Abstract
The recognition of the Aerococcus urinae complex (AUC) as an emerging uropathogen has led to growing concerns due to a limited understanding of its disease spectrum and antibiotic resistance profiles. Here, we investigated the prevalence of macrolide resistance within urinary AUC isolates, shedding light on potential genetic mechanisms. Phenotypic testing revealed a high rate of macrolide resistance: 45%, among a total of 189 urinary AUC isolates. Genomic analysis identified integrative and conjugative elements (ICEs) as carriers of the macrolide resistance gene ermA, suggesting horizontal gene transfer as a mechanism of resistance. Furthermore, comparison with publicly available genomes of related pathogens revealed high ICE sequence homogeneity, highlighting the potential for cross-species dissemination of resistance determinants. Understanding mechanisms of resistance is crucial for developing effective surveillance strategies and improving antibiotic use. Furthermore, the findings underscore the importance of considering the broader ecological context of resistance dissemination, emphasizing the need for community-level surveillance to combat the spread of antibiotic resistance within the urinary microbiome.
1. Introduction
Aerococcus urinae, a Gram-positive coccus that grows in pairs and clusters, was first characterized from human urine in 1992 [1]. Originally described as a rare cause of human infection, a clear rise in diagnoses and case reports has been observed in the years since its first isolation [2,3,4,5]. The range of diseases associated with the bacterium includes malodorous urine, urinary tract infections, urgency urinary incontinence, urosepsis, soft tissue infections, and infective endocarditis, with antibiotic therapy as the primary treatment for infection [6,7,8,9,10,11,12,13]. As more and more strains have been studied, substantial phenotypic and genomic variation between strains has been observed, leading to the formation of subgroups [14,15]. It was from these subgroups that, just recently, new Aerococcus species were proposed, including A. tenax, A. mictus, and A. loyolae [16]. These three species, along with A. urinae, make up the newly described A. urinae complex (AUC) that more accurately describes the species diversity.
Similar to other emerging uropathogens, the AUC is poorly characterized and remains a blind spot in both disease and antibiotic resistance surveillance [17]. Worryingly, studies have been reporting antibiotic resistance, such as to fluoroquinolones, macrolides, and trimethoprim-sulfamethoxazole [18,19,20,21]. The current recommended empiric antimicrobial treatment for A. urinae urinary tract infection is the use of ampicillin, tetracycline, and nitrofurantoin [22]. However, published antibiograms do not regularly include AUC as part of surveillance testing, leaving rates of resistance unknown. These reports of resistance are concerning, especially given the global increase in uropathogenic resistance, particularly during the COVID-19 pandemic [23,24,25]. Resistant uropathogens are of particular concern as they may lead to further disease complications and recurrent urinary tract infections [26].
Within the human urinary microbiome, there is a lack of understanding as to how bacteria acquire antibiotic resistance, especially for emerging uropathogens [17]. The more commonly studied uropathogens, such as Escherichia coli or Streptococcus agalactiae, have been observed to engage in horizontal gene transfer with mobile genetic elements mediating the dissemination of antibiotic resistance genes (ARGs) [27,28]. As for AUC, it is unknown what genetic factors lead to observed resistances and whether horizontal gene transfer plays any role in acquired ARGs. However, the recent complete genome sequencing of the type strains of the new AUC species has enabled investigations into genetic determinants of resistance [29].
Thus, we sought to investigate any relationship between genotypic and phenotypic antimicrobial resistances of AUC isolates from urology and urogynecology patients at a United States quaternary medical center. We tested 189 AUC isolates for macrolide antibiotic susceptibility rates and found a pattern of persistent high resistance over a seven-year period between 2017 and 2023. Furthermore, we obtained evidence for bacterial horizontal gene transfer as the genetic basis for this pattern.
2. Results
2.1. Survey of Macrolide Susceptibility
A total of 158 AUC draft genomes from the National Center for Biotechnology Information (NCBI) were analyzed via comprehensive antibiotic resistance database (CARD) for ARGs. Assessing only perfect hits, 18 (11.4%) hits were predicted to encode macrolide resistance, 12 (7.6%) hits were predicted to encode glycopeptide resistance, and 9 (5.7%) hits were predicted to encode aminoglycoside resistance. We next sought to test whether strains within our local community hospital system demonstrate similar rates of macrolide resistance by characterizing the susceptibility of 189 AUC isolates.
