Fighting Emerging Caspofungin-Resistant Candida Species: Mitigating Fks1-Mediated Resistance and Enhancing Caspofungin Efficacy by Chitosan
Abstract
:1. Introduction
2. Results
2.1. Antifungal Susceptibility of Candida Species Isolates
2.2. Detection of Mutations in FKS Genes
2.3. Antifungal Activity of Chitosan
2.4. Chitosan Increases the Susceptibility of Candida Species to Caspofungin
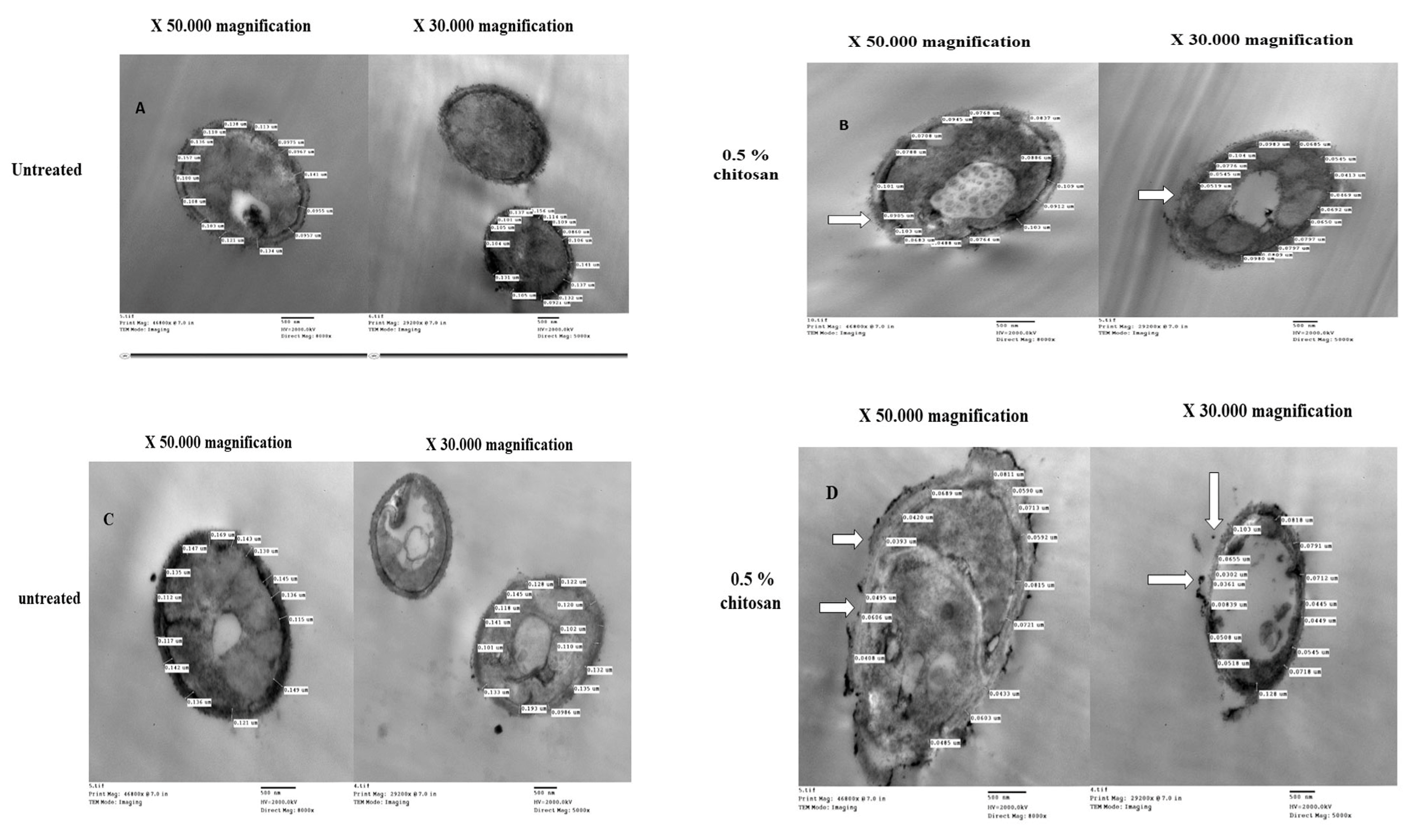
2.5. Chitosan-Induced Changes in Cell Wall Thickness of Caspofungin-Resistant Candida Species
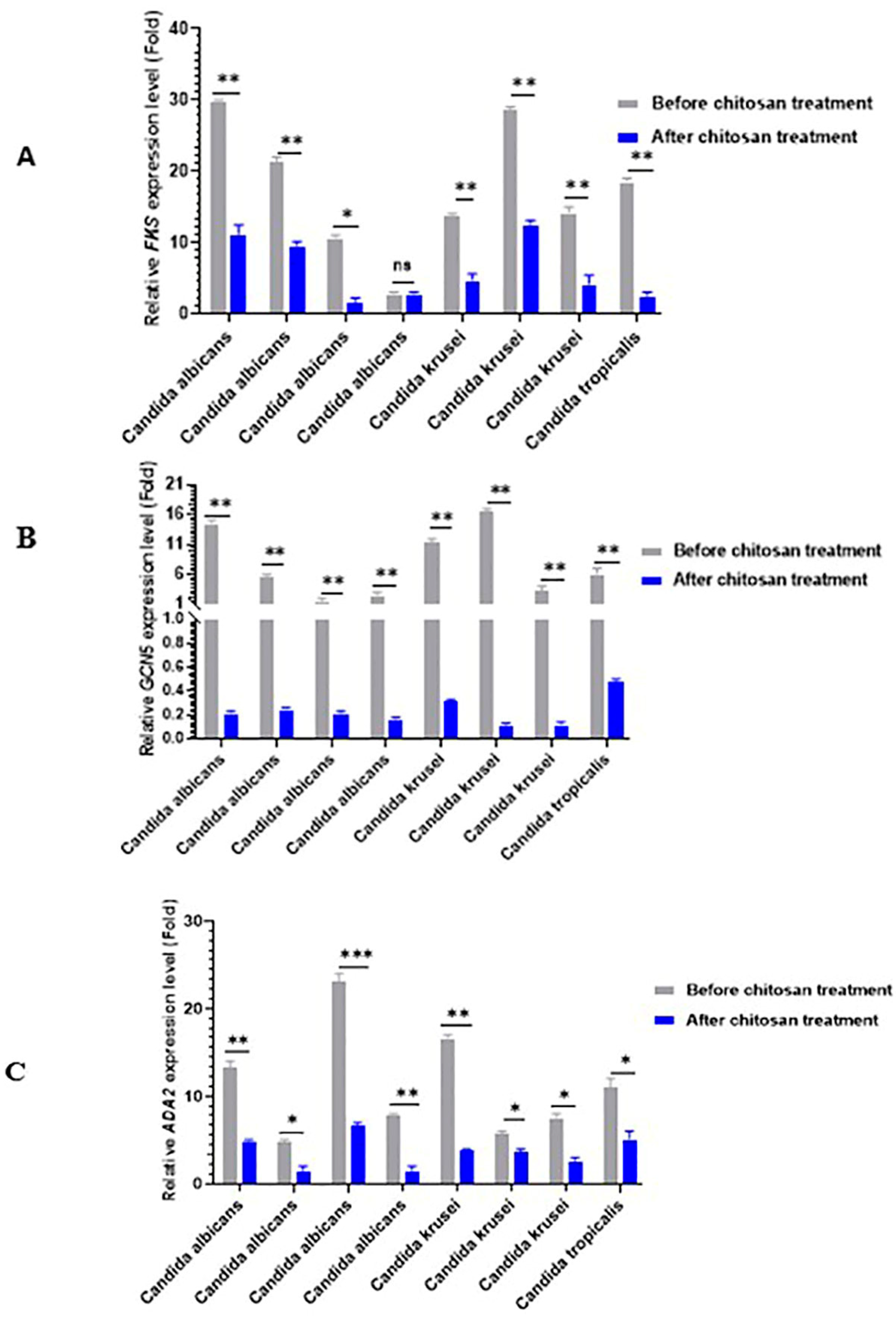
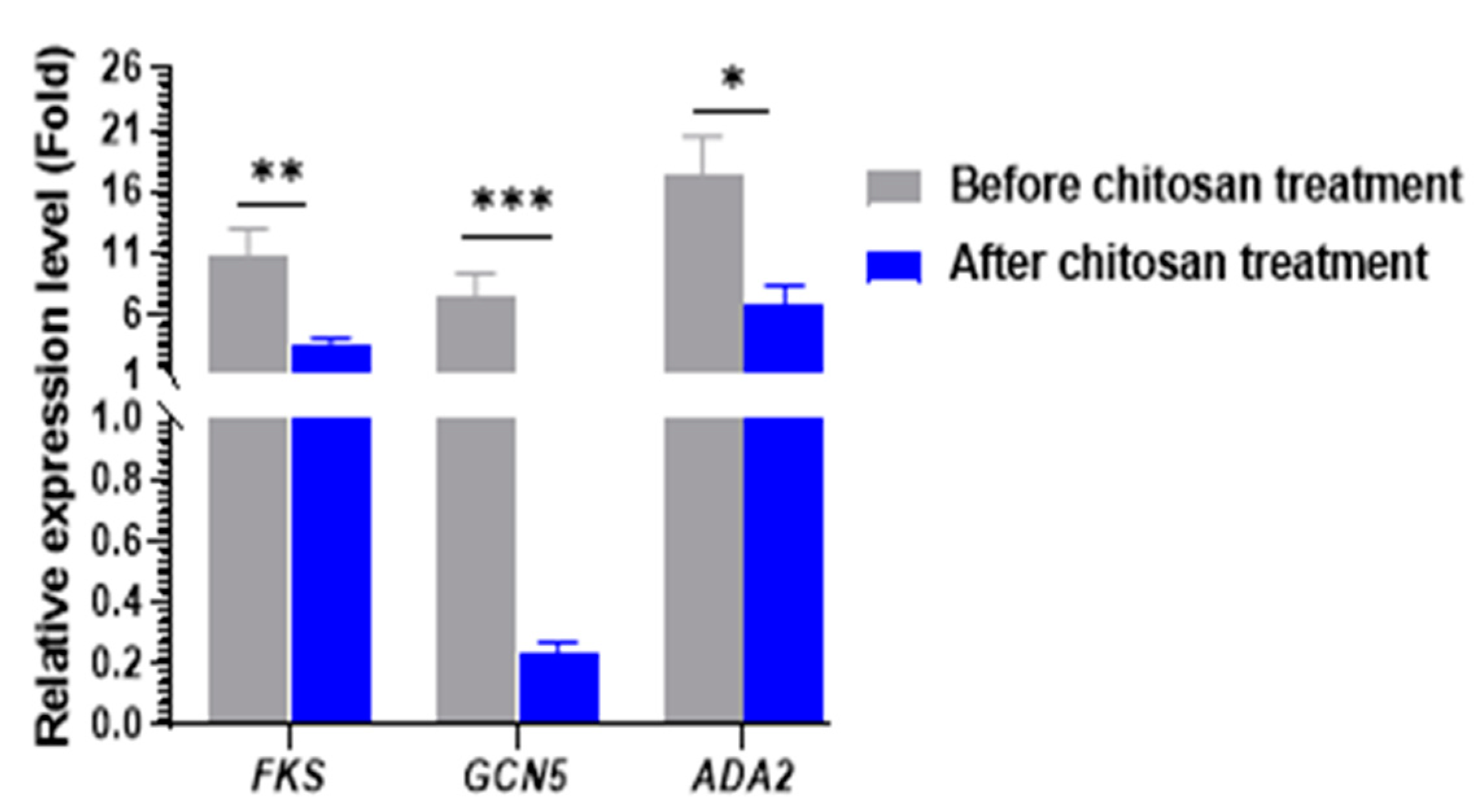
2.6. Real-Time RT-PCR for Gene Expression Analysis
3. Discussion
4. Materials and Methods
4.1. Isolates
4.2. Antifungal Susceptibility Testing
4.3. Determination of Minimum Inhibitory Concentrations
4.4. Sequencing of FKs Gene for Detection of Mutations
4.5. Screening of Antifungal Activity of Chitosan against Candida species Isolates
4.6. Antifungal Susceptibility Testing for Caspofungin after Chitosan Treatment
4.7. Transmission Electron Microscopy Analysis
4.8. Quantitative Reverse Transcription Polymerase Chain Reaction
4.9. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vélez, N.; Argel, A.; Kissmann, A.K.; Alpízar-Pedraza, D.; Escandón, P.; Rosenau, F.; Ständker, L.; Firacative, C. Pore-forming peptide C14R exhibits potent antifungal activity against clinical isolates of Candida albicans and Candida auris. Front. Cell Infect. Microbiol. 2024, 14, 1389020. [Google Scholar] [CrossRef]
- Mboussou, F.; Ndumbi, P.; Ngom, R.; Kassamali, Z.; Ogundiran, O.; Van Beek, J.; Williams, G.; Okot, C.; Hamblion, E.; Impouma, B. Infectious disease outbreaks in the African region: Overview of events reported to the World Health Organization. Epidemiol. Infect. 2019, 147, 299–305. [Google Scholar] [CrossRef]
- Okiro, E.A.; Yahaya, A.A.; Stephen, M.; Bonkoungou, B.; Musa, E.O.; Minkoulou, E.M.; Okeibunor, J.; Impouma, B.; Djingarey, H.M. Spatial and temporal distribution of infectious disease epidemics, disasters and other potential public health emergencies in the World Health Organisation Africa region, 2016. Glob. Health 2020, 16, 1–12. [Google Scholar]
- Tartor, Y.H.; Elmowalid, G.A.; Hassan, M.N.; Shaker, A.; Ashour, D.F.; Saber, T. Promising Anti-Biofilm Agents and Phagocytes Enhancers for the Treatment of Candida albicans Biofilm-Associated Infections. Front. Cell Infect. Microbiol. 2022, 12, 72–80. [Google Scholar] [CrossRef]
- Arastehfar, A.; Gabaldón, T.; Garcia-Rubio, R.; Jenks, J.D.; Hoenigl, M.; Salzer, H.J.F.; Ilkit, M.; Lass-Flörl, C.; Perlin, D.S. Drug-Resistant Fungi: An Emerging Challenge Threatening Our Limited Antifungal Armamentarium. Antibiotics 2020, 9, 877. [Google Scholar] [CrossRef]
