The Genetic Landscape of Antimicrobial Resistance Genes in Enterococcus cecorum Broiler Isolates
Abstract
1. Introduction
2. Results
2.1. Determination of MIC and Epidemiological Cutoff (ECOFF) Values (COWT)
2.2. Identification of Resistance Genes
2.3. Mutations in the E. cecorum GyrA/GyrB/ParC and Genes Encoding Penicillin-Binding Proteins (PBPs) Are Associated with Enrofloxacin and β-Lactam Antimicrobial Resistance
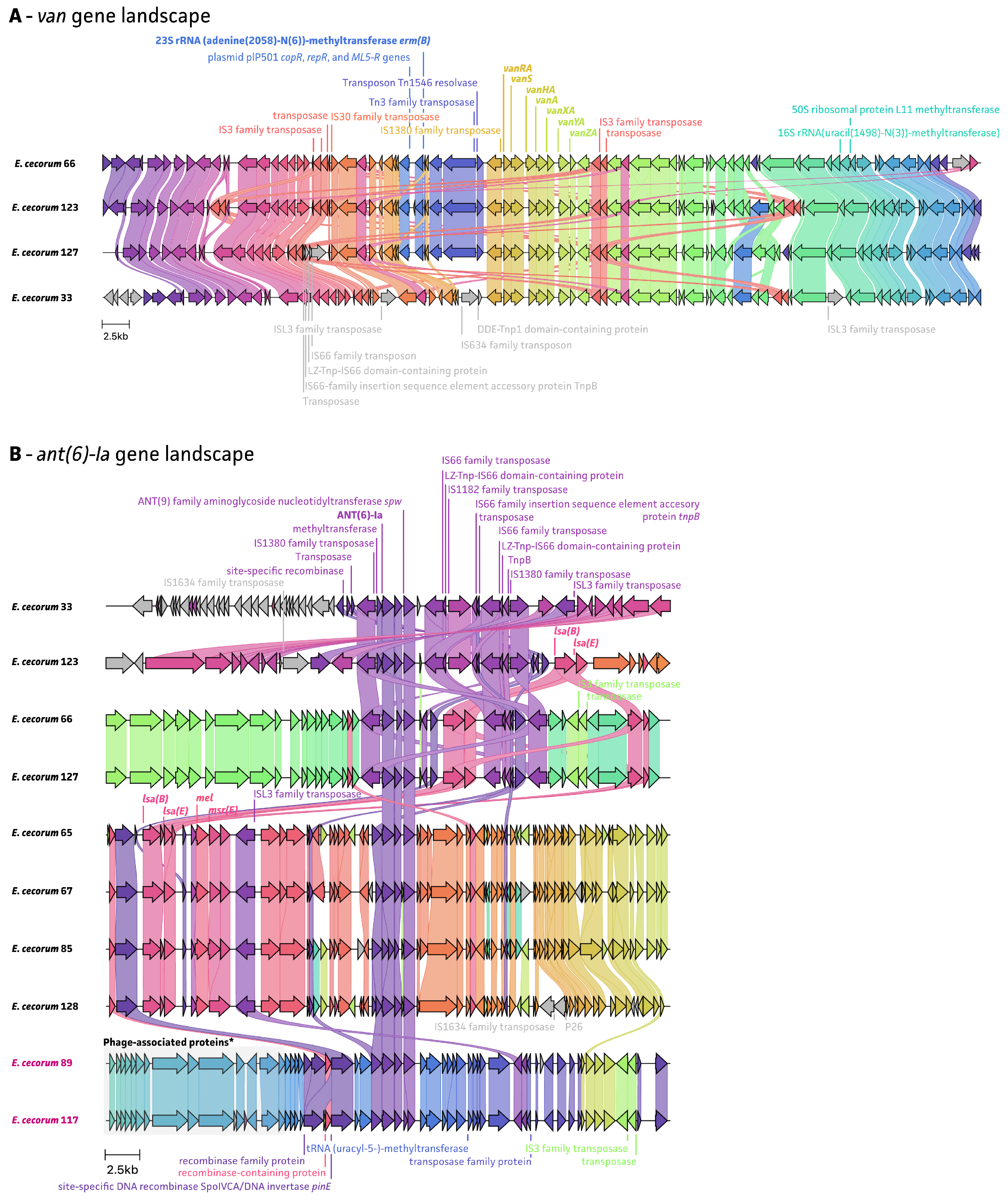
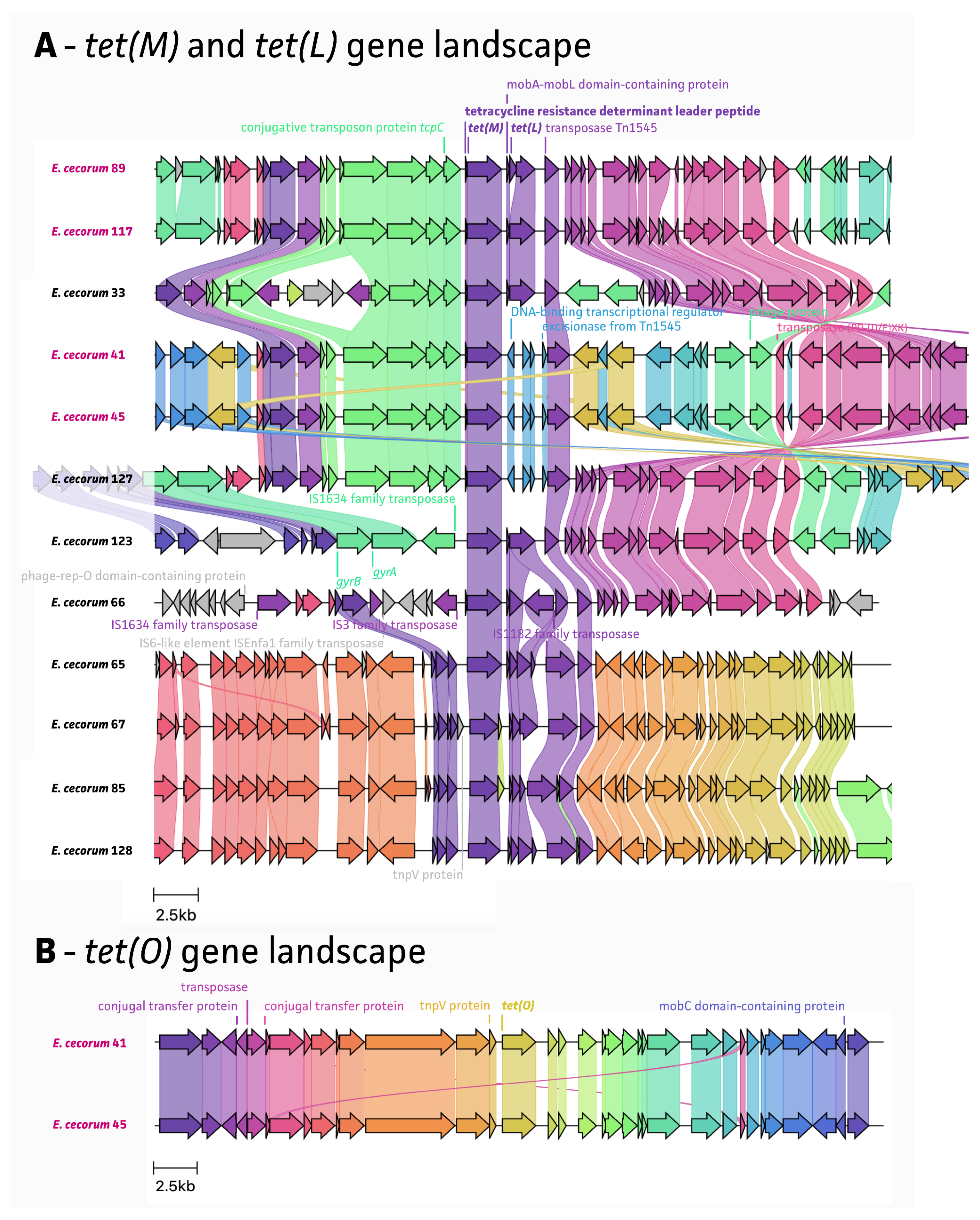
2.4. Mobile Genetic Elements Associated with AMR
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Antimicrobial Susceptibility Testing
4.3. ECOFF Values Determination
4.4. DNA Extraction and Whole-Genome Sequencing
4.5. Phylogenetic Inference, Identification of ARGs, Mobile Genetic Elements, and Point Mutations
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wood, A.M.; MacKenzie, G.; McGiliveray, N.C.; Brown, L.; Devriese, L.A.; Baele, M. Isolation of Enterococcus cecorum from bone lesions in broiler chickens. Vet. Rec. 2002, 150, 27. [Google Scholar] [PubMed]
- Dolka, B.; Chrobak-Chmiel, D.; Czopowicz, M.; Szeleszczuk, P. Characterization of pathogenic Enterococcus cecorum from different poultry groups: Broiler chickens, layers, turkeys, and waterfowl. PLoS ONE 2017, 12, e0185199. [Google Scholar] [CrossRef] [PubMed]
- Wideman, R.F.; Prisby, R.D. Bone circulatory disturbances in the development of spontaneous bacterial chondronecrosis with osteomyelitis: A translational model for the pathogenesis of femoral head necrosis. Front. Endocrinol. 2012, 3, 183. [Google Scholar] [CrossRef] [PubMed]
- Borst, L.B.; Suyemoto, M.M.; Sarsour, A.H.; Harris, M.C.; Martin, M.P.; Strickland, J.D.; Oviedo, E.O.; Barnes, H.J. Pathogenesis of enterococcal Sspondylitis caused by Enterococcus cecorum in broiler chickens. Vet. Pathol. 2017, 54, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Robbins, K.M.; Suyemoto, M.M.; Lyman, R.L.; Martin, M.P.; Barnes, H.J.; Borst, L.B. An outbreak and source investigation of enterococcal spondylitis in broilers caused by Enterococcus cecorum. Avian Dis. 2012, 56, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Schreier, J.; Rychlik, I.; Karasova, D.; Crhanova, M.; Breves, G.; Rautenschlein, S.; Jung, A. Influence of heat stress on intestinal integrity and the caecal microbiota during Enterococcus cecorum infection in broilers. Vet. Res. 2022, 53, 110. [Google Scholar] [CrossRef] [PubMed]