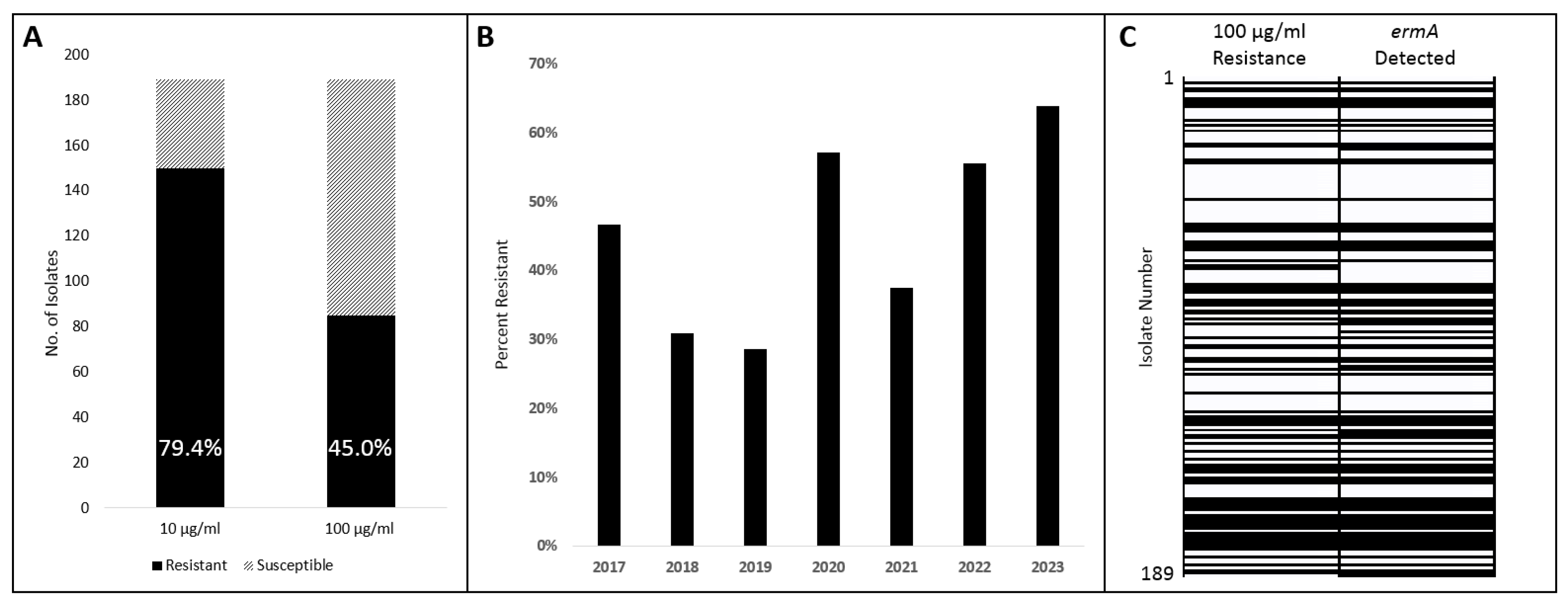
Although the Clinical & Laboratory Standards Institute (CLSI) has published criteria for minimum inhibitory concentration (MIC) interpretation for AUC on several antimicrobial classes, it does not outline criteria for macrolides [30]. Considering that the already existing CLSI criteria were adapted from those of viridans group Streptococcus species, we at first adapted those criteria for azithromycin: susceptibility at ≤0.5 μg/mL and resistance at ≥2 μg/mL. A total of 150 (79.4%) isolates demonstrated resistance at the 2 μg/mL threshold; the same results were obtained at 10 μg/mL azithromycin. However, when we evaluated at our maximum concentration of 100 μg/mL azithromycin, 85 (45.0%) isolates demonstrated resistance (Figure 1A). Using this ≥100 μg/mL criterion, isolates were compared based on isolation year with resistances ranging between 29% and 64% (Figure 1B).
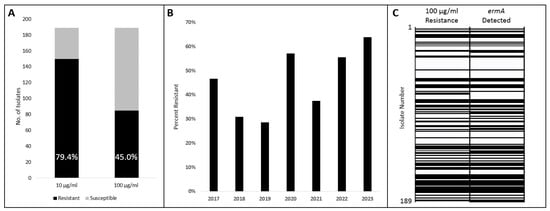
Figure 1.
Azithromycin susceptibility and ermA presence among AUC isolates. (A) Susceptibility of AUC isolates out of a total of 189 tested at 10 μg/mL and 100 μg/mL. (B) Susceptibility of AUC isolates tested at 100 μg/mL compared by isolation year. (C) Concordance of phenotype (100 μg/mL) with genotype (ermA detection) of all 189 isolates. Isolates descending by isolation order with isolate number 1 earliest. Solid black indicates a positive result from either test.
2.2. Survey of Macrolide Resistance Gene ermA
To evaluate a genetic basis for the high rate of macrolide resistance, a PCR test identifying the ermA gene was conducted on the same 189 AUC isolates. The PCR primers were designed based on the ermA sequence detected by the CARD analysis. ermA was detected in 89 (47.1%) isolates, which was similar to the 100 μg/mL Azithromycin resistance result of 85 (45.0%). Comparing these two experiments, 95.8% of isolates matched both the resistance phenotype and ermA genotype (Figure 1C). Of the 8 isolates that did not agree, 2 demonstrated a positive phenotype resistance but a negative ermA genotype, and 6 demonstrated a negative phenotype resistance but a positive ermA genotype (Supplementary Materials Table S1).
2.3. Macrolide Resistance Harbored within Mobile Genetic Elements
An initial analysis with VRprofile2 of the publicly available draft AUC genomes revealed that the macrolide resistance gene ermA was harbored within mobile genetic elements (Supplementary Materials Figure S1). These were identified to be integrative and conjugative elements (ICEs) capable of mobilizing DNA between bacterial chromosomes [31].
