Pseudomonas aeruginosa Infections in Patients with Severe COVID-19 in Intensive Care Units: A Retrospective Study
Abstract
1. Introduction
2. Results
2.1. Demographic and Clinical Data
2.2. Pseudomonas aeruginosa Infections
2.3. Antibiotic Treatments
3. Discussion
4. Materials and Methods
4.1. Study Design and Setting
4.2. Patient Selection
- Adult patients were hospitalized for at least 48 h in an ICU at CHRU-Nancy for ARDS due to COVID-19 (confirmed by reverse transcription polymerase chain reaction or an antigenic test).
- Patients were hospitalized from 1 March 2020 to 31 May 2021.
- Patients developed a HAI caused by P. aeruginosa during their ICU stay.
- Patients < 18 years old.
- Patients without COVID-19 at the ICU admission.
- Patients with P. aeruginosa isolated <48 h following ICU admission.
- Patients who did not want to be included in the study.
4.3. Data Collection and Outcomes
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef]
- Raoofi, S.; Pashazadeh Kan, F.; Rafiei, S.; Hosseinipalangi, Z.; Noorani Mejareh, Z.; Khani, S.; Abdollahi, B.; Seyghalani Talab, F.; Sanaei, M.; Zarabi, F.; et al. Global Prevalence of Nosocomial Infection: A Systematic Review and Meta-Analysis. PLoS ONE 2023, 18, e0274248. [Google Scholar] [CrossRef]
- GBD 2019 Antimicrobial Resistance Collaborators Global Mortality Associated with 33 Bacterial Pathogens in 2019: A Systematic Analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 2221–2248. [CrossRef] [PubMed]
- Kang, C.-I.; Kim, S.-H.; Kim, H.-B.; Park, S.-W.; Choe, Y.-J.; Oh, M.-D.; Kim, E.-C.; Choe, K.-W. Pseudomonas aeruginosa Bacteremia: Risk Factors for Mortality and Influence of Delayed Receipt of Effective Antimicrobial Therapy on Clinical Outcome. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2003, 37, 745–751. [Google Scholar] [CrossRef]
- Lyu, J.; Chen, H.; Bao, J.; Liu, S.; Chen, Y.; Cui, X.; Guo, C.; Gu, B.; Li, L. Clinical Distribution and Drug Resistance of Pseudomonas aeruginosa in Guangzhou, China from 2017 to 2021. J. Clin. Med. 2023, 12, 1189. [Google Scholar] [CrossRef]
- Ng, Q.X.; Ong, N.Y.; Lee, D.Y.X.; Yau, C.E.; Lim, Y.L.; Kwa, A.L.H.; Tan, B.H. Trends in Pseudomonas aeruginosa (P. aeruginosa) Bacteremia during the COVID-19 Pandemic: A Systematic Review. Antibiotics 2023, 12, 409. [Google Scholar] [CrossRef]
- Bongiovanni, M.; Barda, B. Pseudomonas aeruginosa Bloodstream Infections in SARS-CoV-2 Infected Patients: A Systematic Review. J. Clin. Med. 2023, 12, 2252. [Google Scholar] [CrossRef] [PubMed]
- Langford, B.J.; So, M.; Simeonova, M.; Leung, V.; Lo, J.; Kan, T.; Raybardhan, S.; Sapin, M.E.; Mponponsuo, K.; Farrell, A.; et al. Antimicrobial Resistance in Patients with COVID-19: A Systematic Review and Meta-Analysis. Lancet Microbe 2023, 4, e179–e191. [Google Scholar] [CrossRef] [PubMed]
