High Biofilm-Forming Multidrug-Resistant Salmonella Infantis Strains from the Poultry Production Chain
Abstract
1. Introduction
2. Results
2.1. Biofilm Quantification
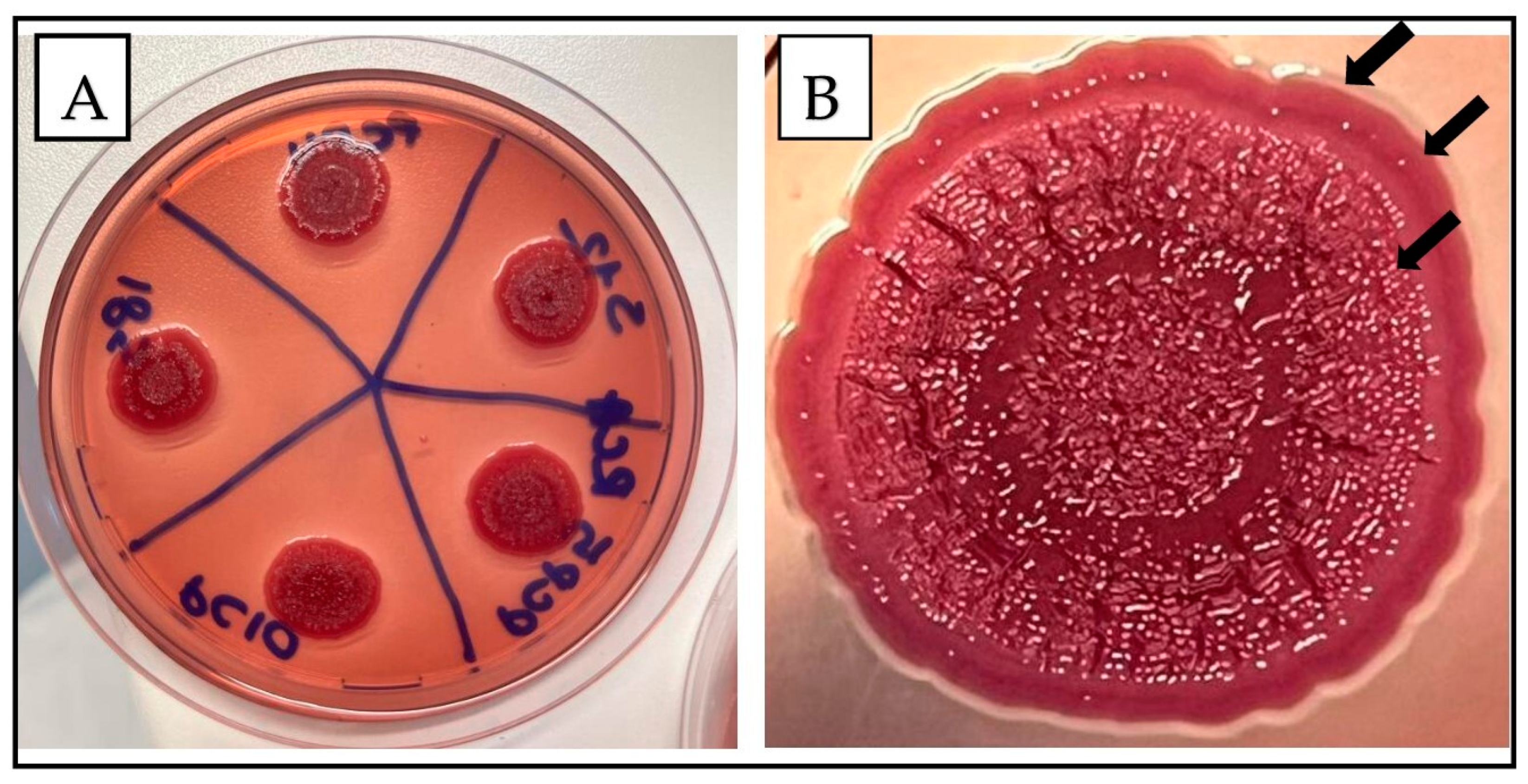
2.2. Morphotype Evaluation
2.3. Molecular Characterization
3. Discussion
4. Materials and Methods
4.1. Collection and Isolate Identification
4.2. Bacterial Cell Adhesion Analysis
4.2.1. Microtiter Plate Assessment
4.2.2. Biofilm Quantification Analysis
4.3. S. Infantis Morphotype (Colony Morphology and Cellulose Production)
4.4. Molecular Characterization
4.4.1. Detection of Biofilm Genes
4.4.2. Whole Genome Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Antunes, P.; Mourão, J.; Campos, J.; Peixe, L. Salmonellosis: The Role of Poultry Meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M.; for the International Collaboration on Enteric Disease “Burden of Illness” Studies. The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [PubMed]
- National Salmonella Surveillance|National Surveillance|CDC. Available online: https://www.cdc.gov/nationalsurveillance/salmonella-surveillance.html (accessed on 15 May 2024).