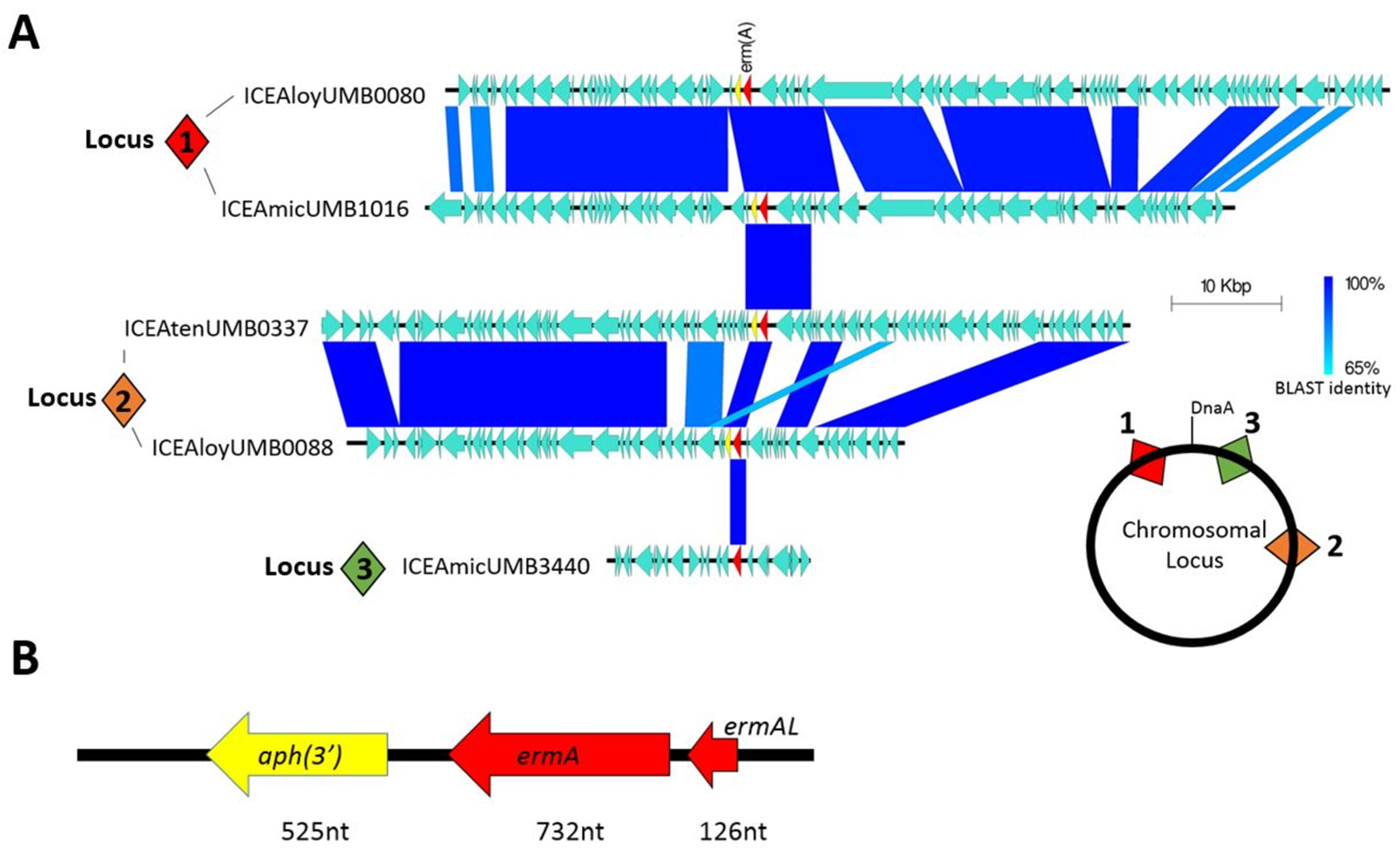
To determine if this pattern of antibiotic resistance inheritance occurred in our isolates, we selected eight isolates that demonstrated resistance at the 100 μg/mL azithromycin threshold for whole genome sequencing. Although all eight genomes were found to actually possess the ermA resistance gene, the gene was not found in the same syntenic locus nor within the same ICE. Comparative genomic analysis revealed that ICEs containing the resistance gene integrated into AUC genomes at three distinct loci (Figure 2A). ICEs inserted at syntenic loci generally shared a high degree of nucleotide identity; however, very little nucleotide identity was observed between ICEs of non-syntenic loci. As one example, all ICEs found to insert within the hsdM type 1 restriction enzyme gene (locus 1) shared nearly 100% gene identity in ICE structural genes. But these ICEs only shared three open reading frames with ICEs inserted at locus 2 and only the ermA gene with ICEs at locus 3. Within the ICEs, the ermA gene was often inherited in combination with an upstream leader peptide and a downstream aminoglycoside 3′-phosphotransferase APH (3′) resistance gene (Figure 2B).
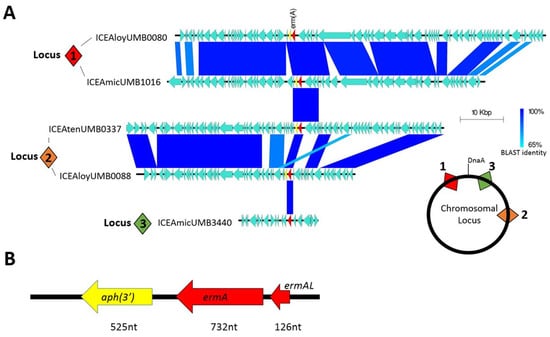
Figure 2.
Macrolide resistance encoded in ICEs. (A) Homology comparison between different ICEs found in AUC isolates. Numbered square diamonds indicate syntenic loci for chromosomal insertion of each ICE. (B) Commonly inherited group of genes containing the ermA resistance gene.
The majority of these ICEs belong to the Tn916 and Tn1806 ICE/dICE superfamily, which are commonly found in Streptococcus species. One ICE in particular, ICEAmicUMB3440, was found to be 97% identical to ICESag066, an ICE first documented in Streptococcus agalactiae (Supplementary Materials Figure S2) [28]. This ICE was the only one not found to contain the leader peptide-ermA-APH(3′) combination, instead only containing the ermA by itself within.
3. Discussion
This study sheds light on the prevalence of macrolide resistance in AUC isolates within a quaternary health system. In 2019, 260,000 global deaths were estimated to have been associated with antimicrobial resistance in urinary tract infections [32]. Although resistance to macrolides is not typically surveilled in organisms isolated from the urinary tract, azithromycin, primarily used for respiratory infections, can reach the bladder during excretion, exposing urinary tract bacteria to azithromycin [33,34]. Long-term use of macrolide antibiotics, such as azithromycin, has been linked to an increase in resistance, particularly in patients with conditions like COPD and cystic fibrosis [35]. Furthermore, a retrospective study found macrolide resistance to be the highest among common uropathogens isolated during the year 2021 [12]. In the current study, we found a persistently high rate of macrolide resistance in AUC isolates over a seven-year study period. Although this study period spanned the COVID-19 pandemic, it is undetermined whether the high rate of azithromycin administration during the COVID-19 pandemic influenced the macrolide resistance rates of uropathogens.
