Impact of Soil Fertilization with Pig Slurry on Antibiotic Residues and Resistance Genes: A Longitudinal Study
Abstract
:1. Introduction
2. Results
2.1. Soil Analysis
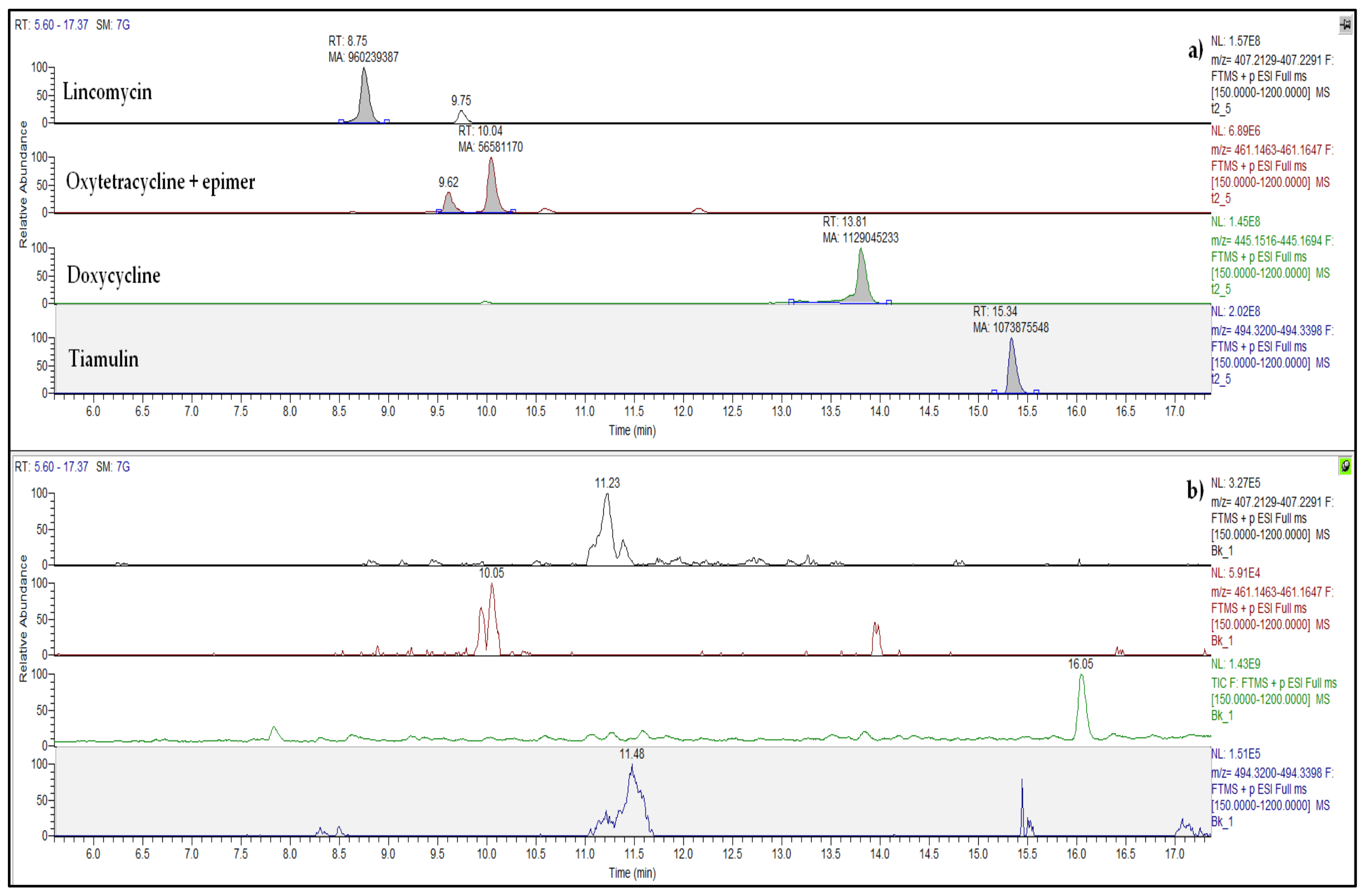
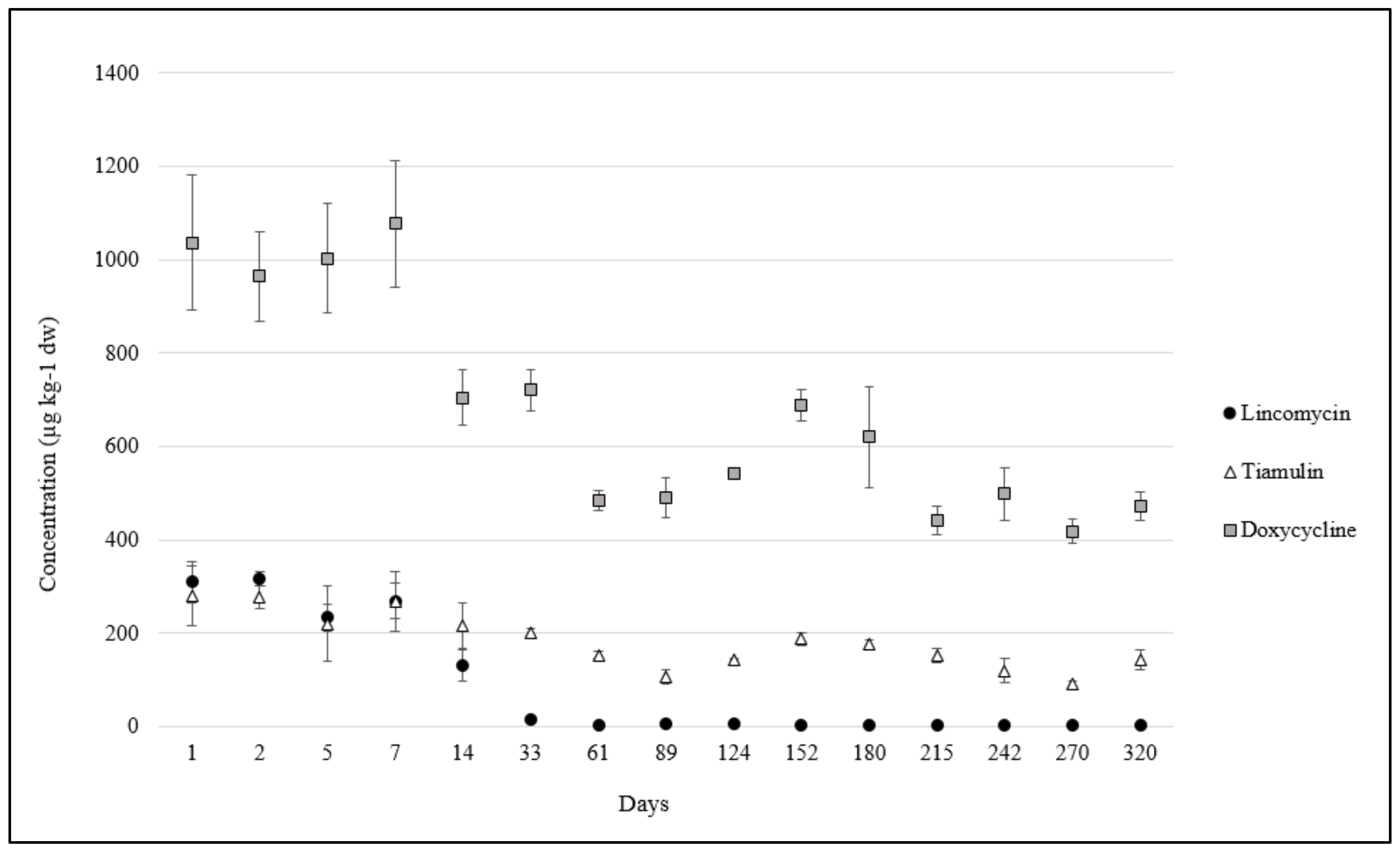
2.2. Antibiotics in Pig Manure and Soil
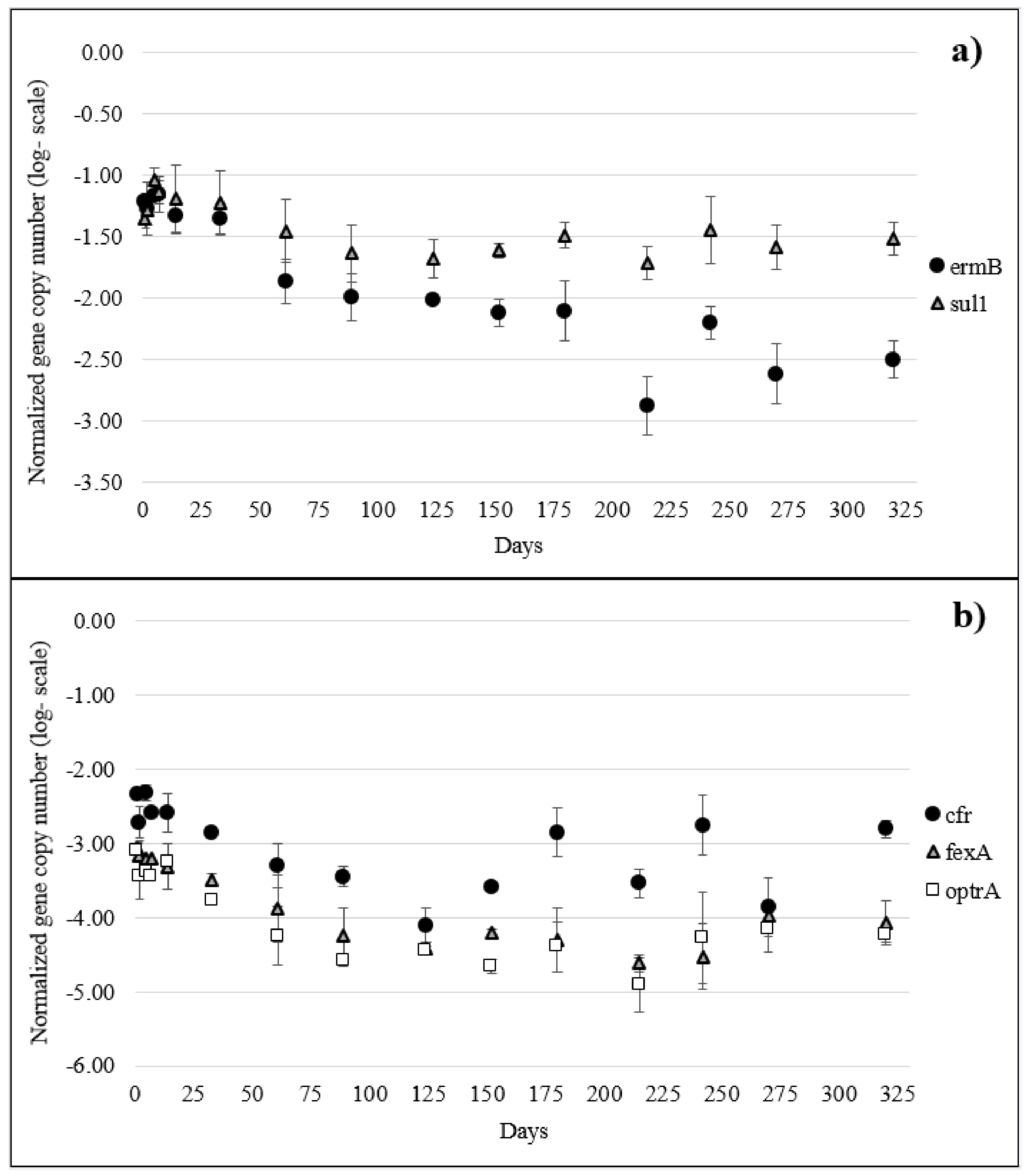
2.3. ARGs in Manure and Soil
2.4. Spearman Correlations and Principal Component Analysis (PCA) Analysis in Soil
3. Discussion
4. Materials and Methods
4.1. Experimental Design and Sampling
4.2. Soil Analyses
4.3. Antibiotic Analysis by LC-HRMS/MS
4.4. Determination of Antibiotics in Pig Slurry and Soil
4.5. DNA Extraction and Quantification of ARGs Using qPCR
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cycoń, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the Soil Environment—Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 2019, 10, 412419. [Google Scholar] [CrossRef] [PubMed]
- Zalewska, M.; Błażejewska, A.; Czapko, A.; Popowska, M. Antibiotics and Antibiotic Resistance Genes in Animal Manure—Consequences of Its Application in Agriculture. Front. Microbiol. 2021, 12, 610656. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, B.J.A.; Wegh, R.S.; Memelink, J.; Zuidema, T.; Stolker, L.A.M. The Analysis of Animal Faeces as a Tool to Monitor Antibiotic Usage. Talanta 2015, 132, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Jechalke, S.; Kopmann, C.; Rosendahl, I.; Groeneweg, J.; Weichelt, V.; Krögerrecklenfort, E.; Brandes, N.; Nordwig, M.; Ding, G.C.; Siemens, J.; et al. Increased Abundance and Transferability of Resistance Genes after Field Application of Manure from Sulfadiazine-Treated Pigs. Appl. Environ. Microbiol. 2013, 79, 1704–1711. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; He, L.Y.; Liu, Y.S.; Zhao, J.L.; Liu, W.R.; Zhang, J.N.; Chen, J.; He, L.K.; Zhang, Q.Q.; Ying, G.G. Fate of Veterinary Antibiotics during Animal Manure Composting. Sci. Total Environ. 2019, 650, 1363–1370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cheng, D.; Xie, J.; Zhang, Y.; Wan, Y.; Zhang, Y.; Shi, X. Impacts of Farmland Application of Antibiotic-Contaminated Manures on the Occurrence of Antibiotic Residues and Antibiotic Resistance Genes in Soil: A Meta-Analysis Study. Chemosphere 2022, 300, 134529. [Google Scholar] [CrossRef] [PubMed]
- Wohde, M.; Berkner, S.; Junker, T.; Konradi, S.; Schwarz, L.; Düring, R.A. Occurrence and Transformation of Veterinary Pharmaceuticals and Biocides in Manure: A Literature Review. Environ. Sci. Eur. 2016, 28, 23. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Ma, L.; Yu, Q.; Yang, J.; Su, W.; Hilal, M.G.; Li, X.; Zhang, S.; Li, H. The Source, Fate and Prospect of Antibiotic Resistance Genes in Soil: A Review. Front. Microbiol. 2022, 13, 976657. [Google Scholar] [CrossRef] [PubMed]
