Antibiotic Resistance Trends in Uropathogens during the COVID-19 Pandemic in Western Romania: A Cross-Sectional Study
Abstract
:1. Introduction
2. Results
2.1. Urinalysis Findings
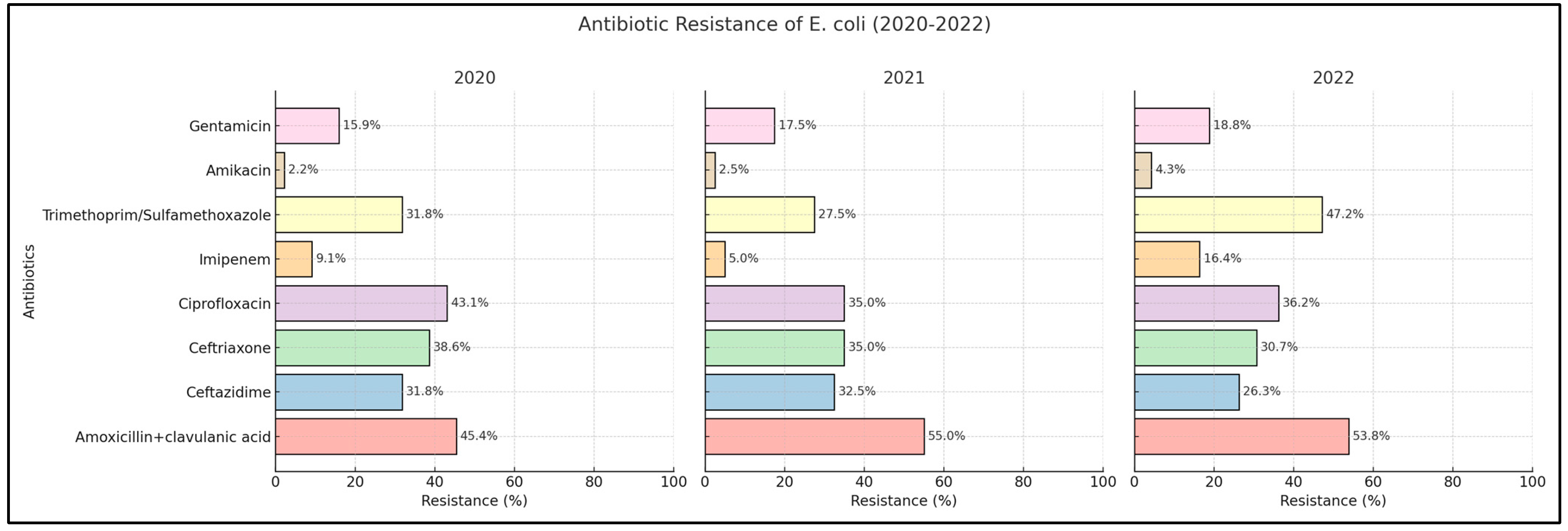
2.2. Escherichia coli
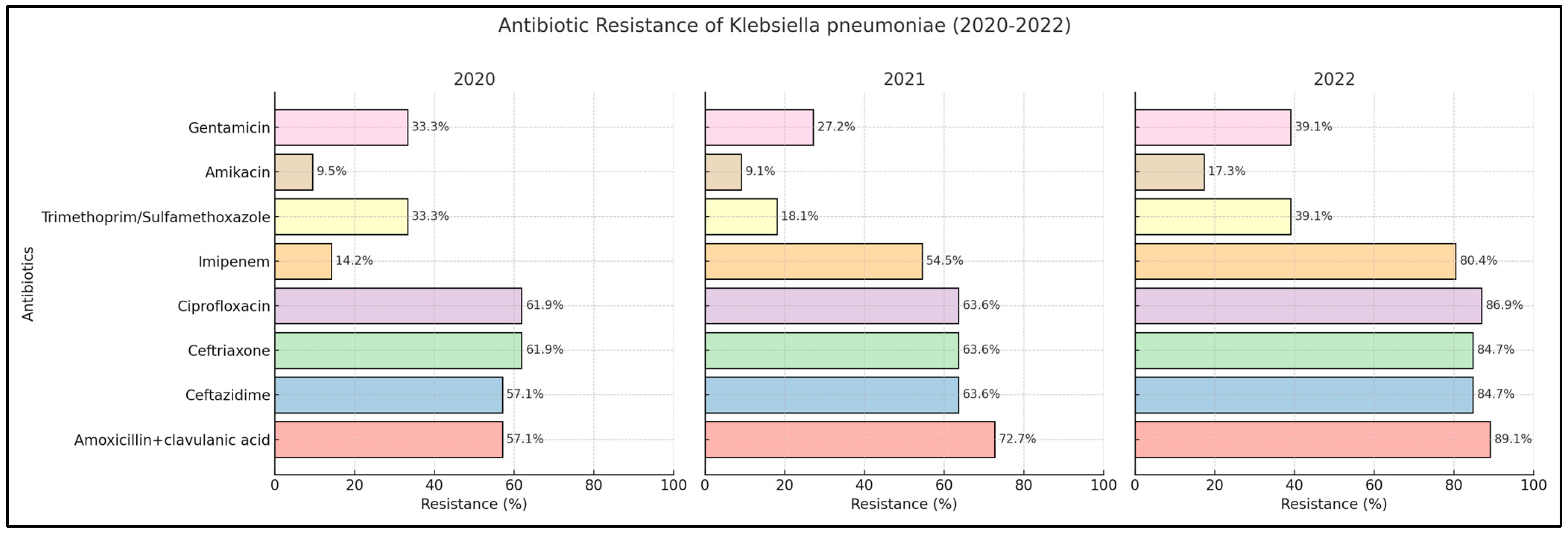
2.3. Klebsiella pneumoniae
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Data Collection and Definition of Variables
4.3. Definitions
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19 (accessed on 7 March 2024).
- Chauhan, A.S.; Singh, K.; Bhatia, R.; Khetrapal, S.; Naskar, A. The Health System’s Response to and the Impact of COVID-19 on Health Services, Providers, and Seekers: A Rapid Review in the Wake of the Pandemic. COVID 2023, 3, 1106–1157. [Google Scholar] [CrossRef]
- World Health Organization. World Health Organization COVID-19 Dashboard. 2023. Available online: https://covid19.who.int (accessed on 7 March 2024).
- Barbosa, E.J.M.; Gefter, W.B.; Ghesu, F.C.; Liu, S.; Mailhe, B.; Mansoor, A.; Grbic, S.; Vogt, S. Automated Detection and Quantification of COVID-19 Airspace Disease on Chest Radiographs: A Novel Approach Achieving Expert Radiologist-Level Performance Using a Deep Convolutional Neural Network Trained on Digital Reconstructed Radiographs from Computed Tomography-Derived Ground Truth. Invest. Radiol. 2021, 56, 471–479. [Google Scholar]
- Das, N.N.; Kumar, N.; Kaur, M.; Kumar, V.; Singh, D. Automated Deep Transfer Learning-Based Approach for Detection of COVID-19 Infection in Chest X-rays. Ing. Rech. Biomed. 2022, 43, 114–119. [Google Scholar]
- Gupta, N.; Zhao, Y.-Y.; Evans, C.E. The stimulation of thrombosis by hypoxia. Thromb. Res. 2019, 181, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in COVID-19. N. Engl. J. Med. 2020, 383, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.P.; de Oliveira, R.B. COVID-19 and Chronic Kidney Disease: A Narrative Study. COVID 2023, 3, 1092–1105. [Google Scholar] [CrossRef]
- Bischoff, S.; Walter, T.; Gerigk, M.; Ebert, M.; Vogelmann, R. Empiric antibiotic therapy in urinary tract infection in patients with risk factors for antibiotic resistance in a German emergency department. BMC Infect. Dis. 2018, 18, 56. [Google Scholar] [CrossRef]
- Zowawi, H.M.; Harris, P.N.A.; Roberts, M.J.; Tambyah, P.A.; Schembri, M.A.; Pezzani, M.D.; Williamson, D.A.; Paterson, D.L. The emerging threat of multidrug-resistant Gram-negative bacteria in urology. Nat. Rev. Urol. 2015, 12, 570–584. [Google Scholar] [CrossRef]
- Kot, B.; Grużewska, A.; Szweda, P.; Wicha, J.; Parulska, U. Antibiotic Resistance of Uropathogens Isolated from Patients Hospitalized in District Hospital in Central Poland in 2020. Antibiotics 2021, 10, 447. [Google Scholar] [CrossRef]
