Emergence of High-Level Gentamicin Resistance in Streptococcus agalactiae Hypervirulent Serotype IV ST1010 (CC452) Strains by Acquisition of a Novel Integrative and Conjugative Element
Abstract
1. Introduction
2. Results
2.1. Antibiotic Susceptibility
2.2. Virulence Factors and Clonality
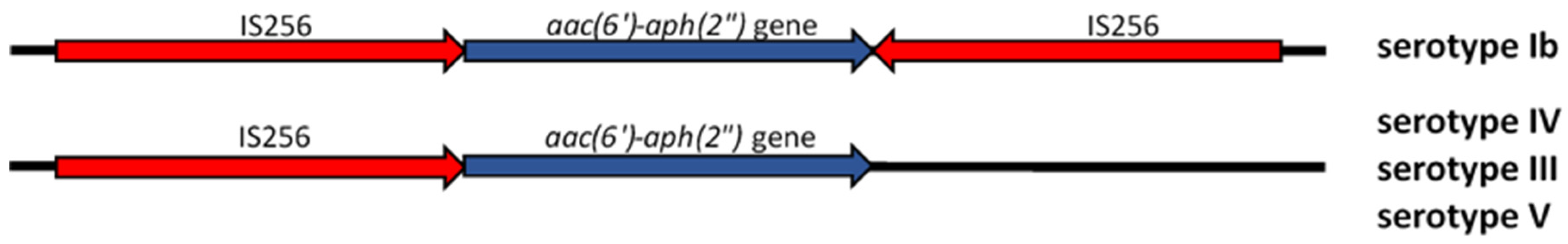
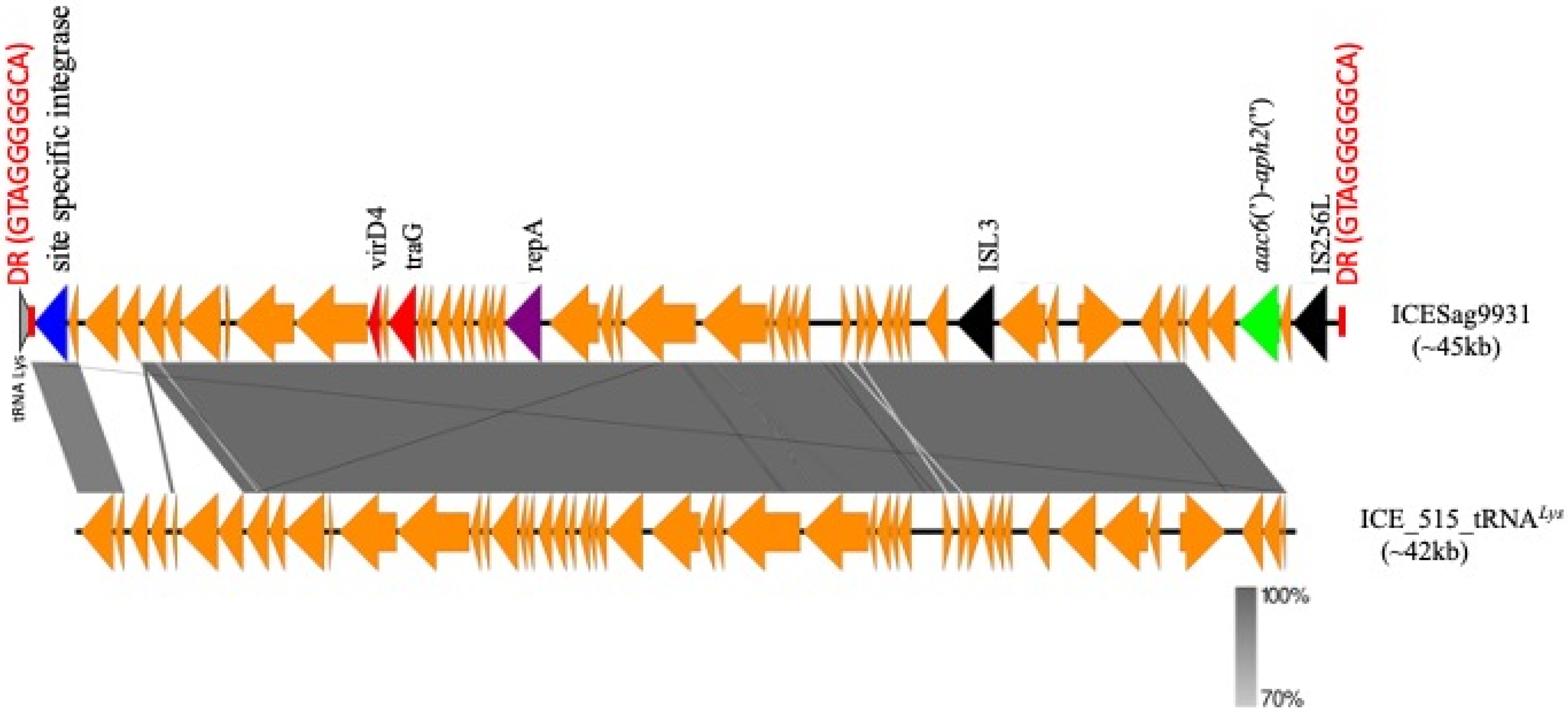
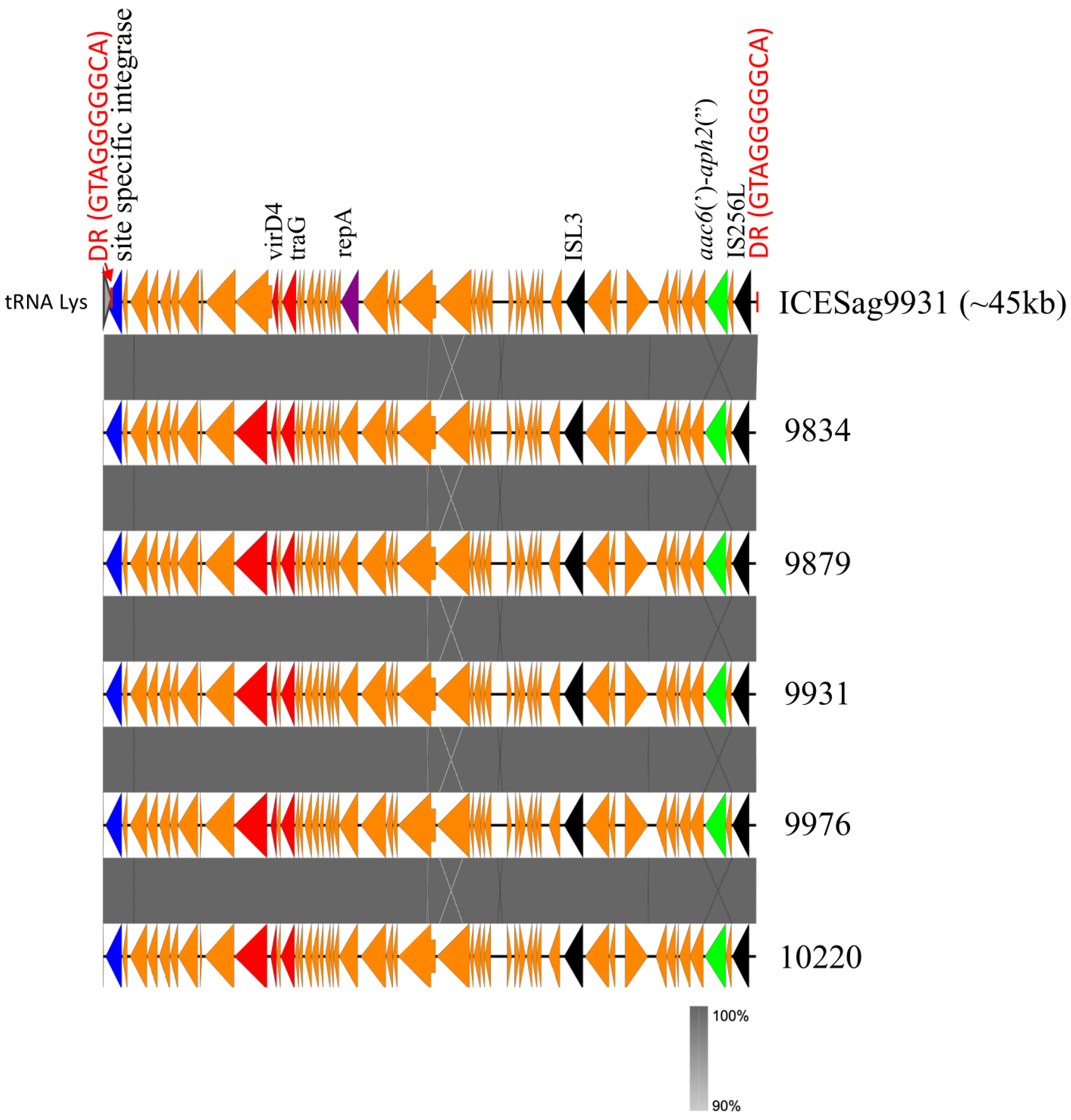
2.3. Transposon Structure of HLGR GBS Strains and New ICE
3. Discussion
4. Materials and Methods
4.1. GBS Strains Dataset
4.2. Bacterial Isolates Typing
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Madrid, L.; Seale, A.C.; Kohli-Lynch, M.; Edmond, K.M.; Lawn, J.E.; Heath, P.T.; Madhi, S.A.; Baker, C.J.; Bartlett, L.; Cutland, C.; et al. Infant GBS Disease Investigator Group. Infant group B streptococcal disease incidence and serotypes worldwide: Systematic review and meta-analyses. Clin. Infect. Dis. 2017, 65, S160–S172. [Google Scholar] [CrossRef] [PubMed]
- Furfaro, L.L.; Chang, B.J.; Payne, M.S. Perinatal Streptococcus agalactiae epidemiology and surveillance targets. Clin. Microbiol. Rev. 2018, 31, e00049-18. [Google Scholar] [CrossRef] [PubMed]
- Schrag, S.J.; Verani, J.R. Intrapartum antibiotic prophylaxis for the prevention of perinatal group B streptococcal disease: Experience in the United States and implications for a potential group B streptococcal vaccine. Vaccine 2013, 31 (Suppl. S4), D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Francois Watkins, L.K.; McGee, L.; Schrag, S.J.; Beall, B.; Jain, J.H.; Pondo, T.; Farley, M.M.; Harrison, L.H.; Zansky, S.M.; Baumbach, J.; et