- Bazaid, A.S.; Barnawi, H.; Qanash, H.; Alsaif, G.; Aldarhami, A.; Gattan, H.; Alharbi, B.; Alrashidi, A.; Al-Soud, W.A.; Moussa, S.; et al. Bacterial Coinfection and Antibiotic Resistance Profiles among Hospitalised COVID-19 Patients. Microorganisms 2022, 10, 495. [Google Scholar] [CrossRef]
- Zhang, G.; Hu, C.; Luo, L.; Fang, F.; Chen, Y.; Li, J.; Peng, Z.; Pan, H. Clinical Features and Short-Term Outcomes of 221 Patients with COVID-19 in Wuhan, China. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2020, 127, 104364. [Google Scholar] [CrossRef]
- RéPIAS Mission Nationale SPIADI. Surveillance des Infections Associées aux Dispositifs Invasifs. Résultats de la Surveillance Menée En 2021; Santé Publique France: Saint-Maurice, France, 2021. [Google Scholar]
- Murgia, F.; Fiamma, M.; Serra, S.; Marras, G.; Argiolas, R.; Mattana, C.; Mattu, M.G.; Garau, M.C.; Doneddu, S.; Olla, S.; et al. The Impact of the Secondary Infections in ICU Patients Affected by COVID-19 during Three Different Phases of the SARS-CoV-2 Pandemic. Clin. Exp. Med. 2023, 23, 1251–1263. [Google Scholar] [CrossRef] [PubMed]
- Nathwani, D.; Raman, G.; Sulham, K.; Gavaghan, M.; Menon, V. Clinical and Economic Consequences of Hospital-Acquired Resistant and Multidrug-Resistant Pseudomonas aeruginosa Infections: A Systematic Review and Meta-Analysis. Antimicrob. Resist. Infect. Control 2014, 3, 32. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Regional Office for Europe; European Centre for Disease Prevention and Control. Antimicrobial Resistance Surveillance in Europe 2022–2020 Data; WHO Regional Office for Europe: Copenhagen, Denmark, 2022; p. 164. [Google Scholar]
- Kariyawasam, R.M.; Julien, D.A.; Jelinski, D.C.; Larose, S.L.; Rennert-May, E.; Conly, J.M.; Dingle, T.C.; Chen, J.Z.; Tyrrell, G.J.; Ronksley, P.E.; et al. Antimicrobial Resistance (AMR) in COVID-19 Patients: A Systematic Review and Meta-Analysis (November 2019–June 2021). Antimicrob. Resist. Infect. Control 2022, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Jeon, K.; Jeong, S.; Lee, N.; Park, M.-J.; Song, W.; Kim, H.-S.; Kim, H.S.; Kim, J.-S. Impact of COVID-19 on Antimicrobial Consumption and Spread of Multidrug-Resistance in Bacterial Infections. Antibiotics 2022, 11, 535. [Google Scholar] [CrossRef] [PubMed]
- Durà-Miralles, X.; Abelenda-Alonso, G.; Bergas, A.; Laporte-Amargós, J.; Sastre-Escolà, E.; Padullés, A.; Carratalà, J.; Gudiol, C. An Ocean between the Waves: Trends in Antimicrobial Consumption in Hospitalized Patients with COVID-19. Antibiotics 2024, 13, 55. [Google Scholar] [CrossRef] [PubMed]
- Abubakar, U.; Al-Anazi, M.; Alanazi, Z.; Rodríguez-Baño, J. Impact of COVID-19 Pandemic on Multidrug Resistant Gram Positive and Gram Negative Pathogens: A Systematic Review. J. Infect. Public Health 2023, 16, 320–331. [Google Scholar] [CrossRef]
- World Health Organization. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Micheli, G.; Sangiorgi, F.; Catania, F.; Chiuchiarelli, M.; Frondizi, F.; Taddei, E.; Murri, R. The Hidden Cost of COVID-19: Focus on Antimicrobial Resistance in Bloodstream Infections. Microorganisms 2023, 11, 1299. [Google Scholar] [CrossRef]