We identified the macrolide resistance gene ermA as a primary determinant of resistance within AUC isolates. Of our 189 isolates, 47.9% tested positive for the ermA gene by PCR, closely matching the 46.1% of isolates that exhibited resistance to high concentrations of macrolide. This gene was not inherited as a part of the AUC core genome but instead must have been introduced into the chromosome by means of horizontal gene transfer via ICEs. Interestingly, 1.8% of ermA-positive isolates remained susceptible to high azithromycin concentrations. This discrepancy could stem from the mobile genetic element itself being transcriptionally silent [36]. The emergence of macrolide resistance due to ICEs has been seen in other related uropathogens, such as Streptococcus agalactiae [17]. In fact, the same ICE characterized in S. agalactiae, ICESag066, was found to be within one of our AUC isolates. This observation implies that either the mobile genetic element can spread directly across species or may be received from a common donor within the bladder microbiome. Horizontal gene transfer of genetic content, not limited to ARGs, has been observed to occur in human-associated bacteria, and it may occur at an even higher rate in these microorganisms compared to environmental microorganisms [37,38]. Thus, emerging uropathogens not under routine resistance surveillance may pose a major health community threat by serving as reservoirs of resistance capable of spreading ARG-harboring ICEs to other species [39,40,41,42].
Contrary to the outdated belief of sterility, the urinary tract is made up of diverse polymicrobial communities [43,44]. The sharing of mobile genetic elements within this space poses a significant challenge to surveillance, highlighting the need to detect antibiotic resistance at a community level rather than cherry-picking select species [45]. Given the level of conservation of the ermA gene sequence between different ICEs and AUC isolates, our PCR test demonstrated a remarkable 95.8% concordance between the ermA genotype and resistance phenotype. As such, a potential future application may be to conduct whole-genome sequencing on bladder metagenomic DNA to evaluate urinary community resistance to macrolides. It would also be of interest to evaluate the movement of these ICEs between urinary bacteria, particularly when these mobile genetic elements move across species.
The implications of finding resistance in urinary bacteria towards an antibiotic class not primarily used in urinary infections provide valuable insight into the impact of off-target antimicrobial exposure. In the instance of macrolides, primarily used in respiratory infections, bladder bacteria are bystander organisms and are unintentionally exposed when the antibiotic is excreted from the body. Antibiotics are only partially broken down or absorbed in the body, with the rest excreted in urine or feces [46]. The concentration of many antibiotics, when excreted, is usually far lower than at the initial route of delivery, potentially priming off-target microorganisms towards resistance as a result of exposure to subinhibitory concentrations [47]. One study even found that subinhibitory antibiotic concentrations can promote horizontal gene transfer [48]. The emergence of off-target resistance is a major threat to patient health, especially when these off-target microorganisms serve as sources of future infections [49]. This speaks to the larger importance of proper antibiotic stewardship; however, a study of the US prescribing practices between 2017 and 2021 found that more than a quarter of prescribed antibiotics went towards conditions for which they are ineffective, providing unnecessary exposure to off-target microbiome bacteria [50].
This study underscores the critical role of mobile genetic elements carrying antibiotic-resistance genes and the emergence of antibiotic resistance. Emerging uropathogens like AUC remain greatly understudied, highlighting the urgency for comprehensive research. The clinical significance and resistance profile of these species are still unknown, emphasizing the need for in-depth exploration [17]. Our study has many features similar to how S. agalactiae has become recalcitrant to tetracycline therapy due to the dissemination of ICEs harboring resistance genes [51]. As such, a similar phenomenon may be occurring in other uropathogens, such as those belonging to the families Aerococcaceae, Actinomycetaceae, and Bifidobacteriaceae, for macrolide and other antibiotic resistances that have yet to be studied.
4. Materials and Methods
4.1. Survey of Publicly Available Genomes for ARGs
All publicly available AUC draft genomes were retrieved from NCBI (n = 158) and analyzed digitally at Loyola University Chicago with the CARD version 3.2.6 [52]. A complete list of genomes analyzed can be found in Supplementary Materials Table S2. Analysis of results was limited to perfect hits with default settings. Perfect hit refers to a perfect match to the curated reference sequences and mutations in the CARD database.
4.2. Study Isolates
The 189 AUC isolates included in this study had been previously recovered from male urology and female urogynecology patients at Loyola University Health Center between the years 2017 and 2023. These isolates were retrieved from a variety of sample sources, including voided urine, transurethral catheter urine, perineal swabs, vaginal swabs, periurethral swabs, kidney stones, and catheter tips as part of the Loyola University Chicago IRB-approved biorepository (LU 215192, 16 August 2021). Bacteria were isolated from urine and swab samples via the expanded quantitative urine culture (EQUC) method (44). A complete list of isolates can be found in Supplementary Materials Table S1.
Bacterial isolates were identified at Loyola University Chicago as AUC via matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) using the direct colony method. In this method, a sample of the bacterial colony is placed on a stainless steel target plate (Bruker Daltonics GmbH, Leipzig, Germany) before being treated with 70% formic acid. Upon drying, the α-cyano-4-hydrocinnamic acid matrix (Bruker Daltonics) is applied to the sample. The prepared sample plate is then analyzed by a MicroFlex LT mass spectrometer (Bruker Daltonics) with the MALDI Biotyper 3.0 software (Bruker Daltonics). E. coli DH5α was used as the quality control strain.
4.3. Phenotypic Screening of Macrolide Susceptibility
Macrolide susceptibility phenotypes of all 189 isolates were determined at Loyola University Chicago by broth microdilution technique in microtiter plates with 10-fold antibiotic dilutions. Overnight bacterial growth was inoculated in NYCIII media containing azithromycin (Sigma, Darmstadt, Germany) with a maximum antibiotic concentration of 100 μg/mL. The microtiter plates were then incubated within a BioTek Epoch 2 Microplate Spectrophotometer (Agilent Technologies, Wood Dale, IL, USA) in aerobic conditions with 5% supplemented CO2 at 37 °C with shaking at 200 rpm for 24 h. Optical density was measured every 30 min at 600 nm, and MIC was determined. For controls, PBS or bacteria was inoculated in NYCIII without antibiotics. All tests were conducted in duplicate.