- Schreier, J.; Karasova, D.; Crhanova, M.; Rychlik, I.; Rautenschlein, S.; Jung, A. Influence of lincomycin-spectinomycin treatment on the outcome of Enterococcus cecorum infection and on the cecal microbiota in broilers. Gut Pathog. 2022, 14, 3. [Google Scholar] [CrossRef] [PubMed]
- Herdt, P.; Defoort, P.; Steelant, J.; Swam, H.; Tanghe, L.; Goethem, S.; Vanrobaeys, M. Enterococcus cecorum osteomyelitis and arthritis in broiler chickens. Vlaams Diergeneeskd. Tijdschr. 2009, 78, 44–48. [Google Scholar] [CrossRef]
- Dolka, B.; Chrobak-Chmiel, D.; Makrai, L.; Szeleszczuk, P. Phenotypic and genotypic characterization of Enterococcus cecorum strains associated with infections in poultry. BMC Vet. Res. 2016, 12, 129. [Google Scholar] [CrossRef]
- Laurentie, J.; Mourand, G.; Grippon, P.; Furlan, S.; Chauvin, C.; Jouy, E.; Serror, P.; Kempf, I. Determination of Epidemiological Cutoff Values for Antimicrobial Resistance of Enterococcus cecorum. J. Clin. Microbiol. 2023, 61, e0144522. [Google Scholar] [CrossRef]
- Borst, L.B.; Suyemoto, M.M.; Robbins, K.M.; Lyman, R.L.; Martin, M.P.; Barnes, H.J. Molecular epidemiology of Enterococcus cecorum isolates recovered from enterococcal spondylitis outbreaks in the southeastern United States. Avian Pathol. 2012, 41, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Authority, European Food Safety. The European Union Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2012. Efsa J. 2014, 12, 3590. [Google Scholar] [CrossRef]
- Jackson, C.R.; Kariyawasam, S.; Borst, L.B.; Frye, J.G.; Barrett, J.B.; Hiott, L.M.; Woodley, T.A. Antimicrobial resistance, virulence determinants and genetic profiles of clinical and nonclinical Enterococcus cecorum from poultry. Lett. Appl. Microbiol. 2015, 60, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Harada, T.; Kawahara, R.; Kanki, M.; Taguchi, M.; Kumeda, Y. Isolation and characterization of vanA genotype vancomycin-resistant Enterococcus cecorum from retail poultry in Japan. Int. J. Food Microbiol. 2012, 153, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Gupta, S.K.; Barrett, J.B.; Hiott, L.M.; Woodley, T.A.; Kariyawasam, S.; Frye, J.G.; Jackson, C.R. Comparison of antimicrobial resistance and pan-genome of clinical and non-clinical Enterococcus cecorum from poultry using whole-genome sequencing. Foods 2020, 9, 686. [Google Scholar] [CrossRef] [PubMed]