- Meyer, N.J.; Gattinoni, L.; Calfee, C.S. Acute Respiratory Distress Syndrome. Lancet 2021, 398, 622–637. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.P.; Gonçalves, L.C.; Oliveira, A.C.C.; Queiroz, P.H.P.; Ito, C.R.M.; Santos, M.O.; Carneiro, L.C. Bacterial Co-Infection in Patients with COVID-19 Hospitalized (ICU and Not ICU): Review and Meta-Analysis. Antibiotics 2022, 11, 894. [Google Scholar] [CrossRef]
- Caiazzo, L.; Temperoni, C.; Canovari, B.; Simonetti, O.; Montalti, R.; Barchiesi, F. Secondary Infections in Critically Ill Patients with COVID-19: A Retrospective Study. Antibiotics 2022, 11, 1598. [Google Scholar] [CrossRef]
- Bardi, T.; Pintado, V.; Gomez-Rojo, M.; Escudero-Sanchez, R.; Azzam Lopez, A.; Diez-Remesal, Y.; Martinez Castro, N.; Ruiz-Garbajosa, P.; Pestaña, D. Nosocomial Infections Associated to COVID-19 in the Intensive Care Unit: Clinical Characteristics and Outcome. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2021, 40, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Venier, A.-G.; Leroyer, C.; Slekovec, C.; Talon, D.; Bertrand, X.; Parer, S.; Alfandari, S.; Guerin, J.-M.; Megarbane, B.; Lawrence, C.; et al. Risk Factors for Pseudomonas aeruginosa Acquisition in Intensive Care Units: A Prospective Multicentre Study. J. Hosp. Infect. 2014, 88, 103–108. [Google Scholar] [CrossRef]
- Loyola-Cruz, M.Á.; Gonzalez-Avila, L.U.; Martínez-Trejo, A.; Saldaña-Padilla, A.; Hernández-Cortez, C.; Bello-López, J.M.; Castro-Escarpulli, G. ESKAPE and Beyond: The Burden of Coinfections in the COVID-19 Pandemic. Pathogens 2023, 12, 743. [Google Scholar] [CrossRef] [PubMed]
- Cureño-Díaz, M.A.; Plascencia-Nieto, E.S.; Loyola-Cruz, M.Á.; Cruz-Cruz, C.; Nolasco-Rojas, A.E.; Durán-Manuel, E.M.; Ibáñez-Cervantes, G.; Gómez-Zamora, E.; Tamayo-Ordóñez, M.C.; Tamayo-Ordóñez, Y.d.J.; et al. Gram-Negative ESKAPE Bacteria Surveillance in COVID-19 Pandemic Exposes High-Risk Sequence Types of Acinetobacter Baumannii MDR in a Tertiary Care Hospital. Pathogens 2024, 13, 50. [Google Scholar] [CrossRef]
- Alcántar-Curiel, M.D.; Huerta-Cedeño, M.; Jarillo-Quijada, M.D.; Gayosso-Vázquez, C.; Fernández-Vázquez, J.L.; Hernández-Medel, M.L.; Zavala-Pineda, M.; Morales-Gil, M.Á.; Hernández-Guzmán, V.A.; Bolaños-Hernández, M.I.; et al. Gram-Negative ESKAPE Bacteria Bloodstream Infections in Patients during the COVID-19 Pandemic. PeerJ 2023, 11, e15007. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, A.; Kreitmann, L.; Nseir, S. ICU-Acquired Colonization and Infection Related to Multidrug-Resistant Bacteria in COVID-19 Patients: A Narrative Review. Antibiotics 2023, 12, 1464. [Google Scholar] [CrossRef]
- Lasater, K.B.; Aiken, L.H.; Sloane, D.M.; French, R.; Martin, B.; Reneau, K.; Alexander, M.; McHugh, M.D. Chronic Hospital Nurse Understaffing Meets COVID-19: An Observational Study. BMJ Qual. Saf. 2021, 30, 639–647. [Google Scholar] [CrossRef]