- Campoy, S.; Adrio, J.L. Antifungals. Biochem. Pharmacol. 2017, 133, 86–96. [Google Scholar] [CrossRef]
- Szymański, M.; Chmielewska, S.; Czyżewska, U.; Malinowska, M.; Tylicki, A. Echinocandins—Structure, mechanism of action and use in antifungal therapy. J. Enzyme Inhib. Med. Chem. 2022, 37, 876–894. [Google Scholar] [CrossRef]
- Perlin, D.S. Echinocandin Resistance in Candida. Clin. Infect. Dis. 2015, 61 (Suppl. S6), 612–617. [Google Scholar] [CrossRef]
- Perlin, D.S. Mechanisms of echinocandin antifungal drug resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 1–11. [Google Scholar] [CrossRef]
- Frazer, C.; Staples, M.I.; Kim, Y.; Hirakawa, M.; Dowell, M.A.; Johnson, N.V.; Hernday, A.D.; Ryan, V.H.; Fawzi, N.L.; Finkelstein, I.J.; et al. Epigenetic cell fate in Candida albicans is controlled by transcription factor condensates acting at super-enhancer-like elements. Nat. Microbiol. 2020, 5, 1374–1378. [Google Scholar] [CrossRef]
- Mallick, E.M.; Bergeron, A.C.; Jones, S.K.; Newman, Z.R.; Brothers, K.M.; Creton, R.; Wheeler, R.T.; Bennett, R.J. Phenotypic Plasticity Regulates Candida albicans Interactions and Virulence in the Vertebrate Host. Front. Microbiol. 2016, 7, 780. [Google Scholar] [CrossRef]
- Conte, M.; Eletto, D.; Pannetta, M.; Petrone, A.M.; Monti, M.C.; Cassiano, C.; Giurato, G.; Rizzo, F.; Tessarz, P.; Petrella, A.; et al. Effects of Hst3p inhibition in Candida albicans: A genome-wide H3K56 acetylation analysis. Front. Cell Infect. Microbiol. 2022, 12, 1031814. [Google Scholar] [CrossRef]
- Yu, S.; Paderu, P.; Lee, A.; Eirekat, S.; Healey, K.; Chen, L.; Perlin, D.S.; Zhao, Y. Histone Acetylation Regulator Gcn5 Mediates Drug Resistance and Virulence of Candida glabrata. Microbiol. Spectr. 2022, 10, e0096322. [Google Scholar] [CrossRef]
- Eberharter, A.; Sterner, D.E.; Schieltz, D.; Hassan, A.; Yates, J.R., 3rd; Berger, S.L.; Workman, J.L. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell Biol. 1999, 19, 6621–6631. [Google Scholar] [CrossRef]
- Helmlinger, D.; Papai, G.; Devys, D.; Tora, L. What do the structures of GCN5-containing complexes teach us about their function? BBA J. 2021, 1864, 194614. [Google Scholar] [CrossRef]
- Shih, P.Y.; Liao, Y.T.; Tseng, Y.K.; Deng, F.S.; Lin, C.H. A Potential Antifungal Effect of Chitosan Against Candida albicans Is Mediated via the Inhibition of SAGA Complex Component Expression and the Subsequent Alteration of Cell Surface Integrity. Front. Microbiol. 2019, 10, 602. [Google Scholar] [CrossRef]