- Ceric, O.; Tyson, G.H.; Goodman, L.B.; Mitchell, P.K.; Zhang, Y.; Prarat, M.; Cui, J.; Peak, L.; Scaria, J.; Antony, L.; et al. Enhancing the one health initiative by using whole genome sequencing to monitor antimicrobial resistance of animal pathogens: Vet-LIRN collaborative project with veterinary diagnostic laboratories in United States and Canada. BMC Vet. Res. 2019, 15, 130. [Google Scholar] [CrossRef] [PubMed]
- Laurentie, J.; Loux, V.; Hennequet-Antier, C.; Chambellon, E.; Deschamps, J.; Trotereau, A.; Furlan, S.; Darrigo, C.; Kempf, F.; Lao, J.; et al. Comparative genome analysis of Enterococcus cecorum reveals intercontinental spread of a lineage of clinical poultry isolates. mSphere 2023, 8, e0049522. [Google Scholar] [CrossRef] [PubMed]
- Bokma, J.; Vereecke, N.; Nauwynck, H.; Haesebrouck, F.; Theuns, S.; Pardon, B.; Boyen, F. Genome-wide association study reveals genetic markers for antimicrobial resistance in Mycoplasma bovis. Microbiol. Spectr. 2021, 9, e0026221. [Google Scholar] [CrossRef]
- Vereecke, N.; Botteldoorn, N.; Brosse, C.; Bonckaert, C.; Nauwynck, H.; Haesebrouck, F.; Boyen, F.; Maes, D.; Theuns, S. Predictive Power of Long-Read Whole-genome sequencing for rapid diagnostics of multidrug-resistant Brachyspira hyodysenteriae Strains. Microbiol. Spectr. 2023, 11, e0412322. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31. [Google Scholar] [CrossRef]
- Vereecke, N.; Van Hoorde, S.; Sperling, D.; Theuns, S.; Devriendt, B.; Cox, E. Virotyping and genetic antimicrobial susceptibility testing of porcine ETEC/STEC strains and associated plasmid types. Front. Microbiol. 2023, 14, 1139312. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Eeckhaut, V.; Goossens, E.; Rasschaert, G.; Van Erum, J.; Roovers, G.; Ducatelle, R.; Antonissen, G.; Van Immerseel, F. Bacterial chondronecrosis with osteomyelitis related Enterococcus cecorum isolates are genetically distinct from the commensal population and are more virulent in an embryo mortality model. Vet. Res. 2023, 54, 13. [Google Scholar] [CrossRef] [PubMed]
- Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 6th ed.; CLSI Supplement VET01S; Clinical Laboratory Standards Institute: Wayne, PA, USA, 2023. [Google Scholar]
- Cochetti, I.; Tili, E.; Mingoia, M.; Varaldo, P.E.; Montanari, M.P. erm(B)-carrying elements in tetracycline-resistant pneumococci and correspondence between Tn1545 and Tn6003. Antimicrob. Agents Chemother. 2008, 52, 1285–1290. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Soge, O.O.; Beck, N.K.; White, T.M.; No, D.B.; Roberts, M.C. A novel transposon, Tn6009, composed of a Tn916 element linked with a Staphylococcus aureus mer operon. J. Antimicrob. Chemother. 2008, 62, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Jung, A.; Chen, L.R.; Suyemoto, M.M.; Barnes, H.J.; Borst, L.B. A review of Enterococcus cecorum infection in poultry. Avian Dis. 2018, 62, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M.; Kruse, H.; Tast, E.; Hammerum, A.M.; Jensen, L.B. Associations between the use of antimicrobial agents for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers and pigs in Denmark, Finland, and Norway. Microb. Drug Resist. 2000, 6, 63–70. [Google Scholar] [CrossRef]
- Jensen, L.B.; Frimodt-Moller, N.; Aarestrup, F.M. Presence of erm gene classes in gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol. Lett. 1999, 170, 151–158. [Google Scholar] [CrossRef]
- Roberts, M.C.; Sutcliffe, J.; Courvalin, P.; Jensen, L.B.; Rood, J.; Seppala, H. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 1999, 43, 2823–2830. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.R.; Fedorka-Cray, P.J.; Barrett, J.B.; Ladely, S.R. Effects of tylosin use on erythromycin resistance in enterococci isolated from swine. Appl. Environ. Microbiol. 2004, 70, 4205–4210. [Google Scholar] [CrossRef]
- Okitsu, N.; Kaieda, S.; Yano, H.; Nakano, R.; Hosaka, Y.; Okamoto, R.; Kobayashi, T.; Inoue, M. Characterization of ermB gene transposition by Tn1545 and Tn917 in macrolide-resistant Streptococcus pneumoniae isolates. J. Clin. Microbiol. 2005, 43, 168–173. [Google Scholar] [CrossRef]
- Chen, Y.G.; Qu, T.T.; Yu, Y.S.; Zhou, J.Y.; Li, L.J. Insertion sequence ISEcp1-like element connected with a novel aph(2″) allele [aph(2″)-Ie] conferring high-level gentamicin resistance and a novel streptomycin adenylyltransferase gene in Enterococcus. J. Med. Microbiol. 2006, 55, 1521–1525. [Google Scholar] [CrossRef]