- Ranney, M.L.; Griffeth, V.; Jha, A.K. Critical Supply Shortages–The Need for Ventilators and Personal Protective Equipment during the COVID-19 Pandemic. N. Engl. J. Med. 2020, 382, e41. [Google Scholar] [CrossRef]
- Magalhães, C.; Lima, M.; Trieu-Cuot, P.; Ferreira, P. To Give or Not to Give Antibiotics Is Not the Only Question. Lancet Infect. Dis. 2021, 21, e191–e201. [Google Scholar] [CrossRef]
- Russell, C.D.; Fairfield, C.J.; Drake, T.M.; Turtle, L.; Seaton, R.A.; Wootton, D.G.; Sigfrid, L.; Harrison, E.M.; Docherty, A.B.; de Silva, T.I.; et al. Co-Infections, Secondary Infections, and Antimicrobial Use in Patients Hospitalised with COVID-19 during the First Pandemic Wave from the ISARIC WHO CCP-UK Study: A Multicentre, Prospective Cohort Study. Lancet Microbe 2021, 2, e354–e365. [Google Scholar] [CrossRef]
- Adams, J.; Ferguson, K.; Hirschy, R.; Konopka, E.; Meckel, J.; Benanti, G.; Kuhrau, S.; Albarillo, F.; Chang, K.; Santarossa, M.; et al. Antimicrobial Stewardship Techniques for Critically Ill Patients with Pneumonia. Antibiotics 2023, 12, 295. [Google Scholar] [CrossRef] [PubMed]
- Baudet, A.; Agrinier, N.; Charmillon, A.; Pulcini, C.; Lozniewski, A.; Aissa, N.; Lizon, J.; Thilly, N.; Demoré, B.; Florentin, A. Evaluating Antibiotic Stewardship and Healthcare-Associated Infections Surveillance Assisted by Computer: Protocol for an Interrupted Time Series Study. BMJ Open 2022, 12, e056125. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Pseudomonas aeruginosa—Calibration of Zone Diameter Breakpoints to MIC Values. 2023. Available online: https://www.eucast.org/ast_of_bacteria/calibration_and_validation (accessed on 20 April 2024).
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Kadri, S.S.; Adjemian, J.; Lai, Y.L.; Spaulding, A.B.; Ricotta, E.; Prevots, D.R.; Palmore, T.N.; Rhee, C.; Klompas, M.; Dekker, J.P.; et al. Difficult-to-Treat Resistance in Gram-Negative Bacteremia at 173 US Hospitals: Retrospective Cohort Analysis of Prevalence, Predictors, and Outcome of Resistance to All First-Line Agents. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2018, 67, 1803–1814. [Google Scholar] [CrossRef]
- Haute Autorité de Santé (HAS). Antibiothérapie des Infections à Entérobactéries et à Pseudomonas aeruginosa Chez l’adulte: Place des Carbapénèmes et de Leurs Alternatives; HAS: Saint-Denis, France, 2019. [Google Scholar]
- Société Française de Pharmacologie et de Thérapeutique; Société Française d’Anesthésie et Réanimation. Optimisation du Traitement par Beta-Lactamines Chez le Patient de Soins Critiques. 2018. Available online: https://sfar.org/optimisation-du-traitement-par-beta-lactamines-chez-le-patient-de-soins-critiques/ (accessed on 20 April 2024).
- AntiobioEst. AntibioGuide. Available online: https://guides.antibioest.org/#/ (accessed on 20 April 2024).