- Mortazavi-Tabatabaei, S.A.R.; Ghaderkhani, J.; Nazari, A.; Sayehmiri, K.; Sayehmiri, F.; Pakzad, I. Pattern of antibacterial resistance in urinary tract infections: A systematic review and meta-analysis. Int. J. Prev. Med. 2019, 10, 169. [Google Scholar] [PubMed]
- Sobel, J.D.; Kaye, D. Urinary tract infections. In Principals and Practice of Infectious Diseases, 5th ed.; Mandell, G.L., Bennett, J.E., Dolin, R., Eds.; Churchill Livingstone: Philadelphia, PA, USA, 2000. [Google Scholar]
- Nicolle, L.E.; Bradley, S.; Colgan, R.; Rice, J.C.; Schaeffer, A.; Hooton, T.M. Infectious Diseases Society of America guidelines for the diagnosis and treatment of asymptomatic bacteriuria in adults. Clin. Infect. Dis. 2005, 40, 643–654. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Point Prevalence Survey of Healthcare-Associated Infections and Antimicrobial Use in European Acute Care Hospitals; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2013. [Google Scholar]
- Chu, C.M.; Lowder, J.L. Diagnosis and treatment of urinary tract infections across age groups. Am. J. Obstet. Gynecol. 2018, 219, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B. Epidemiology of urinary tract infections: Incidence, morbidity, and economic costs. Am. J. Med. 2002, 113 (Suppl. 1A), 5S–13S. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, M.A.; Sadoma, H.H.M.; Mathew, S.; Alghamdi, S.; Malik, J.A.; Anwar, S. Retrospective analysis of antimicrobial susceptibility of Uropathogens isolated from pediatric patients in tertiary Hospital at Al-Baha Region, Saudi Arabia. Healthcare 2021, 9, 1564. [Google Scholar] [CrossRef] [PubMed]
- Schmiemann, G.; Kniehl, E.; Gebhardt, K.; Matejczyk, M.M.; Hummers-Pradier, E. The diagnosis of urinary tract infection: A systematic review. Dtsch. Arztebl. Int. 2010, 107, 361–367. [Google Scholar] [PubMed]
- Wolfe, A.J.; Brubaker, L. “Sterile Urine” and the Presence of Bacteria. Eur. Urol. 2015, 68, 173–174. [Google Scholar] [CrossRef] [PubMed]
- Werneburg, G.T. Catheter-Associated Urinary Tract Infections: Current Challenges and Future Prospects. Res. Rep. Urol. 2022, 14, 109–133. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.; Mediati, D.G.; Duggin, I.G.; Harry, E.J.; Bottomley, A.L. Metabolic adaptations of uropathogenic E. coli in the urinary tract. Front. Cell. Infect. Microbiol. 2017, 7, 241. [Google Scholar] [CrossRef]
- Minardi, D.; d’Anzeo, G.; Cantoro, D.; Conti, A.; Muzzonigro, G. Urinary tract infections in women: Etiology and treatment options. Int. J. Gen. Med. 2011, 4, 333–343. [Google Scholar] [CrossRef]
