No | Yes | Yes | Yes | Yes | No |
| [23] | Yes | Yes | No | No | Yes | No | Yes | Yes |
| [24] | Yes | Yes | Yes | Yes | No | No | Yes | Yes |
| [25] | Yes | Yes | No | Yes | Yes | Yes | Yes | Not specified |
| [26] | Yes | Yes | Not specified | Yes | Yes | No | No | Yes (only ICU patients) |
| [27] | Yes | Yes | No | No | Yes | No | Yes | No |
| [28] | Yes | Yes | No | Yes | Yes | No | Yes | Yes |
| [29] | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes (only ICU patients) |
| [30] | Yes | Yes | No | Yes | Yes | Yes | Yes | Not specified |
| [31] | Yes | Yes | Yes | Yes | Yes | Yes | Not specified | Yes |
| [32] | Yes | Yes | No | Yes | Yes | No | Yes | Not specified |
| [33] | Yes | Yes | Not specified | No | Yes | No | Yes | Not specified |
| [34] | Yes | Yes | No | Yes | Yes | Yes | Yes | No |
| [35] | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes |
| [36] | Yes | No | Yes | Yes | Yes | No | Yes | Yes |
| [37] | Yes | Yes | No | Yes | Yes | Not specified | Yes | Not specified |
| Criteria\ Study | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | Quality Rating |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [20] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | NR | Fair (57.1%) |
| [21] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | No | Fair (57.1%) |
| [22] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | Yes | Fair (64.3%) |
| [23] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | No | Fair (57.1%) |
| [24] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | NR | Fair (57.1%) |
| [25] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | NR | Fair (57.1%) |
| [26] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | Yes | Fair (64.3%) |
| [27] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | NR | Fair (57.1%) |
| [28] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | NR | Fair (57.1%) |
| [29] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | NR | Fair (57.1%) |
| [30] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | NR | Fair (57.1%) |
| [31] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | NR | Fair (57.1%) |
| [32] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | Yes | Fair (64.3%) |
| [33] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | NR | Fair (57.1%) |
| [34] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | NR | Fair (57.1%) |
| [35] | Yes | Yes | NA | Yes | Yes | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | No | Fair (64.3%) |
| [36] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | NR | Fair (57.1%) |
| [37] | Yes | Yes | NA | Yes | NR | Yes | Yes | NA | Yes | Yes | Yes | NR | NA | NR | Fair (57.1%) |
| Study | Risk of Bias | Applicability | Overall | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1. Participants | 2. Predictors | 3. Outcome | 4. Analysis | 1. Participants | 2. Predictors | 3. Outcome | Risk of Bias | Applicability | |
| [20] | - | + | + | ? | ? | ? | + | - | ? |
| [21] | + | ? | + | ? | + | + | + | ? | + |
| [22] | + | ? | + | + | + | + | + | ? | + |
| [23] | + | + | + | ? | + | + | + | ? | + |
| [24] | + | + | + | + | + | + | + | + | + |
| [25] | ? | ? | + | ? | ? | ? | + | ? | ? |
| [26] | + | + | + | ? | + | + | + | ? | + |
| [27] | - | + | + | - | - | + | + | - | - |
| [28] | + | + | + | - | + | ? | + | - | ? |
| [29] | + | ? | + | ? | + | ? | + | ? | ? |
| [30] | + | + | + | - | + | ? | + | - | ? |
| [31] | + | + | + | - | + | + | ? | - | ? |
| [32] | + | + | + | - | + | + | + | - | + |
| [33] | + | + | + | - | + | ? | + | - | ? |
| [34] | + | + | + | - | + | + | + | - | + |
| [35] | + | + | ? | - | + | + | ? | - | ? |
| [36] | + | + | ? | - | + | + | ? | - | ? |
| [37] | + | + | + | - | + | + | ? | - | ? |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto-de-Sá, R.; Sousa-Pinto, B.; Costa-de-Oliveira, S. Brave New World of Artificial Intelligence: Its Use in Antimicrobial Stewardship—A Systematic Review. Antibiotics 2024, 13, 307. https://doi.org/10.3390/antibiotics13040307
Pinto-de-Sá R, Sousa-Pinto B, Costa-de-Oliveira S. Brave New World of Artificial Intelligence: Its Use in Antimicrobial Stewardship—A Systematic Review. Antibiotics. 2024; 13(4):307. https://doi.org/10.3390/antibiotics13040307
Chicago/Turabian StylePinto-de-Sá, Rafaela, Bernardo Sousa-Pinto, and Sofia Costa-de-Oliveira. 2024. "Brave New World of Artificial Intelligence: Its Use in Antimicrobial Stewardship—A Systematic Review" Antibiotics 13, no. 4: 307. https://doi.org/10.3390/antibiotics13040307
APA StylePinto-de-Sá, R., Sousa-Pinto, B., & Costa-de-Oliveira, S. (2024). Brave New World of Artificial Intelligence: Its Use in Antimicrobial Stewardship—A Systematic Review. Antibiotics, 13(4), 307. https://doi.org/10.3390/antibiotics13040307