- Van den Meersche, T.; Rasschaert, G.; Vanden Nest, T.; Haesebrouck, F.; Herman, L.; Van Coillie, E.; Van Weyenberg, S.; Daeseleire, E.; Heyndrickx, M. Longitudinal Screening of Antibiotic Residues, Antibiotic Resistance Genes and Zoonotic Bacteria in Soils Fertilized with Pig Manure. Environ. Sci. Pollut. Res. 2020, 27, 28016–28029. [Google Scholar] [CrossRef]
- Farouk, F.; Azzazy, H.M.E.; Niessen, W.M.A. Challenges in the Determination of Aminoglycoside Antibiotics, a Review. Anal. Chim. Acta 2015, 890, 21–43. [Google Scholar] [CrossRef]
- Berendsen, B.J.A.; Lahr, J.; Nibbeling, C.; Jansen, L.J.M.; Bongers, I.E.A.; Wipfler, E.L.; van de Schans, M.G.M. The Persistence of a Broad Range of Antibiotics during Calve, Pig and Broiler Manure Storage. Chemosphere 2018, 204, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, A.M.; Halling-Sørensen, B.; Ingerslev, F.; Hansen, S.H. Simultaneous Extraction of Tetracycline, Macrolide and Sulfonamide Antibiotics from Agricultural Soils Using Pressurised Liquid Extraction, Followed by Solid-Phase Extraction and Liquid Chromatography-Tandem Mass Spectrometry. J. Chromatogr. A 2004, 1038, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Jansen, L.J.M.; van de Schans, M.G.M.; de Boer, D.; Bongers, I.E.A.; Schmitt, H.; Hoeksma, P.; Berendsen, B.J.A. A New Extraction Procedure to Abate the Burden of Non-Extractable Antibiotic Residues in Manure. Chemosphere 2019, 224, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Mejías, C.; Santos, J.L.; Martín, J.; Aparicio, I.; Alonso, E. Multiresidue Method for the Determination of Critically and Highly Important Classes of Antibiotics and Their Metabolites in Agricultural Soils and Sewage Sludge. Anal. Bioanal. Chem. 2023, 415, 7161–7173. [Google Scholar] [CrossRef] [PubMed]
- Federici, E.; Massaccesi, L.; Pezzolla, D.; Fidati, L.; Montalbani, E.; Proietti, P.; Nasini, L.; Regni, L.; Scargetta, S.; Gigliotti, G. Short-Term Modifications of Soil Microbial Community Structure and Soluble Organic Matter Chemical Composition Following Amendment with Different Solid Olive Mill Waste and Their Derived Composts. Appl. Soil Ecol. 2017, 119, 234–241. [Google Scholar] [CrossRef]
- Chen, J.; Xie, S. Overview of Sulfonamide Biodegradation and the Relevant Pathways and Microorganisms. Sci. Total Environ. 2018, 640, 1465–1477. [Google Scholar] [CrossRef] [PubMed]