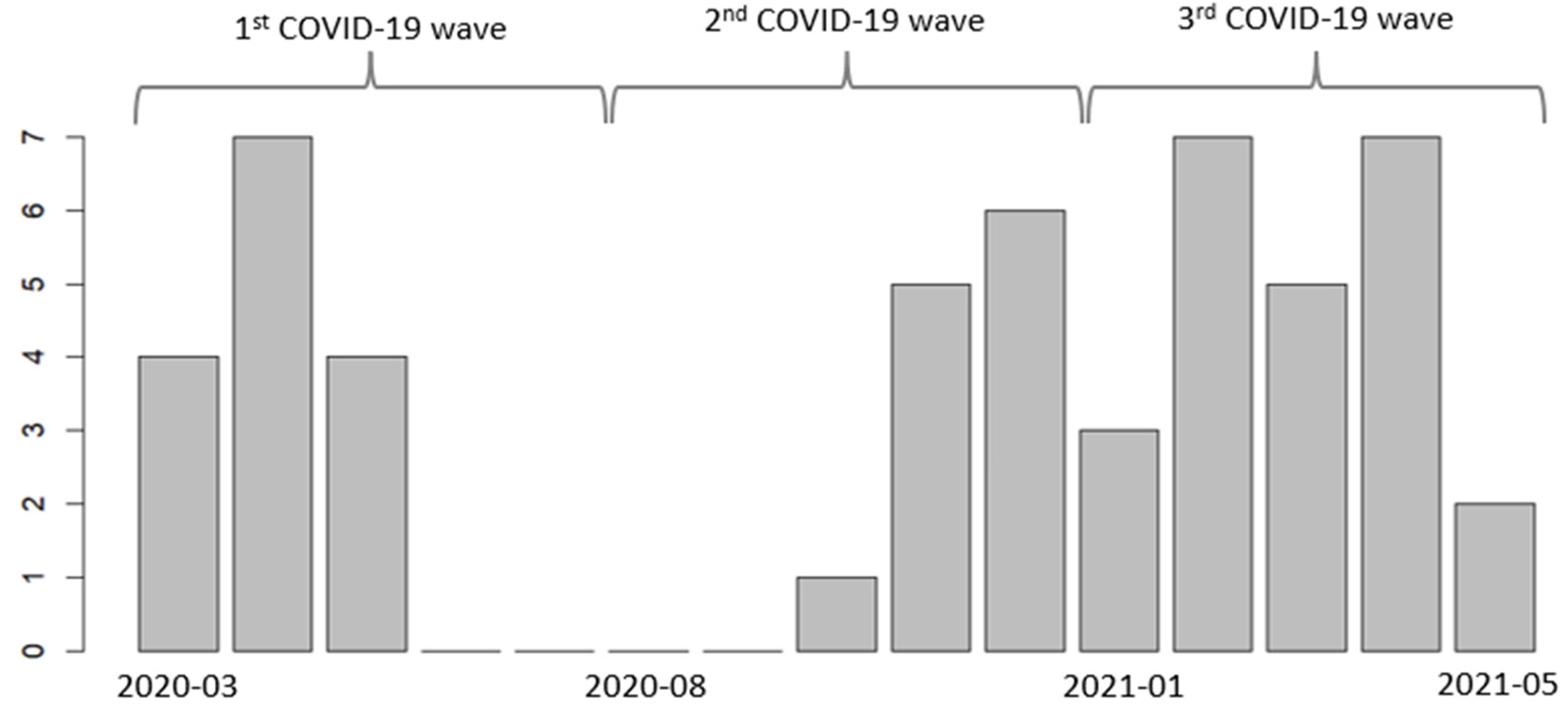
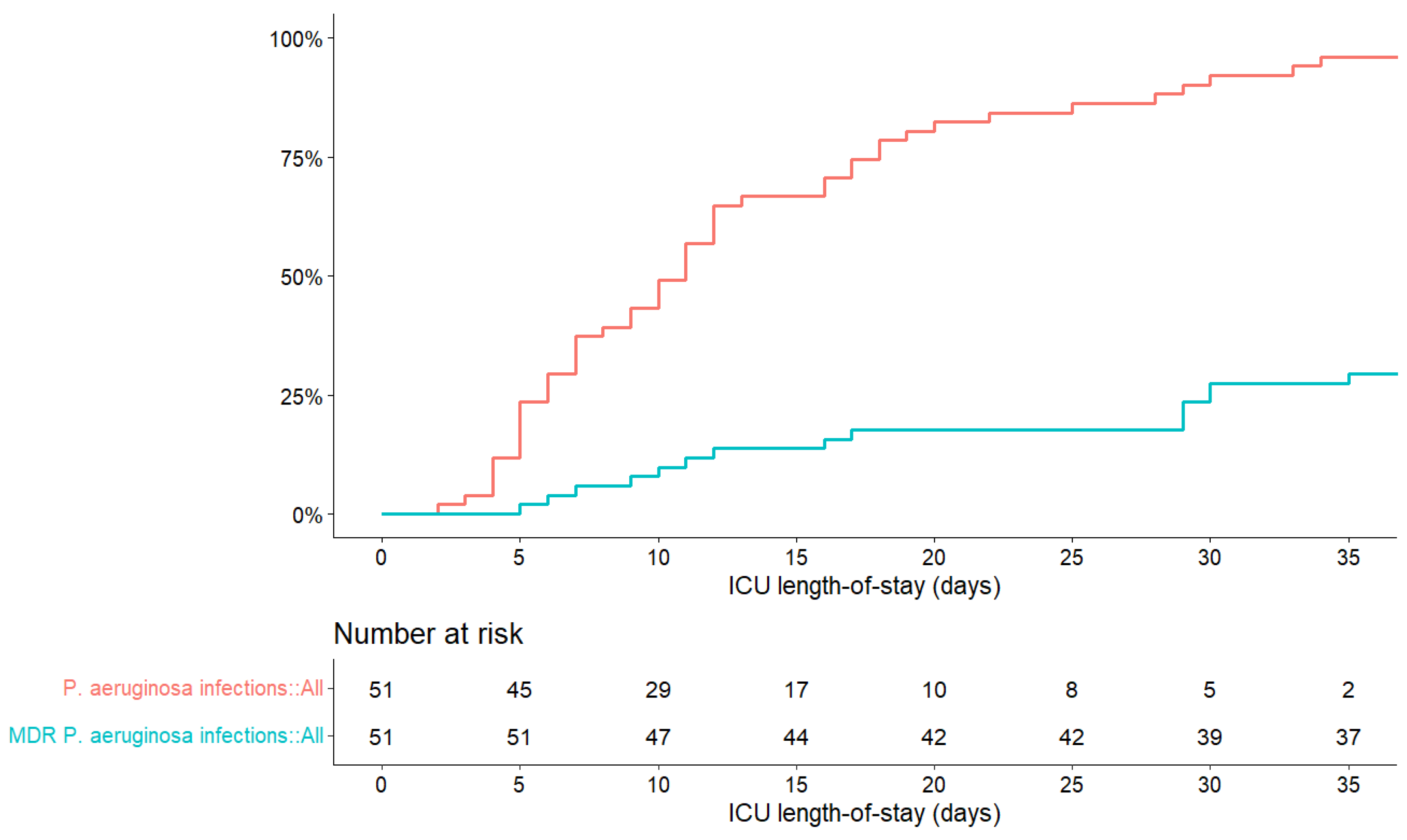
| Patient Characteristics | All Patients (N = 51) | Survivors (n = 29) | Patients Who Died (n = 22) |
|---|---|---|---|
| Demographic characteristics | |||
| Age (years) | 65 ± 12 | 62 ± 12 | 69 ± 11 |
| BMI (kg/m2) | 30 ± 6 | 30 ± 6 | 29 ± 7 |
| Gender (male) | 46 (90%) | 25 (86%) | 21 (95%) |
| Comorbidities | |||
| Arterial hypertension | 31 (61%) | 19 (63%) | 12 (55%) |
| Obesity (BMI ≥ 30) | 23 (45%) | 15 (52%) | 8 (40%) |
| Chronic cardiac disease | 23 (45%) | 13 (42%) | 10 (46%) |
| Kidney insufficiency | 20 (39%) | 9 (29%) | 11 (50%) |
| Obstructive pulmonary disease * | 15 (29%) | 10 (33%) | 5 (23%) |
| Diabetes | 13 (25%) | 8 (27%) | 5 (23%) |
| Malignancy | 3 (6%) | 2 (7%) | 1 (5%) |
| Hospital stays | |||
| Hospital length-of-stay (days) | 36 (27–62) | 42 (32–69) | 31 (21–42) |
| ICU length-of-stay (days) | 27 (20–39) | 27 (20–40) | 27 (21–36) |
| Admission from another hospital | 30 (59%) | 15 (52%) | 15 (68%) |
| Admission from home/emergency | 21 (41%) | 14 (48%) | 7 (32%) |
| Death during stay | 22 (43%) | 0 (0%) | 22 (100%) |
| Discharge to home | 14 (27%) | 14 (48%) | 0 (0%) |
| Discharge to another hospital | 8 (16%) | 8 (28%) | 0 (0%) |
| Discharge to recuperative care center | 7 (14%) | 7 (24%) | 0 (0%) |
| Treatments | |||
| Antibiotics before hospital admission | 21 (41%) | 9 (31%) | 12 (55%) |