- Wilcks, A.; Andersen, S.R.; Licht, T.R. Characterization of transferable tetracycline resistance genes in Enterococcus faecalis isolated from raw food. FEMS Microbiol. Lett. 2005, 243, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; Leon-Sampedro, R.; Del Campo, R.; Coque, T.M. Antimicrobial resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Cauwerts, K.; Decostere, A.; De Graef, E.M.; Haesebrouck, F.; Pasmans, F. High prevalence of tetracycline resistance in Enterococcus isolates from broilers carrying the erm(B) gene. Avian Pathol. 2007, 36, 395–399. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef]
- Haack, B.J.; Andrews, R.E., Jr. Isolation of Tn916-like conjugal elements from swine lot effluent. Can. J. Microbiol. 2000, 46, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Arthur, M.; Courvalin, P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 1993, 37, 1563–1571. [Google Scholar] [CrossRef]
- Bach Knudsen, K.E. Development of antibiotic resistance and options to replace antimicrobials in animal diets. Proc. Nutr. Soc. 2001, 60, 291–299. [Google Scholar] [CrossRef]
- Chan, Y.Y.; Abd Nasir, M.H.; Yahaya, M.A.; Salleh, N.M.; Md Dan, A.D.; Musa, A.M.; Ravichandran, M. Low prevalence of vancomycin- and bifunctional aminoglycoside-resistant enterococci isolated from poultry farms in Malaysia. Int. J. Food Microbiol. 2008, 122, 221–226. [Google Scholar] [CrossRef]
- Casewell, M.; Friis, C.; Marco, E.; McMullin, P.; Phillips, I. The European ban on growth-promoting antibiotics and emerging consequences for human and animal health. J. Antimicrob. Chemother. 2003, 52, 159–161. [Google Scholar] [CrossRef]
- Yoshimura, H.; Ishimaru, M.; Endoh, Y.S.; Suginaka, M.; Yamatani, S. Isolation of glycopeptide-resistant enterococci from chicken in Japan. Antimicrob. Agents Chemother. 1998, 42, 3333. [Google Scholar] [CrossRef] [PubMed]