- Barola, C.; Brambilla, G.; Galarini, R.; Moretti, S.; Morabito, S. Assessment of the Combined Inputs of Antimicrobials from Top Soil Improvers and Irrigation Waters on Green Leafy Vegetable Fields. Food Addit. Contam. Part A 2024, 41, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Nightingale, J.; Carter, L.; Sinclair, C.J.; Rooney, P.; Dickinson, M.; Tarbin, J.; Kay, P. Assessing the Influence of Pig Slurry PH on the Degradation of Selected Antibiotic Compounds. Chemosphere 2022, 290, 133191. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, X.; Fu, Y.; Chen, Y.; Wang, Y.; Ye, D.; Wang, C.; Hu, X.; Zhou, L.; Du, J.; et al. Association of Florfenicol Residues with the Abundance of Oxazolidinone Resistance Genes in Livestock Manures. J. Hazard. Mater. 2020, 399, 123059. [Google Scholar] [CrossRef]
- Fioriti, S.; Coccitto, S.N.; Cedraro, N.; Simoni, S.; Morroni, G.; Brenciani, A.; Mangiaterra, G.; Vignaroli, C.; Vezzulli, L.; Biavasco, F.; et al. Linezolid Resistance Genes in Enterococci Isolated from Sediment and Zooplankton in Two Italian Coastal Areas. Appl. Environ. Microbiol. 2021, 87, e02958-20. [Google Scholar] [CrossRef]
- EMA. ESVAC—Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2019 and 2020. Trends from 2010 to 2020. Eleventh ESVAC Report; (EMA/58183/2021); EMA: Amsterdam, The Netherlands, 2021; ISBN 9789291550685.
- European Food Safety Authority. The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2017/2018. EFSA J. 2020, 18, e06007. [Google Scholar] [CrossRef]
- Österberg, J.; Wingstrand, A.; Nygaard Jensen, A.; Kerouanton, A.; Cibin, V.; Barco, L.; Denis, M.; Aabo, S.; Bengtsson, B. Antibiotic Resistance in Escherichia coli from Pigs in Organic and Conventional Farming in Four European Countries. PLoS ONE 2016, 11, e0157049. [Google Scholar] [CrossRef]
- Nunes, O.C.; Manaia, C.M.; Kolvenbach, B.A.; Corvini, P.F.X. Living with Sulfonamides: A Diverse Range of Mechanisms Observed in Bacteria. Appl. Microbiol. Biotechnol. 2020, 104, 10389–10408. [Google Scholar] [CrossRef] [PubMed]
- Spielmeyer, A. Occurrence and Fate of Antibiotics in Manure during Manure Treatments: A Short Review. Sustain. Chem. Pharm. 2018, 9, 76–86. [Google Scholar] [CrossRef]
- Huygens, J.; Rasschaert, G.; Heyndrickx, M.; Dewulf, J.; Van Coillie, E.; Quataert, P.; Daeseleire, E.; Becue, I. Impact of Fertilization with Pig or Calf Slurry on Antibiotic Residues and Resistance Genes in the Soil. Sci. Total Environ. 2022, 822, 153518. [Google Scholar] [CrossRef] [PubMed]
- EFSA/ECDC. The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2021–2022. EFSA J. 2024, 22, e8583. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Mao, D.; Gao, H.; Zheng, L.; Chen, Z.; Gao, Y.; Duan, Y.; Guo, J.; Luo, Y.; Ren, H. Colonization of Gut Microbiota by Plasmid-Carrying Bacteria Is Facilitated by Evolutionary Adaptation to Antibiotic Treatment. ISME J. 2022, 16, 1284–1293. [Google Scholar] [CrossRef]