al. Epidemiology of invasive group B streptococcal infections among nonpregnant adults in the United States, 2008–2016. JAMA Intern. Med. 2019, 179, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Cieslewicz, M.J.; Chaffin, D.; Glusman, G.; Kasper, D.; Madan, A.; Rodrigues, S.; Fahey, J.; Wessels, M.R.; Rubens, C.E. Structural and genetic diversity of group B streptococcus capsular polysaccharides. Infect. Immun. 2005, 73, 3096–3103. [Google Scholar] [CrossRef] [PubMed]
- Ferrieri, P.; Lynfield, R.; Creti, R.; Flores, A.E. Serotype IV and invasive group B Streptococcus disease in neonates, Minnesota, USA, 2000–2010. Emerg. Infect. Dis. 2013, 19, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Nanduri, S.A.; Petit, S.; Smelser, C.; Apostol, M.; Alden, N.B.; Harrison, L.H.; Lynfield, R.; Vagnone, P.S.; Burzlaff, K.; Spina, N.L.; et al. Epidemiology of invasive early-onset and late-onset group B streptococcal disease in the United States, 2006 to 2015: Multistate laboratory and population-based surveillance. JAMA Pediatr. 2019, 173, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Suzuki, S.; Wachino, J.; Kurokawa, H.; Yamane, K.; Shibata, N.; Nagano, N.; Kato, H.; Shibayama, K.; Arakawa, Y. First molecular characterization of group B streptococci with reduced penicillin susceptibility. Antimicrob. Agents Chemother. 2008, 52, 2890–2897. [Google Scholar] [CrossRef] [PubMed]