4.4. DNA Extraction
To isolate genomic DNA from AUC isolates, 10 mL bacterial cultures were grown overnight in NYCIII media before being treated with lytic enzyme (lysozyme) at 37 °C for 60 min. Cells were lysed using the Wizard Genomic DNA Purification kit (Promega, Madison, WI, USA). The final DNA pellet was rehydrated in nuclease-free water overnight at 4 °C. Extracted bacterial DNA was stored at −20 °C until further processing. Genomic DNA was processed and stored at Loyola University Chicago.
4.5. PCR Survey
The presence of bacterial macrolide resistance gene ermA was detected by PCR and conducted at Loyola University Chicago. Forward primer: 5′-ACA TGA TAT TCC CTG TTT ACC CA-3′. Reverse primer: 5′-TGG AAA TGA GTC AAC GGG TG-3′. Each PCR reaction was carried out in a final volume of 50 μL consisting of molecular grade nuclease-free water (35 μL), 10× Taq buffer (5 μL) (Thermo, Waltham, MA, USA), 25 mM MgCl2 (3 μL), DNA template (3 μL), 10 mM dNTPs (1 μL), forward primer (1 μL), reverse primer (1 μL), and Taq polymerase (1 μL) (Thermo). The PCR reactions were performed in a SimpliAmp thermocycler (Applied Biosystems, Waltham, MA, USA). Thermocycling conditions were as follows: initial denaturation at 94 °C (10 min), 30 cycles of 94 °C (30 s) + 55 °C (1 min) + 72 °C (1 min), and final extension at 72 °C (7 min). PCR products were analyzed by separation on 1.0% agarose gels. A PCR reaction without template DNA was used as a negative control.
4.6. Genome Sequencing
Complete genome sequences of eight AUC isolates demonstrating resistance phenotypes were assembled by combining short-read and long-read sequences. DNA extraction and sequencing of isolates was performed by SeqCenter (Pittsburgh, PA, USA). For short reads, the Qiagen UltraClean Microbial Kit (Qiagen, Hilden, Germany) for extraction and the Nextera DNA Flex Library Prep kit (Illumina, San Diego, CA, USA) for library preparation were used before being run on the Illumina MiSeq or NovaSeq platform. For long reads, the Zymo DNA Miniprep kit (Zymo Research, Irvine, CA, USA) for extraction and Oxford Nanopore Technology Ligation Sequencing kit V14 (Oxford, UK) for library preparation were used before being run on the MiniION platform. Hybrid assembly was performed by combining filtered short- and long-sequence reads using SPAdes v3.15.4 [53], Flye v2.9 [54], and/or Canu v1.5 [55]. Genomes were polished with Pilon v1.24 [56], validated with QUAST v5.2.0 [57], and circularized using Circlator v1.5.5 [58]. Open reading frames were annotated via PGAP v6.6 [59]. Complete genomes were deposited and are publicly available at BioProject PRJNA316969 (Supplementary Materials Table S3).
4.7. Integrative and Conjugative Element Analysis
The identification and classification of ICEs were conducted by the same method used for the identification of Streptococcus species [60,61]. Briefly, genomes were queried via BLAST comparison against reference ICE proteins (e.g., integrases, transposases, Type IV secretion system proteins, and relaxases). Putative ICEs were delineated by syntenic comparison. The resistance gene ermA within ICEs was detected with VRprofile2 [62]. The homology of ICEs and ermA was visualized using EasyFig version 2.2.5 [63].
5. Conclusions
In this study, we characterized a concerning rate of macrolide resistance among AUC uropathogen isolates within a United States quaternary medical center. The identification of the ermA gene as a primary determinant of macrolide resistance highlights the role of horizontal gene transfer via mobile genetic elements, particularly ICEs, in disseminating resistance within bacterial populations. Our findings suggest that emerging uropathogens, such as AUC, may serve as reservoirs of resistance genes capable of receiving resistance determinants from other species within the urinary microbiome. This underscores the importance of understanding the dynamics of antibiotic resistance transmission within polymicrobial communities in the urinary tract.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/antibiotics13050433/s1. Table S1. List of Isolates. Table S2. List of Analyzed Genomes. Table S3. NCBI Deposit of Complete Genomes. Figure S1. VRprofile2 output of ICEAloyUMB0088. Figure S2. Nucleotide homology of AUC ICEAmicUMB3440 and Streptococcus agalactiae ICESag066.
Author Contributions
Conceptualization, J.L. and B.I.C.; Methodology, J.L. and B.I.C.; Software, M.F.N. and N.S.; Validation, J.L., B.I.C. and A.J.W.; Formal analysis, J.L., B.I.C., M.F.N. and N.S.; Investigation, J.L. and B.I.C.; Resources, A.J.W.; Data curation, M.F.N.; Writing—original draft, J.L.; Writing—review and editing, B.I.C. and A.J.W.; Visualization, J.L. and B.I.C.; Supervision, A.J.W.; Funding acquisition, A.J.W. All authors have read and agreed to the published version of the manuscript.
Funding
The authors gratefully acknowledge funding by Pathnostics.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All complete genomes in the study have been deposited in NCBI. Links to the genomes can be found in the Supplementary Materials Table S2.
Acknowledgments
We would like to thank David Hecht for providing valuable clinical insight and support.
Conflicts of Interest
A.J.W. declares membership on the advisory boards of Cerillo, Pathnostics, and Urobiome Therapeutics, and funding from Pathnostics. The other authors declare no conflicts of interest.