- Roy, K.; Islam, M.S.; Paul, A.; Ievy, S.; Talukder, M.; Sobur, M.A.; Ballah, F.M.; Khan, M.S.R.; Rahman, M.T. Molecular Detection and Antibiotyping of Multi-Drug Resistant Enterococcus faecium from Healthy Broiler Chickens in Bangladesh. Vet. Med. Sci. 2022, 8, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.-G.; Li, B.-Y.; Luan, R.-D.; Wang, H.-N.; Lei, C.-W. Whole Genome Sequence of Enterococcus gallinarum EG81, a Porcine Strain Harbouring the Oxazolidinone-Phenicol Resistance Gene OptrA with Chromosomal and Plasmid Location. J. Glob. Antimicrob. Resist. 2020, 22, 598–600. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Yan, J.; Huang, F.; Yang, L. On the Mechanism of Antibiotic Resistance and Fecal Microbiota Transplantation. Math. Biosci. Eng. 2019, 16, 7057–7084. [Google Scholar] [CrossRef]
- Yang, L.-Y.; Zhou, S.-Y.-D.; Lin, C.-S.; Huang, X.-R.; Neilson, R.; Yang, X.-R. Effects of Biofertilizer on Soil Microbial Diversity and Antibiotic Resistance Genes. Sci. Total Environ. 2022, 820, 153170. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Xu, L.-L.; Zhang, X.; Gong, X.-W.; Zhu, D.-L.; Xu, X.-H.; Wang, F.; Yang, X.-L. Three New Phenanthrenes with Antimicrobial Activities from the Aerial Parts of Juncus effusus. Fitoterapia 2018, 130, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, B.J.A.; Roelofs, G.; van Zanten, B.; Driessen-van Lankveld, W.D.M.; Pikkemaat, M.G.; Bongers, I.E.A.; de Lange, E. A Strategy to Determine the Fate of Active Chemical Compounds in Soil; Applied to Antimicrobially Active Substances. Chemosphere 2021, 279, 130495. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Wang, Y.; Wang, S.; Wang, Z.; Du, X.-D.; Jiang, H.; Xia, X.; Shen, Z.; Ding, S.; Wu, C.; et al. Prevalence and Abundance of Florfenicol and Linezolid Resistance Genes in Soils Adjacent to Swine Feedlots. Sci. Rep. 2016, 6, 32192. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Vismans, G.; Yu, K.; Song, Y.; de Jonge, R.; Burgman, W.P.; Burmølle, M.; Herschend, J.; Bakker, P.A.H.M.; Pieterse, C.M.J. Disease-Induced Assemblage of a Plant-Beneficial Bacterial Consortium. ISME J. 2018, 12, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. An Extraction Method for Measuring Soil Microbial Biomass C. Soil Biol. Biochem. 1987, 19, 703–707. [Google Scholar] [CrossRef]
- Agnelli, A.; Corti, G.; Massaccesi, L.; Ventura, S.; D’Acqui, L.P. Impact of Biological Crusts on Soil Formation in Polar Ecosystems. Geoderma 2021, 401, 115340. [Google Scholar] [CrossRef]
- Moretti, S.; Cruciani, G.; Romanelli, S.; Rossi, R.; Saluti, G.; Galarini, R. Multiclass Method for the Determination of 62 Antibiotics in Milk. J. Mass Spectrom. 2016, 51, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Sargenti, M.; Bartolacci, S.; Luciani, A.; Di Biagio, K.D.; Baldini, M.; Galarini, R.; Giusepponi, D.; Capuccella, M. Investigation of the Correlation between the Use of Antibiotics in Aquaculture Systems and Their Detection in Aquatic Environments: A Case Study of the Nera River Aquafarms in Italy. Sustainability 2020, 12, 5176. [Google Scholar] [CrossRef]