- Dahesh, S.; Hensler, M.E.; Van Sorge, N.M.; Gertz, R.E., Jr.; Schrag, S.; Nizet, V.; Beall, B.W. Point mutation in the group B streptococcal pbp2x gene conferring decreased susceptibility to beta-lactam antibiotics. Antimicrob. Agents Chemother. 2008, 52, 2915–2918. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Nagano, N.; Arakawa, Y. Classification of group B streptococci with reduced β-lactam susceptibility (GBS-RBS) based on the amino acid substitutions in PBPs. J. Antimicrob. Chemother. 2015, 70, 1601–1603. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, B.J.; Chochua, S.; Gertz, R.E., Jr.; Hawkins, P.A.; Ricaldi, J.; Li, Z.; Walker, H.; Tran, T.; Rivers, J.; Mathis, S.; et al. Short-read whole genome sequencing for determination of antimicrobial resistance mechanisms and capsular serotypes of current invasive Streptococcus agalactiae recovered in the USA. Clin. Microbiol. Infect. 2017, 23, 574.e7–574.e14. [Google Scholar] [CrossRef] [PubMed]
- Koide, S.; Nagano, Y.; Takizawa, S.; Sakaguchi, K.; Soga, E.; Hayashi, W.; Tanabe, M.; Denda, T.; Kimura, K.; Arakawa, Y.; et al. Genomic traits associated with virulence and antimicrobial resistance of invasive group B Streptococcus isolates with reduced penicillin susceptibility from elderly adults. Microbiol. Spectr. 2022, 10, e0056822. [Google Scholar] [CrossRef] [PubMed]
- American College of Obstetricians and Gynecologists. Prevention of Early-Onset Group B Streptococcal Disease in Newborns. 2019. No. 782. Available online: https://www.acog.org/clinical/clinical-guidance/committee-opinion/articles/2020/02/prevention-of-group-b-streptococcal-early-onset-disease-in-newborns?utm_source=vanity&utm_medium=web&utm_campaign=clinical (accessed on 6 May 2024).