- Stepien-Pysniak, D.; Marek, A.; Banach, T.; Adaszek, L.; Pyzik, E.; Wilczynski, J.; Winiarczyk, S. Prevalence and antibiotic resistance of Enterococcus strains isolated from poultry. Acta Vet. Hung. 2016, 64, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Laurentie, J.; Mourand, G.; Jouy, E.; Bougeard, S.; Keita, A.; Amelot, M.; Serror, P.; Kempf, I. Study of the effect of administration of narasin or antibiotics on in vivo selection of a narasin- and multidrug-resistant Enterococcus cecorum strain. Vet. Microbiol. 2023, 282, 109757. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Williams, J.D. Penicillin-binding proteins in Streptococcus faecalis and S. faecium. J. Med. Microbiol. 1987, 23, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Fontana, R.; Canepari, P.; Lleo, M.M.; Satta, G. Mechanisms of resistance of enterococci to beta-lactam antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 1990, 9, 103–105. [Google Scholar] [CrossRef] [PubMed]
- Moon, T.M.; D’Andréa, É.D.; Lee, C.W.; Soares, A.; Jakoncic, J.; Desbonnet, C.; Garcia-Solache, M.; Rice, L.B.; Page, R.; Peti, W. The structures of penicillin-binding protein 4 (PBP4) and PBP5 from Enterococci provide structural insights into β-lactam resistance. J. Biol. Chem. 2018, 293, 18574–18584. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Tam, D.M.; Lau, S.K.; Fung, A.M.; Yuen, K.Y. Enterococcus cecorum empyema thoracis successfully treated with cefotaxime. J. Clin. Microbiol. 2004, 42, 919–922. [Google Scholar] [CrossRef] [PubMed]
- Nagai, K.; Davies, T.A.; Jacobs, M.R.; Appelbaum, P.C. Effects of amino acid alterations in penicillin-binding proteins (PBPs) 1a, 2b, and 2x on PBP affinities of penicillin, ampicillin, amoxicillin, cefditoren, cefuroxime, cefprozil, and cefaclor in 18 clinical isolates of penicillin-susceptible, -intermediate, and -resistant pneumococci. Antimicrob. Agents Chemother. 2002, 46, 1273–1280. [Google Scholar] [CrossRef]
- Asahi, Y.; Takeuchi, Y.; Ubukata, K. Diversity of substitutions within or adjacent to conserved amino acid motifs of penicillin-binding protein 2X in cephalosporin-resistant Streptococcus pneumoniae isolates. Antimicrob. Agents Chemother. 1999, 43, 1252–1255. [Google Scholar] [CrossRef]
- Hsieh, Y.C.; Su, L.H.; Hsu, M.H.; Chiu, C.H. Alterations of penicillin-binding proteins in pneumococci with stepwise increase in beta-lactam resistance. Pathog. Dis. 2013, 67, 84–88. [Google Scholar] [CrossRef][Green Version]
- Dahiya, S.; Kapil, A.; Lodha, R.; Kumar, R.; Das, B.K.; Sood, S.; Kabra, S.K. Induction of resistant mutants of Salmonella enterica serotype Typhi under ciprofloxacin selective pressure. Indian J. Med. Res. 2014, 139, 746–753. [Google Scholar] [PubMed]