- Levison, M.E.; Kaye, D. Treatment of complicated urinary tract infections with an emphasis on drug-resistant Gram-negative uropathogens. Curr. Infect. Dis. Rep. 2013, 15, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Petca, R.C.; Popescu, R.I.; Mares, C.; Petca, A.; Mehedintu, C.; Sandu, I.; Maru, N. Antibiotic resistance profile of common uropathogens implicated in urinary tract infections in Romania. Farmacia 2019, 67, 994–1004. [Google Scholar] [CrossRef]
- Bouza, E.; San Juan, R.; Muñoz, P.; Voss, A.; Kluytmans, J.; Co-operative Group of the European Study Group on Nosocomial Infections. A European perspective on nosocomial urinary tract infections I. Report on the microbiology workload, etiology and antimicrobial susceptibility (ESGNI-003 study). European Study Group on Nosocomial Infections. Clin. Microbiol. Infect. 2001, 7, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Chibelean, C.B.; Petca, R.-C.; Mares, C.; Popescu, R.-I.; Eniko, B.; Mehedintu, C.; Petca, A. A clinical perspective on the antimicrobial resistance spectrum of uropathogens in a Romanian male population. Microorganisms 2020, 8, 848. [Google Scholar] [CrossRef] [PubMed]
- Bonkat, G.; Bartoletti, R.; Bruyere, F.; Cai, T.; Geerlings, S.E.; Köves, B.; Schubert, S.; Wagenlehner, F. EAU Guidelines on Urological Infections; European Association of Urology: Arnhem, The Netherlands, 2021. [Google Scholar]
- Gunduz, S.; Uludağ Altun, H. Antibiotic resistance patterns of urinary tract pathogens in Turkish children. Glob. Health Res. Policy 2018, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Patwardhan, V.; Kumar, D.; Goel, V.; Singh, S. Changing prevalence and antibiotic drug resistance pattern of pathogens seen in community-acquired pediatric urinary tract infections at a tertiary care hospital of North India. J. Lab. Physicians 2017, 9, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Praseetha, M.J.; Shantala, G.B.; Ambica, R.; Kusuma, G.R. Trends in Antimicrobial Resistance in Commonly Isolated Uropathogens in a Tertiary Care Hospital Before and During COVID-19. RGUHS Natl. J. Public Health 2022, 7, 135–139. [Google Scholar]
- Mareș, C.; Petca, R.-C.; Petca, A.; Popescu, R.-I.; Jinga, V. Does the COVID Pandemic Modify the Antibiotic Resistance of Uropathogens in Female Patients? A New Storm? Antibiotics 2022, 11, 376. [Google Scholar] [CrossRef] [PubMed]