- Sendi, P.; Furitsch, M.; Mauerer, S.; Florindo, C.; Kahl, B.C.; Shabayek, S.; Berner, R.; Spellerberg, B. Chromosomally and extrachromosomally mediated high-level gentamicin resistance in Streptococcus agalactiae. Antimicrob. Agents Chemother. 2016, 60, 1702–1707. [Google Scholar] [CrossRef] [PubMed]
- Vekemans, J.; Crofts, J.; Baker, C.J.; Goldblatt, D.; Heath, P.T.; Madhi, S.A.; Le Doare, K.; Andrews, N.; Pollard, A.J.; Saha, S.K.; et al. The role of immune correlates of protection on the pathway to licensure, policy decision and use of group B Streptococcus vaccines for maternal immunization: Considerations from World Health Organization consultations. Vaccine 2019, 37, 3190–3198. [Google Scholar] [CrossRef] [PubMed]
- Buurman, E.T.; Timofeyeva, Y.; Gu, J.; Kim, J.H.; Kodali, S.; Liu, Y.; Mininni, T.; Moghazeh, S.; Pavliakova, D.; Singer, C.; et al. A novel hexavalent capsular polysaccharide conjugate vaccine (GBS6) for the prevention of neonatal group B streptococcal infections by maternal immunization. J. Infect. Dis. 2019, 220, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Paoletti, L.C.; Kasper, D.L. Surface structures of group B Streptococcus important in human immunity. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Bradley, J.S.; Barnett, E.D.; Cantey, J.B. (Eds.) Nelson’s Pediatric Antimicrobial Therapy 2021, 27th ed.; American Academy of Pediatrics: Itasca, IL, USA, 2021. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020; CLSI supplement M100. [Google Scholar]
- Creti, R.; Imperi, M.; Berardi, A.; Angeletti, S.; Gherardi, G. Laboratory breakpoints for assessing high level gentamicin resistance in Streptococcus agalactiae: It is the time for a consensus. Clin. Microbiol. Infect. 2022, 28, 1050–1052. [Google Scholar] [CrossRef] [PubMed]
- Carreras-Abad, C.; Ramkhelawon, L.; Heath, P.T.; Le Doare, K. A vaccine against group B Streptococcus: Recent advances. Infect. Drug Resist. 2020, 13, 1263–1272. [Google Scholar] [CrossRef] [PubMed]
- McGee, L.; Chochua, S.; Li, Z.; Mathis, S.; Rivers, J.; Metcalf, B.; Ryan, A.; Alden, N.; Farley, M.M.; Harrison, L.H.; et al. Multistate, population-based distributions of candidate vaccine targets, clonal complexes, and resistance features of invasive group B streptococci within the United States, 2015–2017. Clin. Infect. Dis. 2021, 72, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Tazi, A.; Disson, O.; Bellais, S.; Bouaboud, A.; Dmytruk, N.; Dramsi, S.; Mistou, M.Y.; Khun, H.; Mechler, C.; Tardieux, I.; et al. The surface protein HvgA mediates group B streptococcus hypervirulence and meningeal tropism in neonates. J. Exp. Med. 2010, 207, 2313–2322. [Google Scholar] [CrossRef] [PubMed]
- Schindler, Y.; Rahav, G.; Nissan, I.; Treygerman, O.; Prajgrod, G.; Attia, B.Z.; Raz, R.; Valenci, G.Z.; Tekes-Manova, D.; Maor, Y. Group B streptococcus virulence factors associated with different clinical syndromes: Asymptomatic carriage in pregnant women and early-onset disease in the newborn. Front. Microbiol. 2023, 14, 1093288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.M.; Wang, Q.; Han, T.Y.; Liu, J.H.; Hu, X.X.; Qiao, F.; Yang, X.Y.; Li, C.R.; You, X.F. Structure analysis of transposons carrying the aac(6’)-aph(2″) gene in Enterococcus faecalis isolated in Beijing, China, and comparison of their transfer efficiency. Int. J. Antimicrob. Agents. 2018, 52, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Vakulenko, S.B.; Donabedian, S.M.; Voskresenskiy, A.M.; Zervos, M.J.; Lerner, S.A.; Chow, J.W. Multiplex PCR for detection of aminoglycoside resistance genes in enterococci. Antimicrob. Agents Chemother. 2003, 47, 1423–1426. [Google Scholar] [CrossRef] [PubMed]