- Gruger, T.; Nitiss, J.L.; Maxwell, A.; Zechiedrich, E.L.; Heisig, P.; Seeber, S.; Pommier, Y.; Strumberg, D. A mutation in Escherichia coli DNA gyrase conferring quinolone resistance results in sensitivity to drugs targeting eukaryotic topoisomerase II. Antimicrob. Agents Chemother. 2004, 48, 4495–4504. [Google Scholar] [CrossRef] [PubMed]
- Kronvall, G. Normalized resistance interpretation as a tool for establishing epidemiological MIC susceptibility breakpoints. J. Clin. Microbiol. 2010, 48, 4445–4452. [Google Scholar] [CrossRef] [PubMed]
- Reydams, H.; Toledo-Silva, B.; Mertens, K.; Piepers, S.; Vereecke, N.; Souza, F.N.; Haesebrouck, F.; De Vliegher, S. Phenotypic and genotypic assessment of iron acquisition in diverse bovine-associated non-aureus staphylococcal strains. Vet. Res. 2024, 55, 6. [Google Scholar] [CrossRef] [PubMed]
- Belhout, C.; Boyen, F.; Vereecke, N.; Theuns, S.; Taibi, N.; Stegger, M.; de la Fe-Rodriguez, P.Y.; Bouayad, L.; Elgroud, R.; Butaye, P. Prevalence and molecular characterization of methicillin-resistant staphylococci (MRS) and mammaliicocci (MRM) in dromedary camels from Algeria: First Detection of SCCmec-mecC Hybrid in Methicillin-Resistant Mammaliicoccus lentus. Antibiotics 2023, 12, 674. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Cerdeira, L.T.; Hawkey, J.; Meric, G.; Vezina, B.; Wyres, K.L.; Holt, K.E. Trycycler: Consensus long-read assemblies for bacterial genomes. Genome Biol. 2021, 22, 266. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bliss, C.M.; Bennett, J.S.; Bratcher, H.B.; Brehony, C.; Colles, F.M.; Wimalarathna, H.; Harrison, O.B.; Sheppard, S.K.; Cody, A.J.; et al. Ribosomal multilocus sequence typing: Universal characterization of bacteria from domain to strain. Microbiology 2012, 158, 1005–1015. [Google Scholar] [CrossRef]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Matlock, W.; Lipworth, S.; Constantinides, B.; Peto, T.E.A.; Walker, A.S.; Crook, D.; Hopkins, S.; Shaw, L.P.; Stoesser, N. Flanker: A tool for comparative genomics of gene flanking regions. Microb. Genom. 2021, 7, 000634. [Google Scholar] [CrossRef]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef]