- Van den Meersche, T.; Rasschaert, G.; Haesebrouck, F.; Van Coillie, E.; Herman, L.; Van Weyenberg, S.; Daeseleire, E.; Heyndrickx, M. Presence and Fate of Antibiotic Residues, Antibiotic Resistance Genes and Zoonotic Bacteria during Biological Swine Manure Treatment. Ecotoxicol. Environ. Saf. 2019, 175, 29–38. [Google Scholar] [CrossRef]
- Guo, M.-T.; Yuan, Q.-B.; Yang, J. Ultraviolet reduction of erythromycin and tetracycline resistant heterotrophic bacteria and their resistance genes in municipal wastewater. Chemosphere 2013, 93, 2864–2868. [Google Scholar] [CrossRef] [PubMed]
- Pei, R.; Kim, S.-C.; Carlson, K.H.; Pruden, A. Effect of River Landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 2006, 40, 2427–2435. [Google Scholar] [CrossRef]
- He, L.-Y.; Liu, Y.-S.; Su, H.-C.; Zhao, J.-L.; Liu, S.-S.; Chen, J.; Liu, W.-R.; Ying, G.-G. Dissemination of Antibiotic Resistance Genes in Representative Broiler Feedlots Environments: Identification of Indicator ARGs and Correlations with Environmental Variables. Environ. Sci. Technol. 2014, 48, 13120–13129. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shao, B.; Shen, J.; Wang, S.; Wu, Y. Occurrence of Chloramphenicol-Resistance Genes as Environmental Pollutants from Swine Feedlots. Environ. Sci. Technol. 2013, 47, 2892–2897. [Google Scholar] [CrossRef] [PubMed]
- Pholwat, S.; Liu, J.; Taniuchi, M.; Chinli, R.; Pongpan, T.; Thaipisutikul, I.; Ratanakorn, P.; Platts-Mills, J.A.; Fleece, M.; Stroup, S.; et al. Genotypic antimicrobial resistance assays for use on E. coli isolates and stool specimens. PLoS ONE 2019, 14, e0216747. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.-K.; Martin, I.; Alfa, M.; Mulvey, M. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 2001, 15, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Zhi, S.; Zhou, J.; Liu, H.; Wu, H.; Zhang, Z.; Ding, Y.; Zhang, K. Simultaneous extraction and determination of 45 veterinary antibiotics in swine manure by liquid chromatography-tandem mass spectrometry. J. Chromatogr. B 2020, 1154, 122286. [Google Scholar] [CrossRef]
- Argüeso-Mata, M.; Bolado, S.; Jiménez, J.J.; López-Serna, R. Determination of antibiotics and other veterinary drugs in the solid phase of pig manure. Chemosphere 2021, 275, 130039. [Google Scholar] [CrossRef]
- Comprehensive Antibiotic Resistance Database. Available online: https://card.mcmaster.ca (accessed on 13 March 2024).
- World Health Organization. Critically Important Antimicrobials for Human Medicine. 6th Revision. 2018. Available online: https://www.who.int/publications/i/item/9789241515528 (accessed on 13 March 2024).