| Antibiotics during ICU stay | 51 (100%) | 29 (100%) | 22 (100%) |
| Prone position | 44 (86%) | 25 (86%) | 19 (86%) |
| Corticosteroids | 35 (69%) | 18 (62%) | 17 (77%) |
| Neuromuscular blockers | 31 (61%) | 16 (55%) | 15 (68%) |
| Maximal oxygenation support | |||
| Extracorporeal membrane oxygenation | 11 (22%) | 7 (24%) | 4 (18%) |
| Invasive mechanical ventilation | 37 (72%) | 19 (66%) | 18 (82%) |
| Non-invasive ventilation | 2 (4%) | 2 (7%) | 0 (0%) |
| High-flow oxygen | 1 (2%) | 1 (3%) | 0 (0%) |
| Antibiotic Resistance | All Strains (N = 118) | First Strains (n = 51) | Subsequent Strains (n = 67) | p-Value |
|---|---|---|---|---|
| Susceptible | 39 (33%) | 33 (65%) | 6 (9%) | <0.0001 |
| Resistant * | 36 (31%) | 10 (19%) | 26 (39%) | 0.11 |
| MDR | 35 (29%) | 7 (14%) | 28 (42%) | <0.01 |
| XDR | 8 (7%) | 1 (2%) | 7 (10%) | 0.54 |
| PDR | 0 (0%) | 0 (0%) | 0 (0%) | - |
| Antimicrobial Category Antimicrobial Agent | All Strains (N = 118) | First Strains (n = 51) | Subsequent Strains (n = 67) | p-Value |
|---|---|---|---|---|
| Penicillin + β-lactamase inhibitors | 70 (59%) | 13 (25%) | 57 (86%) | <0.0001 |
| Ticarcillin-clavulanic acid | 70 (59%) | 13 (25%) | 57 (86%) | <0.0001 |
| Piperacillin-tazobactam | 61 (52%) | 7 (13%) | 54 (82%) | <0.0001 |
| Cephalosporins | 50 (42%) | 7 (13%) | 43 (65%) | <0.0001 |
| Ceftazidime | 49 (42%) | 6 (11%) | 43 (65%) | <0.0001 |