- Schmidt, B.M.; Davidson, N.M.; Hawkins, A.D.K.; Bartolo, R.; Majewski, I.J.; Ekert, P.G.; Oshlack, A. Clinker: Visualizing fusion genes detected in RNA-seq data. Gigascience 2018, 7, giy079. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]

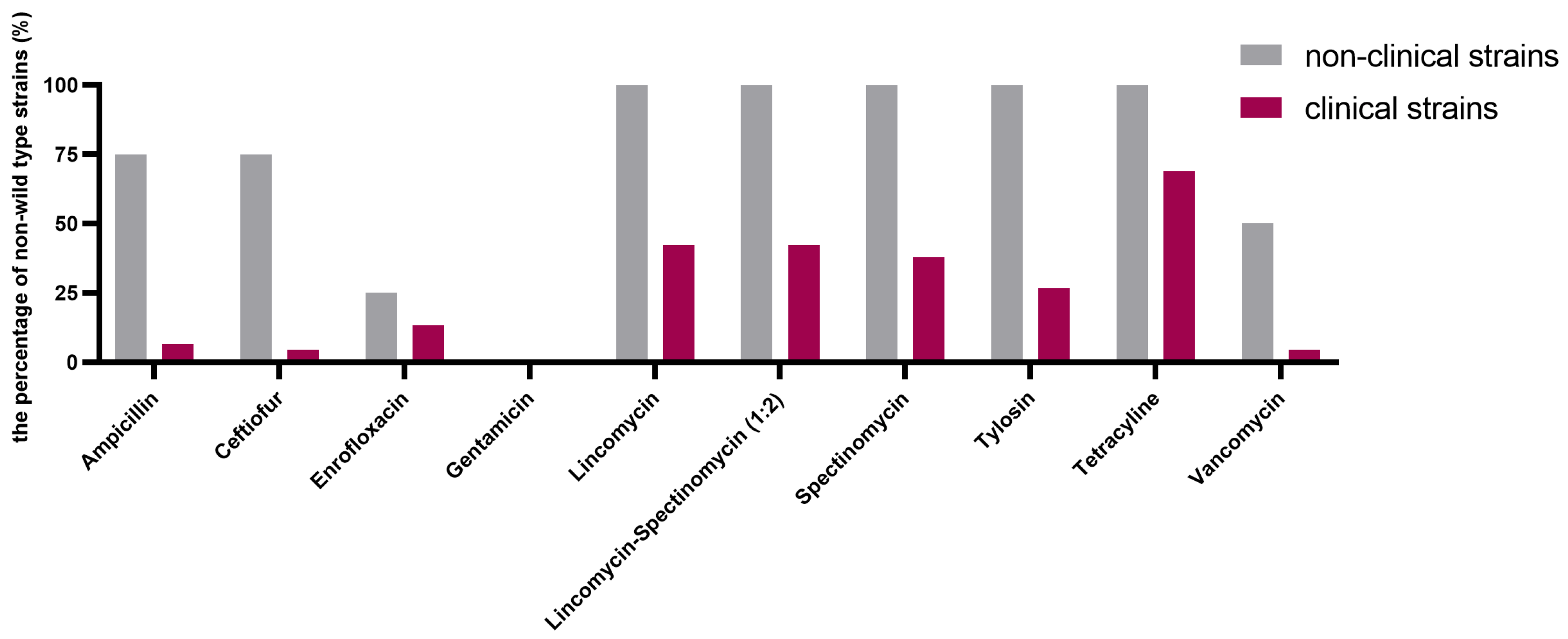



| Antimicrobial Agent | Number of Strains with MIC (µg/mL) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild-Type | Non-Wild-Type | |||||||||||||||||
| ≤0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | >128 | [n] | [%] | [n] | [%] | |
| Ampicillin | 7 | 37 | 3 | 5 | 1 | 44 | 83% | 9 | 17% | |||||||||
| Ceftiofur | 14 | 20 | 10 | 1 | 2 | 3 | 3 | 45 | 85% | 8 | 15% | |||||||
| Enrofloxacin | 2 | 22 | 9 | 12 | 1 | 7 | 45 | 85% | 8 | 15% | ||||||||
| Gentamicin | 2 | 40 | 11 | 53 | 100% | 0 | 0 | |||||||||||
| Lincomycin | 7 | 19 | 3 | 1 | 23 | 26 | 49% | 27 | 51% | |||||||||
| Lincomycin–Spectinomycin (1:2) | 26 | 3 | 1 | 2 | 21 | 26 | 49% | 27 | 51% | |||||||||
| Spectinomycin | 2 | 26 | 25 | 28 | 53% | 25 | 47% | |||||||||||
| Tylosin | 3 | 30 | 20 | 33 | 62% | 20 | 38% | |||||||||||
| Tetracycline | 10 | 4 | 19 | 15 | 5 | 14 | 26% | 39 | 74% | |||||||||
| Vancomycin | 45 | 2 | 3 | 2 | 1 | 47 | 89% | 6 | 11% | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Boyen, F.; Antonissen, G.; Vereecke, N.; Van Immerseel, F. The Genetic Landscape of Antimicrobial Resistance Genes in Enterococcus cecorum Broiler Isolates. Antibiotics 2024, 13, 409. https://doi.org/10.3390/antibiotics13050409
Huang Y, Boyen F, Antonissen G, Vereecke N, Van Immerseel F. The Genetic Landscape of Antimicrobial Resistance Genes in Enterococcus cecorum Broiler Isolates. Antibiotics. 2024; 13(5):409. https://doi.org/10.3390/antibiotics13050409
Chicago/Turabian StyleHuang, Yue, Filip Boyen, Gunther Antonissen, Nick Vereecke, and Filip Van Immerseel. 2024. "The Genetic Landscape of Antimicrobial Resistance Genes in Enterococcus cecorum Broiler Isolates" Antibiotics 13, no. 5: 409. https://doi.org/10.3390/antibiotics13050409
APA StyleHuang, Y., Boyen, F., Antonissen, G., Vereecke, N., & Van Immerseel, F. (2024). The Genetic Landscape of Antimicrobial Resistance Genes in Enterococcus cecorum Broiler Isolates. Antibiotics, 13(5), 409. https://doi.org/10.3390/antibiotics13050409