| Days from Manure Conditioning | Time Points | Treated Soil | |||||||
|---|---|---|---|---|---|---|---|---|---|
| pH | TOC (%) | TN (%) | WSOC (mg kg−1) | TSN (mg kg−1) | Cmic (mg kg−1) | Nmic (mg kg−1) | ΣC-CO2 (µg kg−1) | ||
| 1 | 1 | 6.87 ± 0.12 | 1.58 ± 0.09 | 0.28 ± 0.02 | 17.8 ± 3.8 | 18.0 ± 2.9 | 147 ± 8 | 14 ± 5 | 301 ± 16 |
| 2 | 2 | 6.91 ± 0.11 | 1.58 ± 0.11 | 0.26 ± 0.04 | 18.4 ± 3.1 | 19.0 ± 1.2 | 133 ± 12 | 14 ± 3 | 311 ± 24 |
| 5 | 3 | 7.02 ± 0.08 | 1.53 ± 0.09 | 0.27 ± 0.03 | 16.6 ± 0.7 | 17.8 ± 1.5 | 117 ± 11 | 14 ± 2 | 370 ± 20 |
| 7 | 4 | 7.21 ± 0.08 | 1.49 ± 0.14 | 0.27 ± 0.04 | 17.3 ± 2.3 | 18.4 ± 0.9 | 114 ± 15 | 13 ± 2 | 303 ± 45 |
| 14 | 5 | 7.29 ± 0.04 | 1.50 ± 0.16 | 0.24 ± 0.01 | 16.1 ± 3.2 | 19.1 ± 1.9 | 105 ± 4 | 12 ± 3 | 352 ± 31 |
| 33 | 6 | 7.32 ± 0.08 | 1.50 ± 0.11 | 0.25 ± 0.01 | 15.8 ± 4.2 | 19.9 ± 1.1 | 103 ± 5 | 12 ± 4 | 446 ± 101 |
| 61 | 7 | 7.58 ± 0.04 | 1.53 ± 0.13 | 0.26 ± 0.01 | 16.4 ± 2.2 | 14.5 ± 3.0 | 133 ± 23 | 8.3 ± 1.0 | 433 ± 69 |
| 89 | 8 | 7.37 ± 0.03 | 1.47 ± 0.18 | 0.25 ± 0.01 | 15.4 ± 1.1 | 18.9 ± 0.4 | 125 ± 29 | 5.2 ± 2.0 | 377 ± 84 |
| 124 | 9 | 7.38 ± 0.05 | 1.43 ± 0.09 | 0.25 ± 0.01 | 17.2 ± 0.9 | 18.8 ± 0.6 | 60 ± 6 | 2.1 ± 0.7 | NA b |
| 152 | 10 | 7.54 ± 0.05 | 1.44 ± 0.11 | 0.25 ± 0.02 | 18.9 ± 1.6 | 22.0 ± 0.4 | 73 ± 9 | 3.7 ± 1.2 | 467 ± 52 |
| 180 | 11 | 7.31 ± 0.11 | 1.50 ± 0.10 | 0.28 ± 0.01 | 17.1 ± 0.3 | 20.6 ± 0.8 | 77 ± 4 | 3.1 ± 0.9 | 448 ± 48 |
| 215 | 12 | 7.42 ± 0.02 | 1.48 ± 0.09 | 0.29 ± 0.04 | 16.2 ± 1.8 | 22.6 ± 4.1 | 62 ± 8 | 3.0 ± 0.6 | 597 ± 155 |
| 242 | 13 | 7.44 ± 0.07 | 1.52 ± 0.11 | 0.30 ± 0.03 | 20.5 ± 0.5 | 22.6 ± 0.9 | 58 ± 2 | 2.9 ± 1.0 | 524 ± 26 |
| 270 | 14 | 7.42 ± 0.03 | 1.48 ± 0.16 | 0.28 ± 0.01 | 15.9 ± 2.8 | 18.2 ± 2.3 | 64 ± 7 | 1.9 ± 0.9 | 655 ± 138 |
| 320 | 15 | 7.48 ± 0.01 | 1.52 ± 0.10 | 0.26 ± 0.02 | 17.4 ± 1.8 | 21.3 ± 0.1 | 45 ± 1 | 1.8 ± 0.4 | 621 ± 153 |
| Time Points | Control soil c | ||||||||
| pH | TOC (%) | TN (%) | WSOC (mg kg−1) | TSN (mg kg−1) | Cmic (mg kg−1) | Nmic (mg kg−1) | ΣC-CO2 (µg kg−1) | ||
| - | 1 | 7.90 ± 0.04 | 1.02 ± 0.02 | 0.19 ± 0.01 | 14.7 ± 1.8 | 5.37 ± 1.59 | 92 ± 5.3 | 1.2 ± 0.2 | 550 ± 27 |
| - | 6 | 8.02 ± 0.03 | 1.05 ± 0.06 | 0.19 ± 0.01 | 13.9 ± 1.7 | 3.94 ± 2.59 | 76 ± 5.2 | 2.5 ± 0.4 | 557 ± 35 |
| - | 12 | 7.91 ± 0.04 | 1.04 ± 0.04 | 0.19 ± 0.01 | 12.1 ± 1.0 | 3.95 ± 0.98 | 42 ± 5.8 | 1.2 ± 0.2 | 573 ± 65 |
| - | 15 | 8.07 ± 0.06 | 1.03 ± 0.01 | 0.21 ± 0.01 | 11.1 ± 1.1 | 4.37 ± 1.09 | 56 ± 4.2 | 2.8 ± 0.3 | 594 ± 44 |
| Antibiotic Class | Analyte | Concentration ± SD b (µg kg−1) | DT50/DT90 c (Days) | Time Passed Since the Last Administration d (Days) |
|---|---|---|---|---|
| Sulfonamides | Sulfadimethoxine (SDM) | 5 ± 1 | 3.2/21 | 278 |
| Sulfamethazine (SMT) | 4 ± 1 | 1.8/16 | >650 e | |
| Sulfanilamide (SNA) | 29 ± 6 | - | - | |
| Tetracyclines | Doxycycline (DOX) | 3605 ± 106 | 10/98 | 28 |
| Oxytetracycline (OTC) and epimer | 46 ± 5 | 16/171 | 42 | |
| Chlortetracycline (CTC) and epimer | 20 ± 2 | 19/62 | >650 e | |
| Tetracycline (TC) and epimer | 3 ± 1 | 12/111 | >650 e | |
| Quinolones | Enrofloxacin (ENR) | 13 ± 3 | 6/83 | >650 e |