- Iracane, E.; Vega-Estévez, S.; Buscaino, A. On and Off: Epigenetic Regulation of C. albicans Morphological Switches. Pathogens 2021, 10, 1463. [Google Scholar] [CrossRef]
- Afroz, M.M.; Kashem, M.N.H.; Piash, K.; Islam, N. Saccharomyces Cerevisiae as an Untapped Source of Fungal Chitosan for Antimicrobial Action. Appl. Biochem. Biotechnol. 2021, 193, 3765–3786. [Google Scholar] [CrossRef]
- Zakrzewska, A.; Boorsma, A.; Brul, S.; Hellingwerf, K.J.; Klis, F.M. Transcriptional response of Saccharomyces cerevisiae to the plasma membrane-perturbing compound chitosan. Eukaryot. Cell 2005, 4, 703–715. [Google Scholar] [CrossRef]
- Shields, R.K.; Nguyen, M.H.; Press, E.G.; Kwa, A.L.; Cheng, S.; Du, C.; Clancy, C.J. The presence of an FKS mutation rather than MIC is an independent risk factor for failure of echinocandin therapy among patients with invasive candidiasis due to Candida glabrata. Antimicrob. Agents Chemother. 2012, 56, 4862–4869. [Google Scholar]
- Pogurschi, E.N.; Petcu, C.D.; Mizeranschi, A.E.; Zugravu, C.A.; Cirnatu, D.; Pet, I.; Ghimpețeanu, O.M. Knowledge, Attitudes and Practices Regarding Antibiotic Use and Antibiotic Resistance: A Latent Class Analysis of a Romanian Population. Int. J. Environ. Res. Public Health 2022, 19, 7263. [Google Scholar] [CrossRef] [PubMed]
- de Prado, R.S.; Alves, R.J.; Oliveira, C.M.A.d.; Kato, L.; Silva, R.A.d.; Quintino, G.O.; do Desterro Cunha, S.; de Almeida Soares, C.M.; Pereira, M. Inhibition of Paracoccidioides lutzii Pb 01 isocitrate lyase by the natural compound argentilactone and its semi-synthetic derivatives. PLoS ONE 2014, 9, 94832–94838. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, N.K.; Ramakrishnan, J. In vitro evaluation of antimicrobial activity of crude extracts of medicinal plants against multi drug resistant pathogens. Biyol. Bilim. Araptyrma Derg. 2009, 2, 97–101. [Google Scholar]
- Badiee, P.; Alborzi, A. Susceptibility of clinical Candida species isolates to antifungal agents by E-test, Southern Iran: A five-year study. Iran. J. Microbiol. 2011, 3, 183–188. [Google Scholar] [PubMed]
- Ben-Ami, R. Treatment of Invasive Candidiasis: A Narrative Review. J. Fungi 2018, 4, 97. [Google Scholar] [CrossRef] [PubMed]
- Coste, A.T.; Kritikos, A.; Li, J.; Khanna, N.; Goldenberger, D.; Garzoni, C.; Zehnder, C.; Boggian, K.; Neofytos, D.; Riat, A.; et al. Emerging echinocandin-resistant Candida albicans and glabrata in Switzerland. Infection 2020, 48, 761–766. [Google Scholar] [CrossRef]