- Gavi, F.; Fiori, B.; Gandi, C.; Campetella, M.; Bientinesi, R.; Marino, F.; Fettucciari, D.; Rossi, F.; Moretto, S.; Murri, R.; et al. Prevalence and Antimicrobial Resistance Patterns of Hospital Acquired Infections through the COVID-19 Pandemic: Real-Word Data from a Tertiary Urological Centre. J. Clin. Med. 2023, 12, 7278. [Google Scholar] [CrossRef]
- Gawad, A.M.A.; Ashry, W.M.O.; El-Ghannam, S.; Hussein, M.; Yousef, A. Antibiotic resistance profile of common uropathogens during COVID-19 pandemic: Hospital based epidemiologic study. BMC Microbiol. 2023, 23, 28. [Google Scholar] [CrossRef]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Foxman, B. Urinary tract infection syndromes: Occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect. Dis. Clin. N. Am. 2014, 28, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hooton, T.M.; Scholes, D.; Hughes, J.P.; Winter, C.; Roberts, P.L.; Stapleton, A.E.; Stergachis, A.; Stamm, W.E. A prospective study of risk factors for symptomatic urinary tract infection in young women. N. Engl. J. Med. 1996, 335, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Scholes, D.; Hooton, T.M.; Roberts, P.L.; Stapleton, A.E.; Gupta, K.; Stamm, W.E. Risk factors for recurrent urinary tract infection in young women. J. Infect. Dis. 2000, 182, 1177–1182. [Google Scholar] [CrossRef] [PubMed]
- McEwen, S.A.; Collignon, P.J. Antimicrobial resistance: A one health perspective. Microbiol. Spectr. 2017, 6, ARBA-0009. [Google Scholar]
- Knight, G.M.; E Glover, R.; McQuaid, C.F.; Olaru, I.D.; Gallandat, K.; Leclerc, Q.J.; Fuller, N.M.; Willcocks, S.J.; Hasan, R.; van Kleef, E.; et al. Antimicrobial resistance and COVID-19: Intersections and implications. eLife 2021, 10, e64139. [Google Scholar] [CrossRef] [PubMed]
- Metwally, W.S.; Elnagar, W.M. Multidrug resistant uropathogens among Egyptian pregnant women. Am. J. Infect. Dis. 2019, 15, 115–122. [Google Scholar] [CrossRef]
- Labah, E.A.; Afifi, I.K.; Ahmed, L.M.S. Community-acquired urinary tract infections in Tanta Egypt: Aetiology and antibiotic resistance pattern. Ann. Clin. Microbiol. Antimicrob. 2009, 6, 179–190. [Google Scholar]
- Hummers-Pradier, E.; Ohse, A.M.; Koch, M.; Heizmann, W.R.; Kochen, M.M. Management of urinary tract infections in female general practice patients. Fam. Pract. 2005, 22, 71–77. [Google Scholar] [CrossRef]
- Toner, L.; Papa, N.; Aliyu, S.H.; Dev, H.; Lawrentschuk, N.; Al-Hayek, S. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in hospital urinary tract infections: Incidence and antibiotic susceptibility profile over 9 years. World J. Urol. 2016, 34, 1031–1037. [Google Scholar] [CrossRef]