- Hodel-Christian, S.L.; Murray, B.E. Characterization of the gentamicin resistance transposon Tn5281 from Enterococcus faecalis and comparison to the staphylococcal transposons, Tn4001 and Tn4031. Antimicrob. Agents Chemother. 1991, 35, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Lyon, B.R.; May, J.W.; Skuray, R.A. Tn4001: A gentamicin and kanamycin resistance transposon in Staphylococcus aureus. Mol. Gen. Genet. 1984, 193, 554–556. [Google Scholar] [CrossRef] [PubMed]
- Horaud, T.; de Céspèdes, G.; Trieu-Cuot, P. Chromosomal gentamicin resistance transposon Tn3706 in Streptococcus agalactiae B128. Antimicrob. Agents Chemother. 1996, 40, 1085–1090. [Google Scholar] [CrossRef] [PubMed]
- Nagaoka, K.; Konno, S.; Murase, K.; Kikuchi, T.; Nakagawa, I. Complete Genome Sequence of Streptococcus agalactiae Serotype III, Multilocus Sequence Type 335 Strain HU-GS5823, Isolated from a Human Patient in Japan with Severe Invasive Infection. Microbiol. Resour. Announc. 2018, 7, e01303-18. [Google Scholar] [CrossRef] [PubMed]
- Chuzeville, S.; Puymège, A.; Madec, J.Y.; Haenni, M.; Payot, S. Characterization of a new CAMP factor carried by an integrative and conjugative element in Streptococcus agalactiae and spreading in Streptococci. PLoS ONE 2012, 7, e48918. [Google Scholar] [CrossRef] [PubMed]
- Ku, T.S.N.; Al Mohajer, M.; Newton, J.A.; Wilson, M.H.; Monsees, E.; Hayden, M.K.; Messacar, K.; Kisgen, J.J.; Diekema, D.J.; Morgan, D.J.; et al. Improving antimicrobial use through better diagnosis: The relationship between diagnostic stewardship and antimicrobial stewardship. Infect. Control Hosp. Epidemiol. 2023, 44, 1901–1908. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.S.; Gigante, V.; Sati, H.; Paulin, S.; Al-Sulaiman, L.; Rex, J.H.; Fernandes, P.; Arias, C.A.; Paul, M.; Thwaites, G.E.; et al. Analysis of the Clinical Pipeline of Treatments for Drug-Resistant Bacterial Infections: Despite Progress, More Action Is Needed. Antimicrob. Agents Chemother. 2022, 66, e0199121. [Google Scholar] [CrossRef] [PubMed]
- Doumith, M.; Mushtaq, S.; Martin, V.; Chaudhry, A.; Adkin, R.; Coelho, J.; Chalker, V.; MacGowan, A.; Woodford, N.; Livermore, D.M.; et al. Genomic sequences of Streptococcus agalactiae with high-level gentamicin resistance, collected in the BSAC bacteraemia surveillance. J. Antimicrob. Chemother. 2017, 72, 2704–2707. [Google Scholar] [CrossRef] [PubMed]
- Buu-Hoï, A.; Le Bouguenec, C.; Horaud, T. High-level chromosomal gentamicin resistance in Streptococcus agalactiae (group B). Antimicrob. Agents Chemother. 1990, 34, 985–988. [Google Scholar] [CrossRef] [PubMed]
- Liddy, H.; Holliman, R. Group B Streptococcus highly resistant to gentamicin. J. Antimicrob. Chemother. 2002, 50, 142–143. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhi, Y.; Ji, H.J.; Jung, J.H.; Byun, E.B.; Kim, W.S.; Lin, S.M.; Lim, S.; Jang, A.Y.; Choi, M.J.; Ahn, K.B.; et al. Molecular characteristics of IS1216 carrying multidrug resistance gene cluster in serotype III/sequence type 19 group B streptococcus. mSphere 2021, 6, e00543-21. [Google Scholar] [CrossRef] [PubMed]
- Khan, U.B.; Portal, E.A.R.; Sands, K.; Lo, S.; Chalker, V.J.; Jauneikaite, E.; Spiller, O.B. Genomic Analysis Reveals New Integrative Conjugal Elements and Transposons in GBS Conferring Antimicrobial Resistance. Antibiotics 2023, 12, 544. [Google Scholar] [CrossRef] [PubMed]
- Puymège, A.; Bertin, S.; Guédon, G.; Payot, S. Analysis of Streptococcus agalactiae pan-genome for prevalence, diversity and functionality of integrative and conjugative or mobilizable elements integrated in the tRNA(Lys CTT) gene. Mol. Genet. Genom. 2015, 290, 1727–1740. [Google Scholar] [CrossRef] [PubMed]
- Ruppen, C.; Hemphill, A.; Sendi, P. In vitro activity of gentamicin as an adjunct to penicillin against biofilm group B Streptococcus. J. Antimicrob. Chemother. 2017, 72, 444–447. [Google Scholar] [CrossRef] [PubMed]