- Dannaoui, E.; Desnos-Ollivier, M.; Garcia-Hermoso, D.; Grenouillet, F.; Cassaing, S.; Baixench, M.T.; Bretagne, S.; Dromer, F.; Lortholary, O. Candida spp. with acquired echinocandin resistance, France, 2004. Emerg. Infect. Dis. 2012, 18, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.; Chaudhary, S.; Saxena, R.K.; Talwar, S.; Yadav, S. Evaluation of Antimicrobial and Antifungal efficacy of Chitosan as endodontic irrigant against Enterococcus faecalis and Candida albicans Biofilm formed on tooth substrate. J. Clin. Exp. Dent. 2017, 9, e361–e367. [Google Scholar] [CrossRef] [PubMed]
- Ewais, A.H.; Raslan, M.; Hassanien, R.A.; Zedan, I. Nanoparticles and irradiated chitosan impact on the quality of sweet green bell pepper (Capsicum annuum L.) under cold storage conditions. Arab J. Nucl. Sci. Appl. 2022, 30, 1–12. [Google Scholar]
- Lo, W.H.; Deng, F.S.; Chang, C.J.; Lin, C.H. Synergistic Antifungal Activity of Chitosan with Fluconazole against Candida albicans, Candida tropicalis, and Fluconazole-Resistant Strains. Molecules 2020, 25, 5114. [Google Scholar] [CrossRef]
- Cheung, R.C.F.; Ng, T.B.; Wong, J.H.; Chan, W.Y. Chitosan: An update on potential biomedical and pharmaceutical applications. Mar. Drugs 2015, 13, 5156–5186. [Google Scholar] [CrossRef]
- Kong, M.; Chen, X.G.; Xing, K.; Park, H.J. Antimicrobial properties of chitosan and mode of action: A state of the art review. Int. J. Food Microbiol. 2010, 144, 51–63. [Google Scholar] [CrossRef]
- Poznanski, P.; Hameed, A.; Orczyk, W. Chitosan and chitosan nanoparticles: Parameters enhancing antifungal activity. Molecules 2023, 28, 2996. [Google Scholar] [CrossRef] [PubMed]
- Rabea, E.I.; Badawy, M.E.; Stevens, C.V.; Smagghe, G.; Steurbaut, W. Chitosan as antimicrobial agent: Applications and mode of action. J. Biol. Macromol. 2003, 4, 1457–1465. [Google Scholar] [CrossRef]
- Rathinam, S.; Solodova, S.; Kristjánsdóttir, I.; Hjálmarsdóttir, M.; Másson, M. The antibacterial structure-activity relationship for common chitosan derivatives. Int. J. Biol. Macromol. 2020, 165, 1686–1693. [Google Scholar] [CrossRef]
- Younes, I.; Sellimi, S.; Rinaudo, M.; Jellouli, K.; Nasri, M. Influence of acetylation degree and molecular weight of homogeneous chitosans on antibacterial and antifungal activities. Int. J. Food Microbiol. 2014, 185, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Garcia, L.G.S.; Guedes, G.M.d.M.; da Silva, M.L.Q.; Castelo-Branco, D.S.C.M.; Sidrim, J.J.C.; Cordeiro, R.d.A.; Rocha, M.F.G.; Vieira, R.S.; Brilhante, R.S.N. Effect of the molecular weight of chitosan on its antifungal activity against Candida spp. in planktonic cells and biofilm. Carbohydr. Polym. 2018, 195, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Garcia, L.G.S.; de Melo Guedes, G.M.; Fonseca, X.M.Q.C.; Pereira-Neto, W.A.; Castelo-Branco, D.S.C.M.; Sidrim, J.J.C.; de Aguiar Cordeiro, R.; Rocha, M.F.G.; Vieira, R.S.; Brilhante, R.S.N. Antifungal activity of different molecular weight chitosans against planktonic cells and biofilm of Sporothrix brasiliensis. Int. J. Biol. Macromol. 2020, 143, 341–348. [Google Scholar] [CrossRef]