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, e07666. [Google Scholar] [CrossRef]
- Alba, P.; Leekitcharoenphon, P.; Carfora, V.; Amoruso, R.; Cordaro, G.; Di Matteo, P.; Ianzano, A.; Iurescia, M.; Diaconu, E.L.; ENGAGE-EURL-AR Network Study Group; et al. Molecular Epidemiology of Salmonella Infantis in Europe: Insights into the Success of the Bacterial Host and Its Parasitic pESI-like Megaplasmid. Microb. Genom. 2020, 6, e000365. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, J.; Mallick, S.; Mishra, N.; Patel, S.; Pradhan, J.; Negi, V.D. Chapter 26—Salmonella Biofilm and Its Importance in the Pathogenesis. In Understanding Microbial Biofilms; Das, S., Kungwani, N.A., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 447–459. ISBN 978-0-323-99977-9. [Google Scholar]
- Casagrande Proietti, P.; Musa, L.; Stefanetti, V.; Orsini, M.; Toppi, V.; Branciari, R.; Blasi, F.; Magistrali, C.F.; Capomaccio, S.; Kika, T.S.; et al. Mcr-1-Mediated Colistin Resistance and Genomic Characterization of Antimicrobial Resistance in ESBL-Producing Salmonella Infantis Strains from a Broiler Meat Production Chain in Italy. Antibiotics 2022, 11, 728. [Google Scholar] [CrossRef] [PubMed]
- The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2017. EFSA J. 2019, 17, e05598. [CrossRef]
- Harrell, J.E.; Hahn, M.M.; D’Souza, S.J.; Vasicek, E.M.; Sandala, J.L.; Gunn, J.S.; McLachlan, J.B. Salmonella Biofilm Formation, Chronic Infection, and Immunity within the Intestine and Hepatobiliary Tract. Front. Cell. Infect. Microbiol. 2021, 10, 624622. [Google Scholar] [CrossRef] [PubMed]
- Araújo, D.; Silva, A.R.; Fernandes, R.; Serra, P.; Barros, M.M.; Campos, A.M.; Oliveira, R.; Silva, S.; Almeida, C.; Castro, J. Emerging Approaches for Mitigating Biofilm-Formation-Associated Infections in Farm, Wild, and Companion Animals. Pathogens 2024, 13, 320. [Google Scholar] [CrossRef] [PubMed]
- Giaouris, E.; Nesse, L. Attachment of Salmonella Spp. to Food Contact and Product Surfaces and Biofilm Formation on Them as Stress Adaptation and Survival Strategies. In Salmonella: Prevalence, Risk Factors and Treatment Options; Nova Science Publishers: Hauppauge, NY, USA, 2015; pp. 111–136. ISBN 978-1-63463-651-3. [Google Scholar]
- Simões, M.; Simões, L.C.; Machado, I.; Pereira, M.O.; Vieira, M.J. Control of Flow-Generated Biofilms with Surfactants: Evidence of Resistance and Recovery. Food Bioprod. Process. 2006, 84, 338–345. [Google Scholar] [CrossRef]
- Baugh, S.; Ekanayaka, A.S.; Piddock, L.J.V.; Webber, M.A. Loss of or Inhibition of All Multidrug Resistance Efflux Pumps of Salmonella enterica Serovar Typhimurium Results in Impaired Ability to Form a Biofilm. J. Antimicrob. Chemother. 2012, 67, 2409–2417. [Google Scholar] [CrossRef]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of Bacterial Efflux Pumps in Biofilm Formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020. [Google Scholar] [CrossRef] [PubMed]
- Römling, U. Characterization of the Rdar Morphotype, a Multicellular Behaviour in Enterobacteriaceae. CMLS Cell. Mol. Life Sci. 2005, 62, 1234–1246. [Google Scholar] [CrossRef] [PubMed]