- Campisi, E.; Rinaudo, C.D.; Donati, C.; Barucco, M.; Torricelli, G.; Edwards, M.S.; Baker, C.J.; Margarit, I.; Rosini, R. Serotype IV Streptococcus agalactiae ST-452 has arisen from large genomic recombination events between CC23 and the hypervirulent CC17 lineages. Sci. Rep. 2016, 6, 29799. [Google Scholar] [CrossRef] [PubMed]
- Springman, A.C.; Lacher, D.W.; Waymire, E.A.; Wengert, S.L.; Singh, P.; Zadoks, R.N.; Davies, H.D.; Manning, S.D. Pilus distribution among lineages of group B Streptococcus: An evolutionary and clinical perspective. BMC Microbiol. 2014, 14, 159. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.; Laut, C.; Gaddy, J.A.; Zadoks, R.N.; Davies, H.D.; Manning, S.D. Association between genotypic diversity and biofilm production in group B Streptococcus. BMC Microbiol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Lacasse, M.; Valentin, A.S.; Corvec, S.; Bémer, P.; Jolivet-Gougeon, A.; Plouzeau, C.; Tandé, D.; Mereghetti, L.; Bernard, L.; Lartigue, M.F.; et al. Genotypic Characterization and biofilm production of group B Streptococcus strains isolated from bone and joint infections. Microbiol. Spectr. 2022, 10, e0232921. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Li, J.; Zhou, H.; Wang, Z.; Yi, L.; Liu, N.; Du, J.; Chang, C.Y.; Ji, W. Serotype distribution, virulence determinants and antimicrobial susceptibility of Streptococcus agalactiae isolated from young infants. Pathogens 2022, 11, 1355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ma, L.; Zhu, L.; Zhou, X.H.; Xu, L.J.; Guo, C.; Meng, J.H.; Zhang, X.H.; Liu, Q.H.; Huang, R. Molecular characterization of pathogenic group B streptococcus from a tertiary hospital in Shanxi, China: High incidence of sequence type 10 strains in infants/pregnant women. J. Microbiol. Immunol. Infect. 2021, 54, 1094–1100. [Google Scholar] [CrossRef]
- Nabavinia, M.; Khalili, M.B.; Sadeh, M.; Eslami, G.; Vakili, M.; Azartoos, N.; Mojibiyan, M. Distribution of Pilus island and antibiotic resistance genes in Streptococcus agalactiae obtained from vagina of pregnant women in Yazd, Iran. Iran. J. Microbiol. 2020, 12, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Graux, E.; Hites, M.; Martiny, D.; Maillart, E.; Delforge, M.; Melin, P.; Dauby, N. Invasive group B Streptococcus among non-pregnant adults in Brussels-Capital Region, 2005–2019. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Gherardi, G.; Imperi, M.; Baldassarri, L.; Pataracchia, M.; Alfarone, G.; Recchia, S.; Orefici, G.; Dicuonzo, G.; Creti, R. Molecular epidemiology and distribution of serotypes, surface proteins, and antibiotic resistance among group B streptococci in Italy. J. Clin. Microbiol. 2007, 45, 2909–2916. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Creti, R.; Imperi, M.; Berardi, A.; Pataracchia, M.; Recchia, S.; Alfarone, G.; Baldassarri, L.; Italian Neonatal GBS Infections Working Group. Neonatal group B Streptococcus infections: Prevention strategies, clinical and microbiologic characteristics in 7 years of surveillance. Pediatr. Infect. Dis. J. 2017, 36, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Creti, R.; Imperi, M.; Berardi, A.; Lindh, E.; Alfarone, G.; Pataracchia, M.; Recchia, S. The Italian Network on Neonatal And Infant Gbs Infections. Invasive Group B Streptococcal Disease in Neonates and Infants, Italy, Years 2015–2019. Microorganisms 2021, 9, 2579. [Google Scholar] [CrossRef] [PubMed]
- Imperi, M.; Gherardi, G.; Berardi, A.; Baldassarri, L.; Pataracchia, M.; Dicuonzo, G.; Orefici, G.; Creti, R. Invasive neonatal GBS infections from an area-based surveillance study in Italy. Clin. Microbiol. Infect. 2011, 17, 1834–1839. [Google Scholar] [CrossRef] [PubMed]