- Tandog, Z.; Bartoletti, R.; Cai, T.; Çek, M.; Grabe, M.; Kulchavenya, E.; Köves, B.; Menon, V.; Naber, K.; Perepanova, T.; et al. Antimicrobial resistance in urosepsis: Outcomes from the multinational, multicenter global prevalence of infections in urology (GPIU) study 2003–2013. World J. Urol. 2016, 34, 1193–1200. [Google Scholar] [CrossRef] [PubMed]
- Mirsoleymani, S.R.; Salimi, M.; Shareghi Brojeni, M.; Ranjbar, M.; Mehtarpoor, M. Bacterial pathogens and antimicrobial resistance patterns in pediatric urinary tract infections: A four-year surveillance study (2009–2012). Int. J. Pediatr. 2014, 2014, 126142. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- El Omari, L.; Sakhi, A.; Miloudi, M.; Elkamouni, Y.; Zouhair, S.; Arsalene, L. The impact of the COVID pandemic on the uropathogenic bacterial resistance profile: Experience of the bacteriology lab of the military hospital Avicenne in Marrakech. GSC Adv. Res. Rev. 2023, 14, 59–65. [Google Scholar] [CrossRef]
- Popa, Z.; Rusu, L.; Susan, R.; Pinzaru, I.; Ardelean, E.; Borcan, F.; Voicu, M.; Sas, I.T.; Popovici, R.A.; Lazureanu, V. Obtaining and characterization of a polyurethane carrier used for eugenol as a possible remedy in oral therapies. Mater. Plast. 2018, 55, 9–13. [Google Scholar] [CrossRef]
- Terreni, M.; Taccani, M.; Pregnolato, M. New Antibiotics for Multidrug-Resistant Bacterial Strains: Latest Research Developments and Future Perspectives. Molecules 2021, 26, 2671. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ciuca, I.M.; Dediu, M.; Popin, D.; Pop, L.L.; Tamas, L.A.; Pilut, C.N.; Almajan Guta, B.; Popa, Z.L. Antibiotherapy in Children with Cystic Fibrosis-An Extensive Review. Children 2022, 9, 1258. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| Variables | n | % |
|---|---|---|
| Background information | ||
| Age (mean ± SD) | 53.6 | 16.1 |
| Men | 120 | 31.7% |
| Women | 258 | 68.3% |
| Comorbidities | ||
| Diabetes | 73 | 19.3% |
| Chronic kidney disease | 31 | 8.2% |
| Hypertension | 140 | 37.0% |
| Cardiovascular disease | 94 | 24.9% |
| Other | 22 | 5.8% |
| Bacteria | ||
| Escherichia coli | 175 | 46.3% |
| Klebsiella pneumoniae | 78 | 20.6% |
| Enterococcus spp. | 38 | 10.1% |
| Pseudomonas aeruginosa | 26 | 6.9% |
| Proteus spp. | 19 | 5.0% |
| Acinetobacter baumanii | 7 | 1.9% |
| Other Gram-negative bacteria | 27 | 7.1% |
| Other Gram-positive bacteria | 8 | 2.1% |
| Total | 378 | 100% |
| 2020 | 2021 | 2022 | ||||||
|---|---|---|---|---|---|---|---|---|
| Bacteriuria <1000 CFU/mL | Mixed flora | UTI | Bacteriuria <1000 CFU/mL | Mixed flora | UTI | Bacteriuria <1000 CFU/mL | Mixed flora | UTI |
| 236 | 184 | 93 | 308 | 253 | 89 | 661 | 452 | 196 |
| Total = 513 | Total = 650 | Total = 1309 | ||||||