| Flumequine (FLU) | 9 ± 2 | 44/146 | >650 e | |
| Ciprofloxacin (CIP) | 3 ± 1 | 6/85 | - | |
| Lincosamides | Lincomycin (LIN) | 1196 ± 24 | 269/892 | 23 |
| Pleuromutilins | Tiamulin (TIA) | 369 ± 17 | 101/335 | 65 |
| Time Point | Day | FLOA b | LIN | TIA | DOX | OTC c | CTC c | TC c | ENR | FLU | CIP | SNA | SDM | SMT | TMP e |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 0.4 ± 0.1 | 321 ± 42 | 295 ± 59 | 1095 ± 175 | 31 ± 5.8 | 9.1 ± 3.7 | 1.4 ± 0.5 | 8.8 ± 2.5 | 3.1 ± 0.6 | 1.9 ± 1.3 | 19 ± 2.0 | 5.0 ± 0.8 | 0.4 ± 0.1 | 0.2 ± 0.0 |
| 2 | 2 | 1.0 ± 0.1 | 329 ± 25 | 343 ± 61 | 1212 ± 289 | 49 ± 10 | 14 ± 3.5 | 1.4 ± 0.5 | 9.7 ± 2.0 | 4.0 ± 0.8 | 1.8 ± 0.8 | 26 ± 2.6 | 7.1 ± 1.4 | 0.6 ± 0.1 | 1.1 ± 0.4 |
| 3 | 5 | 1.0 ± 0.1 | 269 ± 37 | 235 ± 64 | 953 ± 159 | 41 ± 8.3 | 11 ± 2.3 | 1.4 ± 0.5 | 9.0 ± 1.7 | 3.4 ± 0.7 | 1.6 ± 0.5 | 27 ± 4.0 | 6.1 ± 1.2 | 0.4 ± 0.1 | 1.0 ± 0.0 |
| 4 | 7 | 1.0 ± 0.1 | 269 ± 53 | 297 ± 91 | 985 ± 384 | 39 ± 15 | 8.5 ± 3.5 | 1.0 ± 0.2 | 9.7 ± 2.9 | 3.0 ± 1.4 | 2.2 ± 0.0 | 24 ± 3.1 | 4.4 ± 1.4 | 0.3 ± 0.1 | 0.7 ± 0.1 |
| 5 | 14 | 1.2 ± 0.1 | 93 ± 38 | 176 ± 56 | 669 ± 81 | 30 ± 4.7 | 8.4 ± 1.2 | 0.9 ± 0.2 | 6.0 ± 0.9 | 2.3 ± 0.3 | 0.9 ± 0.2 | 31 ± 4.1 | 4.4 ± 0.9 | 0.3 ± 0.1 | 1.3 ± 0.1 |
| 6 | 33 | 1.5 ± 0.5 | 15 ± 6.4 | 202 ± 13 | 721 ± 64 | 38 ± 5.7 | 11 ± 1.4 | 1.5 ± 0.7 | 10 ± 0.8 | 4.5 ± 0.7 | 2.5 ± 0.3 | 26 ± 1.8 | 6.5 ± 0.7 | 0.5 ± 0.0 | 0.5 ± 0.0 |
| 7 | 61 | 0.7 ± 0.1 | 3.5 ± 0.7 d | 153 ± 11 | 484 ± 30 | 18 ± 0.7 | 11 ± 0.1 | 0.6 ± 0.1 | 7.5 ± 0.3 | 2.0 ± 0.0 | 1.7 ± 0.3 | 23 ± 6.7 | 3.0 ± 0.0 | 0.2 ± 0.0 | 0.1 ± 0.0 |
| 8 | 89 | 0.8 ± 0.3 | 5.5 ± 3.5 | 107 ± 20 | 491± 61 | 22 ± 0.7 | 7.0 ± 0.0 | 1.0 ± 0.0 | 6.6 ± 0.6 | 3.0 ± 0.0 | 2.1 ± 1.3 | 18 ± 5.2 | 4.0 ± 0.0 | 0.3 ± 0.0 | 0.5 ± 0.0 |
| 9 | 124 | 0.7 ± 0.1 | 5.0 ± 2.8 | 144 ± 1.4 | 543 ± 17 | 21 ± 0.7 | 7.9 ± 0.3 | 0.7 ± 0.1 | 8.8 ± 1.8 | 3.0 ± 0.0 | 1.5 ± 0.5 | 23 ± 4.5 | 4.0 ± 0.0 | 0.3 ± 0.1 | 0.2 ± 0.1 |
| 10 | 152 | 0.7 ± 0.1 | 4.5 ± 0.7 | 189 ± 19 | 689 ± 47 | 24 ± 4.2 | 8.4 ± 1.1 | 1.0 ± 0.0 | 11 ± 1.7 | 3.5 ± 0.7 | 2.0 ± 0.1 | 28 ± 2.7 | 4.5 ± 0.7 | 0.4 ± 0.1 | 0.2 ± 0.0 |
| 11 | 180 | 0.7 ± 0.1 | 3.5 ± 0.7 | 177 ± 11 | 620 ± 152 | 18 ± 1.4 | 7.9 ± 2.3 | 0.9 ± 0.1 | 9.6 ± 1.2 | 3.5 ± 0.7 | 1.5 ± 0.0 | 27 ± 5.7 | 4.5 ± 0.7 | 0.4 ± 0.1 | 0.3 ± 0.1 |
| 12 | 215 | 0.7 ± 0.1 | 4.0 ± 0.0 | 152 ± 22 | 442 ± 44 | 13 ± 1.4 | 6.6 ± 1.6 | 0.6 ± 0.1 | 7.8 ± 2.0 | 2.5 ± 0.7 | 1.2 ± 0.2 | 24 ± 1.5 | 4.0 ± 1.4 | 0.4 ± 0.1 | 0.2 ± 0.0 |
| 13 | 242 | 0.6 ± 0.1 | 3.5 ± 0.7 | 120 ± 36 | 499 ± 79 | 18 ± 5.7 | 6.8 ± 2.2 | 0.8 ± 0.3 | 7.3 ± 2.3 | 2.5 ± 0.7 | 1.1 ± 0.3 | 23 ± 0.5 | 4.0 ± 1.4 | 0.3 ± 0.1 | 0.3 ± 0.1 |
| 14 | 270 | 0.4 ± 0.1 | 3.0 ± 0.0 | 93 ± 7.8 | 418 ± 36 | 18 ± 4.9 | 5.5 ± 0.5 | 0.5 ± 0.1 | 6.8 ± 0.8 | 2.0 ± 0.0 | 2.1 ± 1.3 | 19 ± 4.9 | 3.0 ± 0.0 | 0.3 ± 0.1 | 0.1 ± 0.0 |
| 15 | 320 | 0.8 ± 0.1 | 2.9 ± 0.6 | 121 ± 29 | 408 ± 73 | 23 ± 4.6 | 8.3 ± 1.0 | 1.0 ± 0.1 | 5.6 ± 1.4 | 2.8 ± 0.5 | 0.8 ± 0.3 | 16 ± 3.2 | 4.8 ± 0.7 | 0.3 ± 0.0 | 0.2 ± 0.0 |
| Antibiotic | PCR Target | Primer Sequence (5′–3′) | Annealing Temperature (°C) | Product (bp) | Strain | Insert (µg/mL) | Plasmid | Construct [µg/mL] | Source |