References
- Aguirre, M.; Collins, M.D. Phylogenetic Analysis of Some Aerococcus-like Organisms from Urinary Tract Infections: Description of Aerococcus urinae sp. nov. J. Gen. Microbiol. 1992, 138, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Senneby, E.; Petersson, A.C.; Rasmussen, M. Epidemiology and Antibiotic Susceptibility of Aerococci in Urinary Cultures. Diagn. Microbiol. Infect. Dis. 2015, 81, 149–151. [Google Scholar] [CrossRef] [PubMed]
- Narayanasamy, S.; King, K.; Dennison, A.; Spelman, D.W.; Aung, A.K. Clinical Characteristics and Laboratory Identification of Aerococcus Infections: An Australian Tertiary Centre Perspective. Int. J. Microbiol. 2017, 2017, 5684614. [Google Scholar] [CrossRef] [PubMed]
- Sihvonen, R.; Turunen, M.; Lehtola, L.; Pakarinen, L.; Grönroos, J.O.; Rantakokko-Jalava, K.; Pätäri-Sampo, A. Clinical and Microbiological Characterization of Aerococcus urinae Bacteraemias at Helsinki Metropolitan Area, Finland. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Maezawa, Y.; Nagasaki, K. Aerococcus urinae: An Emerging, Gram-Positive Pathogen Causing Urinary Tract Infection. Am. J. Med. 2024, 137, 89–90. [Google Scholar] [CrossRef] [PubMed]
- Skalidis, T.; Papaparaskevas, J.; Konstantinou, D.; Kapolou, E.; Falagas, M.E.; Legakis, N. Aerococcus urinae, a Cause of Cystitis with Malodorous Urine in a Child: Clinical and Microbiological Challenges. JMM Case Rep. 2017, 4, e005083. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M. Aerococcus: An Increasingly Acknowledged Human Pathogen. Clin. Microbiol. Infect. 2016, 22, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Pearce, M.M.; Hilt, E.E.; Rosenfeld, A.B.; Zilliox, M.J.; Thomas-White, K.; Fok, C.; Kliethermes, S.; Schreckenberger, P.C.; Brubaker, L.; Gai, X.; et al. The Female Urinary Microbiome: A Comparison of Women with and without Urgency Urinary Incontinence. mBio 2014, 5, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Sturm, P.D.; Van Eijk, J.; Veltman, S.; Meuleman, E.; Schülin, T. Urosepsis with Actinobaculum schaalii and Aerococcus urinae. J. Clin. Microbiol. 2006, 44, 652–654. [Google Scholar] [CrossRef]
- Zhang, Q.; Kwoh, C.; Attorri, S.; Clarridge, J.E. 3rd Aerococcus urinae in Urinary Tract Infections. J. Clin. Microbiol. 2000, 38, 1703–1705. [Google Scholar] [CrossRef]
- Ahmed, Y.; Bardia, N.; Judge, C.; Ahmad, S.; Malozzi, C.; Calderon, E. Aerococcus urinae: A Rare Cause of Endocarditis Presenting With Acute Stroke. J. Med. Cases 2021, 12, 65–70. [Google Scholar] [CrossRef]
- Forsvall, A.; Wagenius, M.; Rasmussen, M. Perigenital Necrotizing Soft Tissue Infection Caused by Aerococcus urinae. IDCases 2019, 18, 590. [Google Scholar] [CrossRef] [PubMed]
- Tai, D.B.G.; Go, J.R.; Fida, M.; Saleh, O.A. Management and Treatment of Aerococcus Bacteremia and Endocarditis. Int. J. Infect. Dis. 2021, 102, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Christensen, J.J.; Kilian, M.; Fussing, V.; Andresen, K.; Blom, J.; Korner, B.; Steigerwalt, A.G. Aerococcus urinae: Polyphasic Characterization of the Species. APMIS 2005, 113, 517–525. [Google Scholar] [CrossRef]
- Carkaci, D.; Højholt, K.; Nielsen, X.C.; Dargis, R.; Rasmussen, S.; Skovgaard, O.; Fuursted, K.; Andersen, P.S.; Stegger, M.; Christensen, J.J. Genomic Characterization, Phylogenetic Analysis, and Identification of Virulence Factors in Aerococcus sanguinicola and Aerococcus urinae Strains Isolated from Infection Episodes. Microb. Pathog. 2017, 112, 327–340. [Google Scholar] [CrossRef]
- Choi, B.I.; Ene, A.; Du, J.; Johnson, G.; Putonti, C.; Schouw, C.H.; Dargis, R.; Senneby, E.; Christensen, J.J.; Wolfe, A.J. Taxonomic Considerations on Aerococcus urinae with Proposal of Subdivision into Aerococcus urinae, Aerococcus tenax sp. nov., Aerococcus mictus Sp. Nov., and Aerococcus loyolae sp. nov. Int. J. Syst. Evol. Microbiol. 2023, 73, 006066. [Google Scholar] [CrossRef]
- Moreland, R.B.; Choi, B.; Geaman, W.; Álvarez, C.J.; Hochstedler-Kramer, B.R.; John, J.; Kaindl, J.; Kesav, N.; Lamichhane, J.; Lucio, L.; et al. Beyond the Usual Suspects: Emerging Uropathogens in the Microbiome Age. Front. Urol. 2023, 3, 1212590. [Google Scholar] [CrossRef]
- Ahmadzada, A.; Fuchs, F.; Hamprecht, A. Susceptibility of Aerococcus urinae and Aerococcus sanguinicola to Standard Antibiotics and to Nitroxoline. Microbiol. Spectr. 2023, 11, e0276322. [Google Scholar] [CrossRef]
- Krishnan, A.; Nadeau, L. 1468. Determination of Antibiotic Susceptibilities in Aerococcus urinae Urinary Isolates. Open Forum Infect. Dis. 2019, 6, S536. [Google Scholar] [CrossRef]
- Humphries, R.M.; Hindler, J.A. In Vitro Antimicrobial Susceptibility of Aerococcus urinae. J. Clin. Microbiol. 2014, 52, 2177–2180. [Google Scholar] [CrossRef]