| Cefepime | 26 (22%) | 6 (11%) | 20 (30%) | 0.03 |
| Monobactams | 38 (32%) | 8 (15%) | 30 (45%) | 0.002 |
| Aztreonam | 38 (32%) | 8 (15%) | 30 (45%) | 0.002 |
| Carbapenems | 32 (27%) | 6 (11%) | 26 (39%) | 0.001 |
| Meropenem | 31 (26%) | 6 (11%) | 26 (39%) | <0.001 |
| Imipenem | 29 (25%) | 5 (9%) | 24 (36%) | 0.002 |
| Cephalosporins + β-lactamase inhibitors | 21 (18%) | 5 (9%) | 16 (24%) | 0.08 |
| Ceftazidime-avibactam | 19 (16%) | 3 (6%) | 16 (24%) | 0.02 |
| Ceftolozane-tazobactam | 15 (13%) | 4 (8%) | 11 (17%) | 0.27 |
| Fluoroquinolones | 13 (11%) | 4 (8%) | 9 (14%) | 0.51 |
| Ciprofloxacin | 13 (11%) | 4 (8%) | 9 (14%) | 0.51 |
| Aminoglycosides | 5 (4%) | 3 (6%) | 2 (3%) | 0.75 |
| Gentamicin | 5 (4%) | 3 (6%) | 2 (3%) | 0.75 |
| Tobramycin | 1 (1%) | 1 (2%) | 0 (0%) | 0.90 |
| Amikacin | 0 (0%) | 0 (0%) | 0 (0%) | - |
| Molecule | Patients n (%) | Duration in Days Median (Q1–Q3) | Plasma Dosage Compliance n/N (%) | Plasma Dosage in mg/L Median (Q1–Q3) |
|---|---|---|---|---|
| Piperacillin-tazobactam | 39 (76%) | 4 (2–7) | 17/41 * (41%) | 83.9 (42.4–125.2) |
| Cefotaxime | 33 (65%) | 5 (3–7) | 2/6 (33%) | 21.5 (17.7–24.9) |
| Ceftazidime | 25 (49%) | 7 (5–10) | 18/31 (58%) | 62.0 (44.6–86.0) |
| Amikacin | 24 (47%) | 1 (1–1) | 13/28 (46%) | 41.6 (4.7–84.2) |
| Cefepime | 21 (41%) | 6 (3–9) | 5/15 (33%) | 42.9 (33.6–64.6) |
| Spiramycin | 20 (39%) | 4 (3–5) | 0/0 (NA) | NA |