|---|---|---|---|---|---|---|---|---|---|
| MLSB | ermB | CCGTGCGTCTGACATCTATCT | 57/55 | 189 | E. coli R4287 | 26 | pCRTM 2.1-TOPO® | 479 | Guo et al. [42] |
| GTGGTATGGCGGGTAAGTTTT | |||||||||
| Sulfonamides | sul1 | CGCACCGGAAACATCGCTGCAC | 56/55 | 163 | E. coli R4276 | 147 | pCRTM 2.1-TOPO® | 425 | Pei et al. [43] |
| TGAAGTTCCGCCGCAAGGCTCG | |||||||||
| / | intl1 | GGCTTCGTGATGCCTGCTT | 55/55 | 146 | E. coli R4730 | 69 | pCRTM 2.1-TOPO® | 455 | He et al. [44] |
| CATTCCTGGCCGTGGTTCT | |||||||||
| Chloramphenicol | cfr | GTTGGGAGTCATTTTGTATATC | 55/55 | 179 | E. faecium V375 | 51 | pCRTM 2.1-TOPO® | 370 | This work |
| CTTCWCCCATTCCCATAAAAG | |||||||||
| Florfenicol | fexA | ATTCTCCCGCAAATAACG | 52/55 | 156 | E. faecalis V307 | 71 | pCRTM 2.1-TOPO® | 793 | Li et al. [45] |
| TCGGCTCAGTAGCATCACG | |||||||||
| Oxazolidinone | optrA | GCTATTGTTGGTAGAAATGG | 55/55 | 160 | E. faecalis V307 | 51 | pCRTM 2.1-TOPO® | 507 | This work |
| CTTTCATCTTCAAAAGGCATC | |||||||||
| Tetracycline | tetA | GCTTCATGAGCGCCTGTTT | 60/55 | 706 | E. coli R4730 | 51 | pCRTM 2.1-TOPO® | 759 | Pholwat et al. [46] |
| CACCCGTTCCACGTTGTTAT | |||||||||
| tetG | GCTCGGTGGTATCTCTGCTC | 55/55 | 468 | S. typhimurim DT104 | 69 | pCRTM 2.1-TOPO® | 645 | Ng et al. [47] | |
| AGCAACAGAATCGGGAACAC | |||||||||
| tetM | GTGGACAAAGGTACAACGAG | 55/55 | 406 | S. delphini 2567 | 112 | pCRTM 2.1-TOPO® | 378 | Ng et al. [47] | |
| CGGTAAAGTTCGTCACACAC | |||||||||
| / | 16s rRNA | ACTCCTACGGGAGGCAG | 60/55 | 473 | E. coli R4829 | 75 | pCRTM 2.1-TOPO® | 508 | Van den Meersche et al. [41] |
| ATTACCGCGGCTGCTGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Massaccesi, L.; Albini, E.; Massacci, F.R.; Giusepponi, D.; Paoletti, F.; Sdogati, S.; Morena, F.; Agnelli, A.; Leccese, A.; Magistrali, C.F.; et al. Impact of Soil Fertilization with Pig Slurry on Antibiotic Residues and Resistance Genes: A Longitudinal Study. Antibiotics 2024, 13, 486. https://doi.org/10.3390/antibiotics13060486
Massaccesi L, Albini E, Massacci FR, Giusepponi D, Paoletti F, Sdogati S, Morena F, Agnelli A, Leccese A, Magistrali CF, et al. Impact of Soil Fertilization with Pig Slurry on Antibiotic Residues and Resistance Genes: A Longitudinal Study. Antibiotics. 2024; 13(6):486. https://doi.org/10.3390/antibiotics13060486
Chicago/Turabian StyleMassaccesi, Luisa, Elisa Albini, Francesca Romana Massacci, Danilo Giusepponi, Fabiola Paoletti, Stefano Sdogati, Francesco Morena, Alberto Agnelli, Angelo Leccese, Chiara Francesca Magistrali, and et al. 2024. "Impact of Soil Fertilization with Pig Slurry on Antibiotic Residues and Resistance Genes: A Longitudinal Study" Antibiotics 13, no. 6: 486. https://doi.org/10.3390/antibiotics13060486
APA StyleMassaccesi, L., Albini, E., Massacci, F. R., Giusepponi, D., Paoletti, F., Sdogati, S., Morena, F., Agnelli, A., Leccese, A., Magistrali, C. F., & Galarini, R. (2024). Impact of Soil Fertilization with Pig Slurry on Antibiotic Residues and Resistance Genes: A Longitudinal Study. Antibiotics, 13(6), 486. https://doi.org/10.3390/antibiotics13060486