| Escherichia coli ESBL | Klebsiella pneumoniae ESBL | Klebsiella pneumoniae carba+ XDR | Pseudomonas aeruginosa carba+ | Acinetobacter baumanii carba+ | Enterococcus spp. VRE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 48 | 27.4% | n = 19 | 24.3% | n = 41 | 52.5% | n = 18 | 69.2% | n = 6 | 85.7% | n = 1 | 2.6% |
| % | Escherichia coli | Klebsiella pneumoniae | ||||||
|---|---|---|---|---|---|---|---|---|
| 2020 | 2021 | 2022 | p-Value | 2020 | 2021 | 2022 | p-Value | |
| Amoxicillin + clavulanic acid | 45.4% | 55.0% | 53.8% | 0.334 | 57.1% | 72.7% | 89.1% | <0.001 * |
| Ceftazidime | 31.8% | 32.5% | 26.3% | 0.579 | 57.1% | 63.6% | 84.7% | <0.001 * |
| Ceftriaxone | 38.6% | 35.0% | 30.7% | 0.502 | 61.9% | 63.6% | 84.7% | <0.001 * |
| Ciprofloxacin | 43.1% | 35.0% | 36.2% | 0.445 | 61.9% | 63.6% | 86.9% | <0.001 * |
| Imipenem | 9.09% | 5.0% | 16.4% | 0.026 * | 14.2% | 54.5% | 80.4% | <0.001 * |
| Trimethoprim/sulfamethoxazole | 31.8% | 27.5% | 47.2% | 0.009 * | 33.3% | 18.1% | 39.1% | 0.004 * |
| Amikacin | 2.2% | 2.5% | 4.3% | 0.642 | 9.5% | 9.09% | 17.3% | 0.131 |
| Gentamicin | 15.9% | 17.5% | 18.8% | 0.863 | 33.3% | 27.2% | 39.1% | 0.203 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hogea, E.; Muntean, A.C.; Bratosin, F.; Bogdan, I.G.; Plavitu, O.; Fratutu, A.; Oancea, C.; Bica, M.C.; Muntean, D.; Hrubaru, I.; et al. Antibiotic Resistance Trends in Uropathogens during the COVID-19 Pandemic in Western Romania: A Cross-Sectional Study. Antibiotics 2024, 13, 512. https://doi.org/10.3390/antibiotics13060512
Hogea E, Muntean AC, Bratosin F, Bogdan IG, Plavitu O, Fratutu A, Oancea C, Bica MC, Muntean D, Hrubaru I, et al. Antibiotic Resistance Trends in Uropathogens during the COVID-19 Pandemic in Western Romania: A Cross-Sectional Study. Antibiotics. 2024; 13(6):512. https://doi.org/10.3390/antibiotics13060512
Chicago/Turabian StyleHogea, Elena, Alexandra Cristina Muntean, Felix Bratosin, Iulia Georgiana Bogdan, Oana Plavitu, Adelina Fratutu, Cristian Oancea, Mihai Calin Bica, Delia Muntean, Ingrid Hrubaru, and et al. 2024. "Antibiotic Resistance Trends in Uropathogens during the COVID-19 Pandemic in Western Romania: A Cross-Sectional Study" Antibiotics 13, no. 6: 512. https://doi.org/10.3390/antibiotics13060512
APA StyleHogea, E., Muntean, A. C., Bratosin, F., Bogdan, I. G., Plavitu, O., Fratutu, A., Oancea, C., Bica, M. C., Muntean, D., Hrubaru, I., Popa, Z. L., & Ilie, A. C. (2024). Antibiotic Resistance Trends in Uropathogens during the COVID-19 Pandemic in Western Romania: A Cross-Sectional Study. Antibiotics, 13(6), 512. https://doi.org/10.3390/antibiotics13060512