| Meropenem | 18 (35%) | 4 (3–11) | 9/26 (35%) | 15.9 (10.4–39.7) |
| Imipenem | 13 (25%) | 3 (2–7) | 0/7 (0%) | 7.2 (2.0–15.1) |
| Vancomycin | 9 (18%) | 5 (4–7) | 35/40 (88%) | 27.3 (22.0–33.8) |
| Linezolid | 8 (1%) | 4 (2–7) | 0/0 (NA) | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baudet, A.; Regad, M.; Gibot, S.; Conrath, É.; Lizon, J.; Demoré, B.; Florentin, A. Pseudomonas aeruginosa Infections in Patients with Severe COVID-19 in Intensive Care Units: A Retrospective Study. Antibiotics 2024, 13, 390. https://doi.org/10.3390/antibiotics13050390
Baudet A, Regad M, Gibot S, Conrath É, Lizon J, Demoré B, Florentin A. Pseudomonas aeruginosa Infections in Patients with Severe COVID-19 in Intensive Care Units: A Retrospective Study. Antibiotics. 2024; 13(5):390. https://doi.org/10.3390/antibiotics13050390
Chicago/Turabian StyleBaudet, Alexandre, Marie Regad, Sébastien Gibot, Élodie Conrath, Julie Lizon, Béatrice Demoré, and Arnaud Florentin. 2024. "Pseudomonas aeruginosa Infections in Patients with Severe COVID-19 in Intensive Care Units: A Retrospective Study" Antibiotics 13, no. 5: 390. https://doi.org/10.3390/antibiotics13050390
APA StyleBaudet, A., Regad, M., Gibot, S., Conrath, É., Lizon, J., Demoré, B., & Florentin, A. (2024). Pseudomonas aeruginosa Infections in Patients with Severe COVID-19 in Intensive Care Units: A Retrospective Study. Antibiotics, 13(5), 390. https://doi.org/10.3390/antibiotics13050390