- Simm, R.; Ahmad, I.; Rhen, M.; Le Guyon, S.; Römling, U. Regulation of Biofilm Formation in Salmonella enterica Serovar Typhimurium. Future Microbiol. 2014, 9, 1261–1282. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.; Stoodley, P. Bacterial Biofilms: From the Natural Environment to Infectious Diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Tang, Q.; Ding, Y.; Tan, P.; Zhang, Y.; Wang, T.; Zhou, C.; Xu, S.; Lyu, M.; Bai, Y.; et al. Staphylococcus Aureus and Biofilms: Transmission, Threats, and Promising Strategies in Animal Husbandry. J. Anim. Sci. Biotechnol. 2024, 15, 44. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, P.; Belluco, S.; Mancin, M.; Losasso, C.; Ricci, A. Genes Conferring Resistance to Critically Important Antimicrobials in Salmonella enterica Isolated from Animals and Food: A Systematic Review of the Literature, 2013–2017. Res. Vet. Sci. 2019, 126, 59–67. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control (ECDC); European Food Safety Authority (EFSA); European Medicines Agency (EMA). Third Joint Inter-Agency Report on Integrated Analysis of Consumption of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Humans and Food-Producing Animals in the EU/EEA. EFSA J. 2021, 19, e06712. [Google Scholar] [CrossRef]
- Alvarez, D.M.; Barrón-Montenegro, R.; Conejeros, J.; Rivera, D.; Undurraga, E.A.; Moreno-Switt, A.I. A Review of the Global Emergence of Multidrug-Resistant Salmonella enterica Subsp. Enterica Serovar Infantis. Int. J. Food Microbiol. 2023, 403, 110297. [Google Scholar] [CrossRef]
- Kalaba, V.; Golić, B.; Sladojević, Ž.; Kalaba, D. Incidence of Salmonella Infantis in Poultry Meat and Products and the Resistance of Isolates to Antimicrobials. IOP Conf. Ser. Earth Environ. Sci. 2017, 85, 012082. [Google Scholar] [CrossRef]
- Pate, M.; Mičunovič, J.; Golob, M.; Vestby, L.K.; Ocepek, M. Salmonella Infantis in Broiler Flocks in Slovenia: The Prevalence of Multidrug Resistant Strains with High Genetic Homogeneity and Low Biofilm-Forming Ability. Biomed. Res. Int. 2019, 2019, 4981463. [Google Scholar] [CrossRef]
- Lamas, A.; Fernandez-No, I.C.; Miranda, J.M.; Vázquez, B.; Cepeda, A.; Franco, C.M. Biofilm Formation and Morphotypes of Salmonella enterica Subsp. Arizonae Differs from Those of Other Salmonella enterica Subspecies in Isolates from Poultry Houses. J. Food Prot. 2016, 79, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Karaca, B.; Akcelik, N.; Akcelik, M. Biofilm-Producing Abilities of Salmonella Strains Isolated from Turkey. Biologia 2013, 68, 1–10. [Google Scholar] [CrossRef]
- Schonewille, E.; Nesse, L.L.; Hauck, R.; Windhorst, D.; Hafez, H.M.; Vestby, L.K. Biofilm Building Capacity of Salmonella enterica Strains from the Poultry Farm Environment. FEMS Immunol. Med. Microbiol. 2012, 65, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Galié, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the Food Industry: Health Aspects and Control Methods. Front. Microbiol. 2018, 9, 315815. [Google Scholar] [CrossRef] [PubMed]
- Moraes, J.O.; Cruz, E.A.; Souza, E.G.F.; Oliveira, T.C.M.; Alvarenga, V.O.; Peña, W.E.L.; Sant’Ana, A.S.; Magnani, M. Predicting Adhesion and Biofilm Formation Boundaries on Stainless Steel Surfaces by Five Salmonella enterica Strains Belonging to Different Serovars as a Function of pH, Temperature and NaCl Concentration. Int. J. Food Microbiol. 2018, 281, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.D.N.; Yang, Y.S.; Yuk, H.G. Biofilm Formation of Salmonella Typhimurium on Stainless Steel and Acrylic Surfaces as Affected by Temperature and pH Level. LWT Food Sci. Technol. 2014, 55, 383–388. [Google Scholar] [CrossRef]