- Yuan, G.; Lv, H.; Tang, W.; Zhang, X.; Sun, H. Effect of chitosan coating combined with pomegranate peel extract on the quality of Pacific white shrimp during iced storage. Food Control. 2016, 59, 818–823. [Google Scholar] [CrossRef]
- Tsai, G.-J.; Su, W.-H. Antibacterial Activity of Shrimp Chitosan against Escherichia coli. J. Food Prot. 1999, 62, 239–243. [Google Scholar] [CrossRef]
- Másson, M.; Holappa, J.; Hjálmarsdóttir, M.; Rúnarsson, Ö.V.; Nevalainen, T.; Järvinen, T. Antimicrobial activity of piperazine derivatives of chitosan. Carbohydr. Polym. 2008, 74, 566–571. [Google Scholar] [CrossRef]
- Ke, C.L.; Deng, F.S.; Chuang, C.Y.; Lin, C.H. Antimicrobial actions and applications of chitosan. Polymers 2021, 13, 904. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.G.; Specht, C.A.; Donlin, M.J.; Lodge, J.K. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot. Cell 2007, 6, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Peña, A.; Sánchez, N.S.; Calahorra, M. Effects of chitosan on Candida albicans: Conditions for its antifungal activity. Biomed. Res. Int. 2013, 2013, 5–15. [Google Scholar] [CrossRef]
- De Cesare, G.B.; Hafez, A.; Stead, D.; Llorens, C.; Munro, C.A. Biomarkers of caspofungin resistance in Candida albicans isolates: A proteomic approach. Virulence 2022, 13, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- Aranda-Martinez, A.; Lopez-Moya, F.; Lopez-Llorca, L.V. Cell wall composition plays a key role on sensitivity of filamentous fungi to chitosan. J. Basic. Microbiol. 2016, 56, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- Verlee, A.; Mincke, S.; Stevens, C.V. Recent developments in antibacterial and antifungal chitosan and its derivatives. Carbohydr. Polym. 2017, 164, 268–283. [Google Scholar] [CrossRef]
- Näär, A.M.; Lemon, B.D.; Tjian, R. Transcriptional coactivator complexes. Annu. Rev. Biochem. 2001, 70, 475–501. [Google Scholar] [CrossRef]
- Sellam, A.; Askew, C.; Epp, E.; Lavoie, H.; Whiteway, M.; Nantel, A. Genome-wide mapping of the coactivator AdA2p yields insight into the functional roles of SAGA/ADA complex in Candida albicans. J. Mol. Cell Biol. 2009, 20, 2389–2400. [Google Scholar] [CrossRef]
- Marcus, G.A.; Silverman, N.; Berger, S.L.; Horiuchi, J.; Guarente, L. Functional similarity and physical association between GCN5 and ADA2: Putative transcriptional adaptors. EMBO J. 1994, 13, 4807–4815. [Google Scholar]