- Giovanetti, E.; Montanari, M.P.; Mingoia, M.; Varaldo, P.E. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob. Agents Chemother. 1999, 43, 1935–1940. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Afshar, B.; Broughton, K.; Creti, R.; Decheva, A.; Hufnagel, M.; Kriz, P.; Lambertsen, L.; Lovgren, M.; Melin, P.; Orefici, G.; et al. International external quality assurance for laboratory identification and typing of Streptococcus agalactiae (Group B streptococci) J. Clin. Microbiol. 2011, 49, 1475–1482. [Google Scholar] [CrossRef] [PubMed]
- Slotved, H.C.; Hoffmann, S. Evaluation of procedures for typing of group B Streptococcus: A retrospective study. PeerJ 2017, 5, e3105. [Google Scholar] [CrossRef] [PubMed]
- Imperi, M.; Pataracchia, M.; Alfarone, G.; Baldassarri, L.; Orefici, G.; Creti, R. A multiplex PCR assay for the direct identification of the capsular type (Ia to IX) of Streptococcus agalactiae. J. Microbiol. Methods 2010, 80, 212–214. [Google Scholar] [CrossRef] [PubMed]
- Creti, R.; Fabretti, F.; Orefici, G.; von Hunolstein, C. Multiplex PCR assay for direct identification of group B streptococcal alphaprotein-like protein genes. J. Clin. Microbiol. 2004, 42, 1326–1329. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Martins, E.R.; Melo-Cristino, J.; Ramirez, M. Evidence for rare capsular switching in Streptococcus agalactiae. J. Bacteriol. 2010, 192, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Lamy, M.C.; Dramsi, S.; Billoët, A.; Réglier-Poupet, H.; Tazi, A.; Raymond, J.; Guérin, F.; Couvé, E.; Kunst, F.; Glaser, P.; et al. Rapid detection of the “highly virulent” group B Streptococcus ST-17 clone. Microbes Infect. 2006, 8, 1714–1722. [Google Scholar] [CrossRef] [PubMed]
- Meehan, M.; Eogan, M.; McCallion, N.; Cunney, R.; Bray, J.E.; Jolley, K.A.; Unitt, A.; Maiden, M.C.J.; Harrison, O.B.; Drew, R.J. Genomic epidemiology of group B streptococci spanning 10 years in an Irish maternity hospital, 2008–2017. J. Infect. 2021, 83, 37–45. [Google Scholar] [CrossRef] [PubMed]
| ID | Strain Type | Year | st | CLI | ERY | TET | CHL | LZD | LVX | PEN G | VAN | TEC | Macrolide R Genotype/Phenotype | TET R Genotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC (mg/L), Category (S, I, R) | ||||||||||||||
| 9543 | A-invD | 2016 | Ib | >0.5, R | >0.5, R | >4, R | ≤2, S | 1, S | 1, I | 0.06, S | ≤0.5, S | ≤1, S | erm(B)/cMLSB | tet(O) |
| 9646 | N-invD | 2016 | III | 0.06, S | ≤0.06, S | >4, R | ≤2, S | 1, S | ≤0.5, I | ≤0.03, S | ≤0.5, S | ≤1, S | - | tet(M) |
| 9716 | N-invD | 2016 | III | 0.125, S | ≤0.06, S | >4, R | ≤2, S | 1, S | 1, I | ≤0.03, S | ≤0.5, S | ≤1, S | - | tet(M) |
| 9834 | A-invD | 2018 | IV | 0.06, S | ≤0.06, S | ≤0.5, S | ≤2, S | 1, S | ≤0.5, I | ≤0.03, S | ≤0.5, S | ≤1, S | - | - |
| 9879 | A-invD | 2019 | IV | 0.06, S | ≤0.06, S | ≤0.5, S | ≤2, S | 1, S | ≤0.5, I | 0.06, S | ≤0.5, S | ≤1, S | - | - |
| 9907 | A-invD | 2019 | IV | 0.06, S | ≤0.06, S | ≤0.5, S | ≤2, S | 1, S | ≤0.5, I | 0.06, S | ≤0.5, S | ≤1, S | - | - |
| 9912 | A-invD | 2019 | IV | 0.06, S | ≤0.06, S | ≤0.5, S | ≤2, S | 1, S | ≤0.5, I | 0.06, S | ≤0.5, S | ≤1, S | - | - |
| 9931 | N-invD | 2021 | IV | 0.06, S | ≤0.06, S | ≤0.5, S | ≤2, S | 1, S | ≤0.5, I | 0.06, S | ≤0.5, S | ≤1, S | - | - |
| 10027 | A-carr | 2020 | IV | 0.06, S | ≤0.06, S | ≤0.5, S | ≤2, S | 1, S | ≤0.5, I | 0.06, S | ≤0.5, S | ≤1, S | - | - |
| 10042 | A-carr | 2020 | IV | 0.06, S | ≤0.06, S | ≤0.5, S | ≤2, S | 1, S | ≤0.5, I | 0.06, S | ≤0.5, S | ≤1, S | - | - |
| 10051 | A-carr | 2020 | IV | 0.06, S | ≤0.06, S | ≤0.5, S | ≤2, S | 1, S | ≤0.5, I | 0.06, S | ≤0.5, S | ≤1, S | - | - |
| 10066 | A-carr | 2020 | IV | 0.06, S | ≤0.06, S | ≤0.5, S | ≤2, S | 1, S | ≤0.5, I | 0.06, S | ≤0.5, S | ≤1, S | - | - |
| 10220 | A-carr | 2020 | IV | 0.06, S | ≤0.06, S | ≤0.5, S | ≤2, S | 1, S | ≤0.5, I | 0.06, S | ≤0.5, S | ≤1, S | - | - |