- Stepanović, S.; Ćirković, I.; Mijač, V.; Švabić-Vlahović, M. Influence of the Incubation Temperature, Atmosphere and Dynamic Conditions on Biofilm Formation by Salmonella Spp. Food Microbiol. 2003, 20, 339–343. [Google Scholar] [CrossRef]
- Zogaj, X.; Nimtz, M.; Rohde, M.; Bokranz, W.; Römling, U. The Multicellular Morphotypes of Salmonella Typhimurium and Escherichia Coli Produce Cellulose as the Second Component of the Extracellular Matrix. Mol. Microbiol. 2001, 39, 1452–1463. [Google Scholar] [CrossRef] [PubMed]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G.; Choroszy-Krol, I. Virulence Factors, Prevalence and Potential Transmission of Extraintestinal Pathogenic Escherichia Coli Isolated from Different Sources: Recent Reports. Gut. Pathog. 2019, 11, 10. [Google Scholar] [CrossRef]
- Hedlund, M.; Wachtler, C.; Johansson, E.; Hang, L.; Somerville, J.E.; Darveau, R.P.; Svanborg, C. P Fimbriae-Dependent, Lipopolysaccharide-Independent Activation of Epithelial Cytokine Responses. Mol. Microbiol. 1999, 33, 693–703. [Google Scholar] [CrossRef]
- Yan, C.-H.; Chen, F.-H.; Yang, Y.-L.; Zhan, Y.-F.; Herman, R.A.; Gong, L.-C.; Sheng, S.; Wang, J. The Transcription Factor CsgD Contributes to Engineered Escherichia Coli Resistance by Regulating Biofilm Formation and Stress Responses. Int. J. Mol. Sci. 2023, 24, 13681. [Google Scholar] [CrossRef] [PubMed]
- Azam, M.W.; Khan, A.U. CRISPRi-Mediated Suppression of E. Coli Nissle 1917 Virulence Factors: A Strategy for Creating an Engineered Probiotic Using csgD Gene Suppression. Front. Nutr. 2022, 9, 938989. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Frye, J.G.; McClelland, M.; Harshey, R.M. Gene Expression Patterns during Swarming in Salmonella Typhimurium: Genes Specific to Surface Growth and Putative New Motility and Pathogenicity Genes. Mol. Microbiol. 2004, 52, 169–187. [Google Scholar] [CrossRef]
- Li, W.; Li, Y.; Liu, Y.; Shi, X.; Jiang, M.; Lin, Y.; Qiu, Y.; Zhang, Q.; Chen, Q.; Zhou, L.; et al. Clonal Expansion of Biofilm-Forming Salmonella Typhimurium ST34 with Multidrug-Resistance Phenotype in the Southern Coastal Region of China. Front. Microbiol. 2017, 8, 2090. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, M.; Xu, L.; Shi, C.; Shi, X. Characterization of Antibiotic Resistance Genes, Plasmids, Biofilm Formation, and In Vitro Invasion Capacity of Salmonella Enteritidis Isolates from Children with Gastroenteritis. Microb. Drug Resist. 2019, 25, 1191–1198. [Google Scholar] [CrossRef]
- Narasanna, R.; Chavadi, M.; Oli, A.; Chandrakanth, K. Effect of Subinhibitory Concentration of Cefetoxime on Biofilm Formation. J. Microbiol. Infect. Dis. 2017, 7. [Google Scholar] [CrossRef]
- Lorusso, A.B.; Carrara, J.A.; Barroso, C.D.N.; Tuon, F.F.; Faoro, H. Role of Efflux Pumps on Antimicrobial Resistance in Pseudomonas Aeruginosa. Int. J. Mol. Sci. 2022, 23, 15779. [Google Scholar] [CrossRef] [PubMed]
- Proietti, P.C.; Stefanetti, V.; Musa, L.; Zicavo, A.; Dionisi, A.M.; Bellucci, S.; Mensa, A.L.; Menchetti, L.; Branciari, R.; Ortenzi, R.; et al. Genetic Profiles and Antimicrobial Resistance Patterns of Salmonella Infantis Strains Isolated in Italy in the Food Chain of Broiler Meat Production. Antibiotics 2020, 9, 814. [Google Scholar] [CrossRef] [PubMed]
- Pasteur Institut; WHO. WHO Collaborating Centre for Reference and Research on Salmonella. Available online: https://www.pasteur.fr/sites/default/files/veng_0.pdf (accessed on 1 June 2024).