- Yien, L.; Zin, N.M.; Sarwar, A.; Katas, H. Antifungal activity of chitosan nanoparticles and correlation with their physical properties. Int. J. Biomater. 2012, 632698. [Google Scholar] [CrossRef]
- Shivarathri, R.; Tscherner, M.; Zwolanek, F.; Singh, N.K.; Chauhan, N.; Kuchler, K. The fungal histone acetyl transferase Gcn5 controls virulence of the human pathogen Candida albicans through multiple pathways. Sci. Rep. 2019, 9, 9445–9450. [Google Scholar] [CrossRef] [PubMed]
- Bader, O.; Weig, M.; Taverne-Ghadwal, L.; Lugert, R.; Gross, U.; Kuhns, M. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 2011, 17, 1359–1365. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Method for Antifungal Disk Diffusion Susceptibility Testing of Yeasts, (M44-ED3), 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- CLSI. Performance Standards for Antifungal Susceptibility Testing of Yeasts, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022. [Google Scholar]
- NCCLS. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 2nd ed.; NCCLS: Wayne, PA, USA, 2002. [Google Scholar]
- Thompson, G.R., III; Wiederhold, N.P.; Vallor, A.C.; Villareal, N.C.; Lewis, J.S.; Patterson, T.F. Development of caspofungin resistance following prolonged therapy for invasive candidiasis secondary to Candida glabrata infection. Antimicrob. Agents Chemother. 2008, 52, 3783–3785. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Bhattacharyya, S.; Gupta, P.; Banerjee, G.; Singh, M. Comparative Analysis of Disc Diffusion and E-test with Broth Micro-dilution for Susceptibility Testing of Clinical Candida Isolates Against Amphotericin B, Fluconazole, Voriconazole and Caspofungin. J. Clin. Diagn. Res. 2015, 9, Dc01-4. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Muñoz, R.; Avalos-Borja, M.; Castro-Longoria, E. Ultrastructural analysis of Candida albicans when exposed to silver nanoparticles. PLoS ONE 2014, 9, e108876. [Google Scholar]
- Chang, W.-H.; Liang, S.-H.; Deng, F.-S.; Lin, C.-H. The conserved dual phosphorylation sites of the Candida albicans Hog1 protein are crucial for white–opaque switching, mating, and pheromone-stimulated cell adhesion. Sabouraudia 2016, 54, 628–640. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y. Statistical notes for clinical researchers: Assessing normal distribution (1). Restor. Dent. Endod. 2012, 37, 245–248. [Google Scholar] [CrossRef]
- Cary, N. SAS/STAT statistics user’s guide. In Statistical Analytical System, 5th ed.; SAS Institute Inc.: Cary, NC, USA, 2012; pp. 1–10. [Google Scholar]