- Humphries, R.M.; Lee, C.; Hindler, J.A. Aerococcus urinae and Trimethoprim-Sulfamethoxazole. J. Clin. Microbiol. 2011, 49, 3934–3935. [Google Scholar] [CrossRef]
- Saad, A.; Hailes, J.; Jacobs, M.R.; Navas, M.E. Antimicrobial Susceptibility Profile of Aerococcus urinae: Recommendations for Empirical Therapy. Infect. Dis. Clin. Pract. 2023, 31, 1176. [Google Scholar] [CrossRef]
- CDC. COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022; CDC: Atlanta, GA, USA, 2022. [CrossRef]
- Abdel Gawad, A.M.; Ashry, W.M.O.; El-Ghannam, S.; Hussein, M.; Yousef, A. Antibiotic Resistance Profile of Common Uropathogens during COVID-19 Pandemic: Hospital Based Epidemiologic Study. BMC Microbiol. 2023, 23, 28. [Google Scholar] [CrossRef]
- Mareș, C.; Petca, R.-C.; Petca, A.; Popescu, R.-I.; Jinga, V. Does the COVID Pandemic Modify the Antibiotic Resistance of Uropathogens in Female Patients? A New Storm? Antibiotics 2022, 11, 376. [Google Scholar] [CrossRef]
- Thänert, R.; Reske, K.A.; Hink, T.; Wallace, M.A.; Wang, B.; Schwartz, D.J.; Seiler, S.; Cass, C.; Burnham, C.-A.D.; Dubberke, E.R.; et al. Comparative Genomics of Antibiotic-Resistant Uropathogens Implicates Three Routes for Recurrence of Urinary Tract Infections. mBio 2019, 10, e01977-19. [Google Scholar] [CrossRef]
- Harris, M.; Fasolino, T.; Ivankovic, D.; Davis, N.J.; Brownlee, N. Genetic Factors That Contribute to Antibiotic Resistance through Intrinsic and Acquired Bacterial Genes in Urinary Tract Infections. Microorganisms 2023, 11, 1407. [Google Scholar] [CrossRef]
- Khan, U.B.; Portal, E.A.R.; Sands, K.; Lo, S.; Chalker, V.J.; Elita, J.; Spiller, O.B. Genomic Analysis Reveals New Integrative Conjugal Elements and Transposons in GBS Conferring Antimicrobial Resistance. Antibiotics 2023, 12, 544. [Google Scholar] [CrossRef]
- Choi, B.I.; Fontes Noronha, M.; Kaindl, J.; Wolfe, A.J. Complete Genome Sequences of Aerococcus loyolae ATCC TSD-300T, Aerococcus mictus ATCC TSD-301T, and Aerococcus tenax ATCC TSD-302T. Microbiol. Resour. Announc. 2024. Advance online publication. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, M45, 3rd ed.; Clinical and Laboratory Standards Institute: Berwyn, PA, USA, 2016; Available online: https://clsi.org/standards/products/microbiology/documents/m45/ (accessed on 19 December 2023).
- Johnson, C.M.; Grossman, A.D. Integrative and Conjugative Elements (ICEs): What They Do and How They Work. Annu. Rev. Genet. 2015, 49, 577–601. [Google Scholar] [CrossRef]
- Li, X.; Fan, H.; Zi, H.; Hu, H.; Li, B.; Huang, J.; Luo, P.; Zeng, X. Global and Regional Burden of Bacterial Antimicrobial Resistance in Urinary Tract Infections in 2019. J. Clin. Med. 2022, 11, 2817. [Google Scholar] [CrossRef] [PubMed]
- Wildfeuer, A.; Laufen, H.; Leitold, M.; Zimmermann, T. Comparison of the pharmacokinetics of three-day and five-day regimens of azithromycin in plasma and urine. J. Antimicrob. Chemother. 1993, 31, 51–56. [Google Scholar] [CrossRef]
- Kim, M.; Welch, T. Update on Azithromycin and Cardiac Side Effects. Southwest Respir. Crit. Care Chron. 2014, 2, 48–51. [Google Scholar] [CrossRef]
- Gallacher, D.J.; Zhang, L.; Aboklaish, A.F.; Mitchell, E.; Wach, R.; Marchesi, J.R.; Kotecha, S. Baseline Azithromycin Resistance in the Gut Microbiota of Preterm Born Infants. Pediatr. Res. 2024, 95, 205–212. [Google Scholar] [CrossRef]
- Lipszyc, A.; Szuplewska, M.; Bartosik, D. How Do Transposable Elements Activate Expression of Transcriptionally Silent Antibiotic Resistance Genes? Int. J. Mol. Sci. 2022, 23, 8063. [Google Scholar] [CrossRef]
- Jeong, H.; Arif, B.; Caetano-Anollés, G.; Kim, K.M.; Nasir, A. Horizontal Gene Transfer in Human-associated Microorganisms Inferred by Phylogenetic Reconstruction and Reconciliation. Sci. Rep. 2019, 9, 5953. [Google Scholar] [CrossRef]
- Smillie, C.S.; Smith, M.B.; Friedman, J.; Cordero, O.X.; David, L.A.; Alm, E.J. Ecology Drives a Global Network of Gene Exchange Connecting the Human Microbiome. Nature 2011, 480, 241–244. [Google Scholar] [CrossRef]
- Davies, M.G.; Shera, J.; Van Domselaar, G.H.; Sriprakash, K.S.; McMillan, D.J. A Novel Integrative Conjugative Element Mediates Genetic Transfer from Group G Streptococcus to Other β-Hemolytic Streptococci. J. Bacteriol. 2009, 191, 2257–2265. [Google Scholar] [CrossRef]
- Brenciani, A.; Tiberi, E.; Bacciaglia, A.; Petrelli, D.; Varaldo, P.E.; Giovanetti, E. Two Distinct Genetic Elements Are Responsible Forerm(TR)-Mediated Erythromycin Resistance in Tetracycline-Susceptible and Tetracycline-Resistant Strains of Streptococcus Pyogenes. Antimicrob. Agents Chemother. 2011, 55, 2106–2112. [Google Scholar] [CrossRef]
- Wang, H.; Zhuang, H.; Ji, S.; Sun, L.; Zhao, F.; Wu, D.; Shen, P.; Jiang, Y.; Yu, Y.; Chen, Y. Distribution of Erm Genes among MRSA Isolates with Resistance to Clindamycin in a Chinese Teaching Hospital. Infect. Genet. Evol. 2021, 96, 105127. [Google Scholar] [CrossRef]