- Stepanović, S.; Cirković, I.; Ranin, L.; Svabić-Vlahović, M. Biofilm Formation by Salmonella spp. and Listeria monocytogenes on Plastic Surface. Lett. Appl. Microbiol. 2004, 38, 428–432. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Cirković, I.; Ruzicka, F. Quantification of Biofilm in Microtiter Plates: Overview of Testing Conditions and Practical Recommendations for Assessment of Biofilm Production by Staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Karaca, B.; Buzrul, S.; Tato, V.; Akçelik, N.; Akçelik, M. Modeling and Predicting the Biofilm Formation of Different Salmonella Strains. J. Food Saf. 2013, 33, 503–508. [Google Scholar] [CrossRef]
- Yin, L.; Dai, Y.; Chen, H.; He, X.; Ouyang, P.; Huang, X.; Sun, X.; Ai, Y.; Lai, S.; Zhu, L.; et al. Cinnamaldehyde Resist Salmonella Typhimurium Adhesion by Inhibiting Type I Fimbriae. Molecules 2022, 27, 7753. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Liao, X.; Li, C.-Z.; Abdel-Samie, M.; Cui, H. Inhibitory Effect of Cold Nitrogen Plasma on Salmonella Typhimurium Biofilm and Its Application on Poultry Egg Preservation. LWT 2020, 126, 109340. [Google Scholar] [CrossRef]
- Wang, H.; Dong, Y.; Wang, G.; Xu, X.; Zhou, G. Effect of Growth Media on Gene Expression Levels in Salmonella Typhimurium Biofilm Formed on Stainless Steel Surface. Food Control 2016, 59, 546–552. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]

| Target Genes | Sequence of Primers (5′–3′) | Product Length | References |
|---|---|---|---|
| csgD | F: TCCTGGTCTTCAGTAGCGTAA R: TATGATGGAAGCGGATAAGAA | 168 bp | [48] |
| csgB | F: ATCAGGCGGCCATTATTGGTCAAG R: TGCTGTTTTCTGCGTACCGTACTG | 275 bp | [47] |
| fimA | F: TGCCTTTCTCCATCGTCC R: TGCGGTAGTGCTATTGTCC | 134 bp | [46] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Musa, L.; Toppi, V.; Stefanetti, V.; Spata, N.; Rapi, M.C.; Grilli, G.; Addis, M.F.; Di Giacinto, G.; Franciosini, M.P.; Casagrande Proietti, P. High Biofilm-Forming Multidrug-Resistant Salmonella Infantis Strains from the Poultry Production Chain. Antibiotics 2024, 13, 595. https://doi.org/10.3390/antibiotics13070595
Musa L, Toppi V, Stefanetti V, Spata N, Rapi MC, Grilli G, Addis MF, Di Giacinto G, Franciosini MP, Casagrande Proietti P. High Biofilm-Forming Multidrug-Resistant Salmonella Infantis Strains from the Poultry Production Chain. Antibiotics. 2024; 13(7):595. https://doi.org/10.3390/antibiotics13070595
Chicago/Turabian StyleMusa, Laura, Valeria Toppi, Valentina Stefanetti, Noah Spata, Maria Cristina Rapi, Guido Grilli, Maria Filippa Addis, Giacomo Di Giacinto, Maria Pia Franciosini, and Patrizia Casagrande Proietti. 2024. "High Biofilm-Forming Multidrug-Resistant Salmonella Infantis Strains from the Poultry Production Chain" Antibiotics 13, no. 7: 595. https://doi.org/10.3390/antibiotics13070595
APA StyleMusa, L., Toppi, V., Stefanetti, V., Spata, N., Rapi, M. C., Grilli, G., Addis, M. F., Di Giacinto, G., Franciosini, M. P., & Casagrande Proietti, P. (2024). High Biofilm-Forming Multidrug-Resistant Salmonella Infantis Strains from the Poultry Production Chain. Antibiotics, 13(7), 595. https://doi.org/10.3390/antibiotics13070595