- Desnos-Ollivier, M.; Bretagne, S.; Raoux, D.; Hoinard, D.; Dromer, F.; Dannaoui, E. Mutations in the fks1 gene in Candida albicans, C. tropicalis, and C. krusei correlate with elevated caspofungin MICs uncovered in AM3 medium using the method of the European Committee on Antibiotic Susceptibility Testing. Antimicrob. Agents Chemother. 2008, 52, 3092–3098. [Google Scholar] [CrossRef]
- Khan, Z.; Ahmad, S.; Mokaddas, E.; Meis, J.F.; Joseph, L.; Abdullah, A.; Vayalil, S. Development of Echinocandin Resistance in Candida tropicalis following Short-Term Exposure to Caspofungin for Empiric Therapy. Antimicrob. Agents Chemother. 2018, 62, 201–211. [Google Scholar] [CrossRef] [PubMed]

| Isolate No. | Species | Source | Resistance Pattern | Caspofungin MIC (µg/mL) | Caspofungin MIC after 0.5% Chitosan Treatment (µg/mL) |
|---|---|---|---|---|---|
| C1 | C. albicans | Calf, Diarrhea | FLZ, CLT, TERB, KTZ, ITC, CAS | 2 | 0.25 |
| C2 | C. krusei | Urine, Human | FLZ, CLT, KTZ, ITC, TERB, MCZ VRZ, CAS | 4 | 0.5 |
| C3 | C. krusei | Onychomycosis, Human | FLZ, CLT, ITC, AMB, MCZ, VRZ, TERB, CAS | 8 | 0.25 |
| C4 | C. albicans | Urine, Human | FLZ, CLT, AMB, TERB, KTZ, ITC, MCZ, VRZ, CAS | 2 | 1 |
| C5 | C. krusei | Urine, Human | FLZ, AMB, ITC, TERB, VRZ, CAS | 8 | 0.125 |
| C6 | C. albicans | Calf, diarrhea | FLZ, CLT, TERB, KTZ, ITC, VRZ, CAS | 2 | 0.0625 |
| C7 | C. krusei | Calf, diarrhea | FLZ, CLT, KTZ, CAS | 2 | 0.25 |
| C8 | C. albicans | Urine, Human | FLZ, CLT, AMB, KTZ, TERB, VRZ, MCZ, CASP | 4 | 0.25 |
| C9 | C. tropicalis | Sputum, Human | CLT, AMB, TERB, KTZ, ITC, VRZ, MCZ, CAS | 8 | 0.125 |
| C10 | C. albicans | Urine, Human | FLZ, CLT, KTZ, ITC, VRZ, MCZ | 0.25 | 0.125 |
| C11 | C. albicans | Onychomycosis, Human | FLZ, CLT, TERB, KTZ, AMB | 0.125 | 0.0625 |
| C12 | C. krusei | Urine, Human | FLZ, CLT, VRZ, MCZ, AMB, TERB | 0.25 | 0.125 |
| C13 | C. krusei | Urine, Human | FLZ, CLT, TERB, KTZ, ITC, VRZ, MCZ | 0.25 | 0.125 |
| C14 | C. krusei | Urine, Human | FLZ, TERB, KTZ, MCZ | 0.125 | 0.0625 |
| C15 | C. krusei | Onychomycosis, Human | FLZ, CLT, KTZ, TERB, MCZ | 0.25 | 0.125 |
| C16 | C. krusei | Vaginal swab, Human | FLZ, CLT, KTZ | 0.125 | 0.065 |
| C17 | C. krusei | Urine, Human | FLZ, CLT, TERB, KTZ, ITC, MCZ | 0.25 | 0.125 |
| C18 | C. albicans | Calf, diarrhea | CLT, AMB, KTZ, ITC, MCZ | 0.0625 | 0.03125 |
| C19 | C. krusei | Sputum, Human | AMB, KTZ, MCZ | 0.125 | 0.0625 |
| C20 | C. krusei | Calf, diarrhea | AMB, ITC, MCZ | 0.0625 | 0.03125 |
| C21 | C. krusei | Calf, diarrhea | FLZ, AMB, KTZ | 0.125 | 0.03125 |
| C22 | C. krusei | Human, Urine | FLZ, CLT, TERB, KT, VRZ | 0.25 | 0.03125 |
| C23 | C. albicans | Urine, Human | CLT, AMB, TERB, KTZ, ITC | 0.25 | 0.0625 |