- Giovanetti, E. Conjugative Transfer of the Erm(A) Gene from Erythromycin-Resistant Streptococcus Pyogenes to Macrolide-Susceptible, S. Pyogenes, Enterococcus Faecalis and Listeria Innocua. J. Antimicrob. Chemother. 2002, 50, 249–252. [Google Scholar] [CrossRef][Green Version]
- Wolfe, A.J.; Toh, E.; Shibata, N.; Rong, R.; Kenton, K.; FitzGerald, M.; Mueller, E.R.; Schreckenberger, P.; Dong, Q.; Nelson, D.E.; et al. Evidence of Uncultivated Bacteria in the Adult Female Bladder. J. Clin. Microbiol. 2012, 50, 1376–1383. [Google Scholar] [CrossRef]
- Hilt, E.E.; McKinley, K.; Pearce, M.M.; Rosenfeld, A.B.; Zilliox, M.J.; Mueller, E.R.; Brubaker, L.; Gai, X.; Wolfe, A.J.; Schreckenberger, P.C. Urine Is Not Sterile: Use of Enhanced Urine Culture Techniques to Detect Resident Bacterial Flora in the Adult Female Bladder. J. Clin. Microbiol. 2013, 52, 871–876. [Google Scholar] [CrossRef]
- Vollstedt, A.; Baunoch, D.; Wolfe, A.; Luke, N.; Wojno, K.J.; Cline, K.; Belkoff, L.; Milbank, A.; Sherman, N.; Haverkorn, R.; et al. Bacterial Interactions as Detected by Pooled Antibiotic Susceptibility Testing (P-AST) in Polymicrobial Urine Specimens. J. Surg. Urol. 2020, 1, 101. [Google Scholar]
- Zhou, X.; Cuasquer, G.J.P.; Li, Z.; Mang, H.P.; Lv, Y. Occurrence of Typical Antibiotics, Representative Antibiotic-resistant Bacteria, and Genes in Fresh and Stored Source-separated Human Urine. Environ. Int. 2021, 146, 106280. [Google Scholar] [CrossRef]
- Wistrand-Yuen, E.; Knopp, M.; Hjort, K.; Koskiniemi, S.; Berg, O.G.; Andersson, D.I. Evolution of High-level Resistance during Low-level Antibiotic Exposure. Nat. Commun. 2018, 9, 1599. [Google Scholar] [CrossRef]
- Ding, M.; Ye, Z.; Liu, L.; Wang, W.; Chen, Q.; Zhang, F.; Wang, Y.; Sjöling, Å.; Martín-Rodríguez, A.J.; Hu, R.; et al. Subinhibitory Antibiotic Concentrations Promote the Horizontal Transfer of Plasmid-borne Resistance Genes from Klebsiellae pneumoniae to Escherichia coli. Front. Microbiol. 2022, 13, 1017092. [Google Scholar] [CrossRef]
- Morley, V.J.; Woods, R.J.; Read, A.F. Bystander Selection for Antimicrobial Resistance: Implications for Patient Health. Trends Microbiol. 2019, 27, 864–877. [Google Scholar] [CrossRef]
- Chua, K.P.; Fischer, M.A.; Rahman, M.; Linder, J.A. Changes in the Appropriateness of US Outpatient Antibiotic Prescribing After the Coronavirus Disease 2019 Outbreak: An Interrupted Time Series Analysis of 2016-2021 Data. Clin. Infect. Dis. 2024, ciae135. [Google Scholar] [CrossRef]
- Da Cunha, V.; Davies, M.R.; Douarre, P.-E.; Rosinski-Chupin, I.; Margarit, I.; Spinali, S.; Perkins, T.; Lechat, P.; Dmytruk, N.; Sauvage, E.; et al. Streptococcus agalactiae Clones Infecting Humans Were Selected and Fixed through the Extensive Use of Tetracycline. Nat. Commun. 2014, 5, 4544. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Antipov, D.; Korobeynikov, A.; McLean, J.S.; Pevzner, P.A. HybridSPAdes: An Algorithm for Hybrid Assembly of Short and Long Reads. Bioinformatics 2015, 32, 1009–1015. [Google Scholar] [CrossRef]
- Lin, Y.; Yuan, J.; Kolmogorov, M.; Shen, M.W.; Chaisson, M.; Pevzner, P.A. Assembly of Long Error-Prone Reads Using de Bruijn Graphs. Proc. Natl. Acad. Sci. USA 2016, 113, E8396–E8405. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and Accurate Long-Read Assembly via Adaptivek-Mer Weighting and Repeat Separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Hunt, M.; Silva, N.D.; Otto, T.D.; Parkhill, J.; Keane, J.A.; Harris, S.R. Circlator: Automated Circularization of Genome Assemblies Using Long Sequencing Reads. Genome Biol. 2015, 16, 294. [Google Scholar] [CrossRef]
- Tatusova, T.; Di Cuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Lao, J.; Guédon, G.; Lacroix, T.; Charron-Bourgoin, F.; Libante, V.; Loux, V.; Chiapello, H.; Payot, S.; Leblond-Bourget, N. Abundance, Diversity and Role of ICEs and IMEs in the Adaptation of Streptococcus salivarius to the Environment. Genes 2020, 11, 999. [Google Scholar] [CrossRef]
- Ambroset, C.; Coluzzi, C.; Guédon, G.; Devignes, M.; Loux, V.; Lacroix, T.; Payot, S.; Leblond-Bourget, N. New Insights into the Classification and Integration Specificity of Streptococcus Integrative Conjugative Elements through Extensive Genome Exploration. Front. Microbiol. 2016, 6, 1483. [Google Scholar] [CrossRef]
- Wang, M.; Goh, Y.-X.; Tai, C.; Wang, H.; Deng, Z.; Ou, H.-Y. VRprofile2: Detection of Antibiotic Resistance-Associated Mobilome in Bacterial Pathogens. Nucleic Acids Res. 2022, 50, W768–W773. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A Genome Comparison Visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).