| 10239 | A-carr | 2020 | IV | 0.06, S | ≤0.06, S | ≤0.5, S | ≤2, S | 1, S | ≤0.5, I | 0.06, S | ≤0.5, S | ≤1, S | - | - |
| 10251 | A-carr | 2020 | IV | 0.06, S | ≤0.06, S | ≤0.5, S | ≤2, S | 1, S | ≤0.5, I | 0.06, S | ≤0.5, S | ≤1, S | - | - |
| 10272 | A-carr | 2020 | IV | 0.06, S | ≤0.06, S | ≤0.5, S | ≤2, S | 1, S | ≤0.5, I | 0.06, S | ≤0.5, S | ≤1, S | - | - |
| 10273 | A-carr | 2020 | IV | 0.06, S | ≤0.06, S | ≤0.5, S | ≤2, S | 1, S | ≤0.5, I | 0.06, S | ≤0.5, S | ≤1, S | - | - |
| 9714 | A-inv | 2016 | V | 0.06, S | 0.125, S | >4, R | ≤2, S | 1, S | >4, R | ≤0.03, S | ≤0.5, S | ≤1, S | - | tet(M) |
| 9976 | A-carr | 2020 | V | >0.5, R | >0.5, R | >4, R | ≤2, S | 1, S | >4, R | ≤0.03, S | ≤0.5, S | ≤1, S | erm(A)/iMLSB | tet(M) |
| 10039 | A-carr | 2020 | V | 0.06, S | >0.5, R | >4, R | ≤2, S | 1, S | >4, R | ≤0.03, S | ≤0.5, S | ≤1, S | erm(A)/iMLSB | tet(M) |
| Serotype | Year (n) | Surface Proteins Genes (n) | hvgA Gene (n) | PI (n) | ST (n) | CC |
|---|---|---|---|---|---|---|
| IV | 2018 (1 A-invD); 2019 (3 A-invD); 2020 (9 A-carr); 2021 (N-invD) | alpha-C (14) | Positive (14) | PI-2b (14) | 1010 (14) | 452 |
| V | 2016 (1 A-invD), 2020 (2 A-carr) | alp1 (3) | Negative (3) | PI-1 + 2a (3) | 19 (3) | 19 |
| III | 2016 (2 N-invD) | rib (2) | Positive (2) | PI-1 + 2b (2) | 17 (2) | 17 |
| Ib | 2016 (1 A-invD) | alpha-C | Negative | PI-2a | 12 | 12 |
| Serotype (n) | Transposon Structure | |||
|---|---|---|---|---|
| aac(6′)-aph(2″) Gene | IS256L | 3′-inter | 5′-inter | |
| Ib (1) | + | + | + | + |
| III (2) | + | + | − | + |
| IV (14) | + | + | − | + |
| V (3) | + | + | − | + |
| Fragment Name | Primers | Amplicon Size (bp) |
|---|---|---|
| aac(6′)-aph(2″) | F1: 5′-CAGAGCCTTGGGAAGATGAAG-3′ | 348 (from Vakulenko et al. AAC 2003; 47:1423) [26] |
| R1: 5′-CCTCGTGTAATTCATGTTCTGGC-3′ | ||
| 3′-inter | F3: 5′-GATATATTAAGAATGTATGG-3′ | 371 (from Zhang et al. IJAA 2018; 52:799) [25] |
| R3: 5′-GAGCCGTTCTTATGGACCTAC-3’ | ||
| 5′-inter | F2: 5′-GAGCCGTTCTTATGGACCTAC-3’ | 628 (from Zhang et al. IJAA 2018; 52:799) [25] |
| R2: 5′-CCACCATAAAATTCTAATAC-3’ | ||
| IS256L | F5: 5′-TGAAAAGCGAAGAGATTCAAA GC-3′ | 1103 (from Zhang et al. IJAA 2018; 52:799) [25] |
| R5: 5′-ATGTAGGTCCATAAGAACGGC-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Creti, R.; Imperi, M.; Khan, U.B.; Berardi, A.; Recchia, S.; Alfarone, G.; Gherardi, G. Emergence of High-Level Gentamicin Resistance in Streptococcus agalactiae Hypervirulent Serotype IV ST1010 (CC452) Strains by Acquisition of a Novel Integrative and Conjugative Element. Antibiotics 2024, 13, 491. https://doi.org/10.3390/antibiotics13060491
Creti R, Imperi M, Khan UB, Berardi A, Recchia S, Alfarone G, Gherardi G. Emergence of High-Level Gentamicin Resistance in Streptococcus agalactiae Hypervirulent Serotype IV ST1010 (CC452) Strains by Acquisition of a Novel Integrative and Conjugative Element. Antibiotics. 2024; 13(6):491. https://doi.org/10.3390/antibiotics13060491
Chicago/Turabian StyleCreti, Roberta, Monica Imperi, Uzma Basit Khan, Alberto Berardi, Simona Recchia, Giovanna Alfarone, and Giovanni Gherardi. 2024. "Emergence of High-Level Gentamicin Resistance in Streptococcus agalactiae Hypervirulent Serotype IV ST1010 (CC452) Strains by Acquisition of a Novel Integrative and Conjugative Element" Antibiotics 13, no. 6: 491. https://doi.org/10.3390/antibiotics13060491
APA StyleCreti, R., Imperi, M., Khan, U. B., Berardi, A., Recchia, S., Alfarone, G., & Gherardi, G. (2024). Emergence of High-Level Gentamicin Resistance in Streptococcus agalactiae Hypervirulent Serotype IV ST1010 (CC452) Strains by Acquisition of a Novel Integrative and Conjugative Element. Antibiotics, 13(6), 491. https://doi.org/10.3390/antibiotics13060491