| C24 | C. albicans | Vaginal swab, Human | FLZ, AMB, KTZ, ITC, VRZ | 0.125 | 0.0625 |
| C25 | C. krusei | Urine, Human | CLT, TERB, KTZ, ITC | 0.25 | 0.0625 |
| C26 | C. krusei | Onychomycosis, Human | FLZ, CLT, KTZ, ITC | 0.25 | 0.125 |
| C27 | C. albicans | Onychomycosis, Human | KTZ, ITC, VRZ, MCZ | 0.125 | 0.03125 |
| C28 | C. krusei | Urine, Human | CLT, AmB, KTZ, ITC, MCZ | 0.0625 | 0.03125 |
| C29 | C. krusei | Calf, Diarrhea | AmB, TERB | 0.25 | 0.0625 |
| C30 | C. albicans | Calf, Diarrhea | AmB, KTZ, MCZ | 0.125 | 0.03125 |
| C31 | C. albicans | Calf, Diarrhea | KTZ, TERB, MCZ | 0.25 | 0.0625 |
| C32 | C. albicans | Calf, Diarrhea | TERB, AMP, KTZ | 0.25 | 0.0625 |
| C33 | C. albicans | Vaginal swab, Human | AmB, TERB, KTZ, ITC | 0.0625 | 0.03125 |
| C34 | C. krusei | Sputum, Human | AmB, TERB, KTZ, ITC | 0.25 | 0.125 |
| C35 | C. krusei | Urine, Human | FLZ, MCZ, AmB | 0.125 | 0.0325 |
| Isolates No. | Species | Caspofungin MIC (µg/mL) | Caspofungin MIC after 0.5% Chitosan Treatment | Mutation (s) in FKS1 | Amino Acid Change | Accession No. |
|---|---|---|---|---|---|---|
| C1 | C. albicans | 2 | 0.25 | A1929T T1933C | S645P | PP663620 |
| C2 | C. krusei | 4 | 0.5 | C1934T | S645F | PP663621 |
| C3 | C. krusei | 8 | 0.25 | A1929T A1988G | E663G | PP663622 |
| C4 | C. albicans | 2 | 1 | T1922C | F641S | PP663623 |
| C5 | C. krusei | 8 | 0.125 | A1929T T2062A, A2064G | L688M | PP663624 |
| C6 | C. albicans | 2 | 0.0625 | C1934A | S645Y | PP663625 |
| C7 | C. krusei | 2 | 0.25 | T1933C | S645P | PP663626 |
| C8 | C. albicans | 4 | 0.25 | T1933C | S645P | PP663627 |
| C9 | C. tropicalis | 8 | 0.125 | G1932T | L644F | PP663628 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tarek, A.; Tartor, Y.H.; Hassan, M.N.; Pet, I.; Ahmadi, M.; Abdelkhalek, A. Fighting Emerging Caspofungin-Resistant Candida Species: Mitigating Fks1-Mediated Resistance and Enhancing Caspofungin Efficacy by Chitosan. Antibiotics 2024, 13, 578. https://doi.org/10.3390/antibiotics13070578
Tarek A, Tartor YH, Hassan MN, Pet I, Ahmadi M, Abdelkhalek A. Fighting Emerging Caspofungin-Resistant Candida Species: Mitigating Fks1-Mediated Resistance and Enhancing Caspofungin Efficacy by Chitosan. Antibiotics. 2024; 13(7):578. https://doi.org/10.3390/antibiotics13070578
Chicago/Turabian StyleTarek, Aya, Yasmine H. Tartor, Mohamed N. Hassan, Ioan Pet, Mirela Ahmadi, and Adel Abdelkhalek. 2024. "Fighting Emerging Caspofungin-Resistant Candida Species: Mitigating Fks1-Mediated Resistance and Enhancing Caspofungin Efficacy by Chitosan" Antibiotics 13, no. 7: 578. https://doi.org/10.3390/antibiotics13070578







