Isolation and Characterization of Two Novel Genera of Jumbo Bacteriophages Infecting Xanthomonas vesicatoria Isolated from Agricultural Regions in Mexico
Abstract
1. Introduction
2. Results
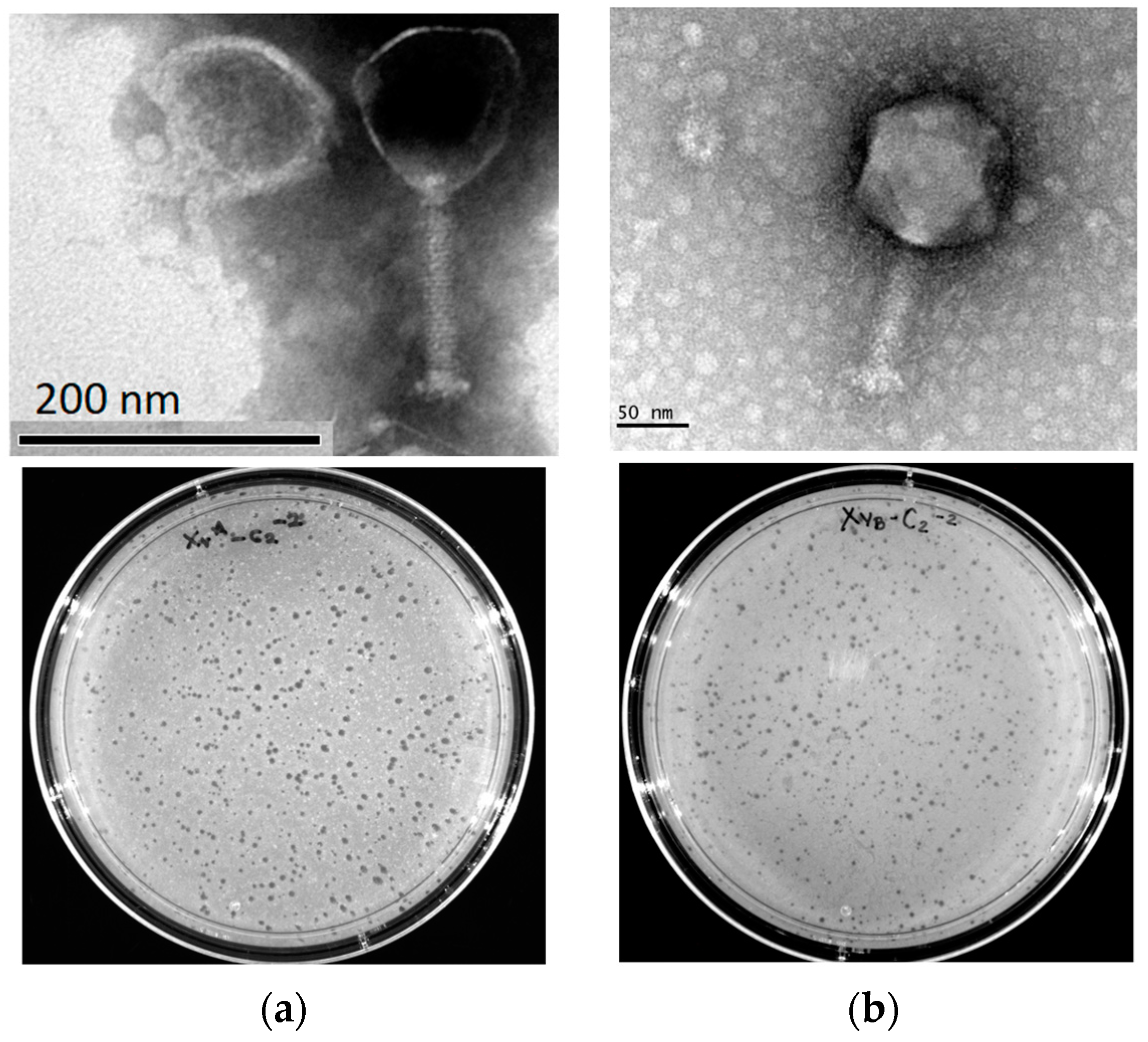
2.1. Isolation and Morphology of X. vesicatoria Phages
2.2. Host Range in X. vesicatoria, X. euvesicatoria pv. euvesivatoria and X. hortorum pv. garneri
2.3. Restriction Analysis of Phage
2.4. General Genome Features
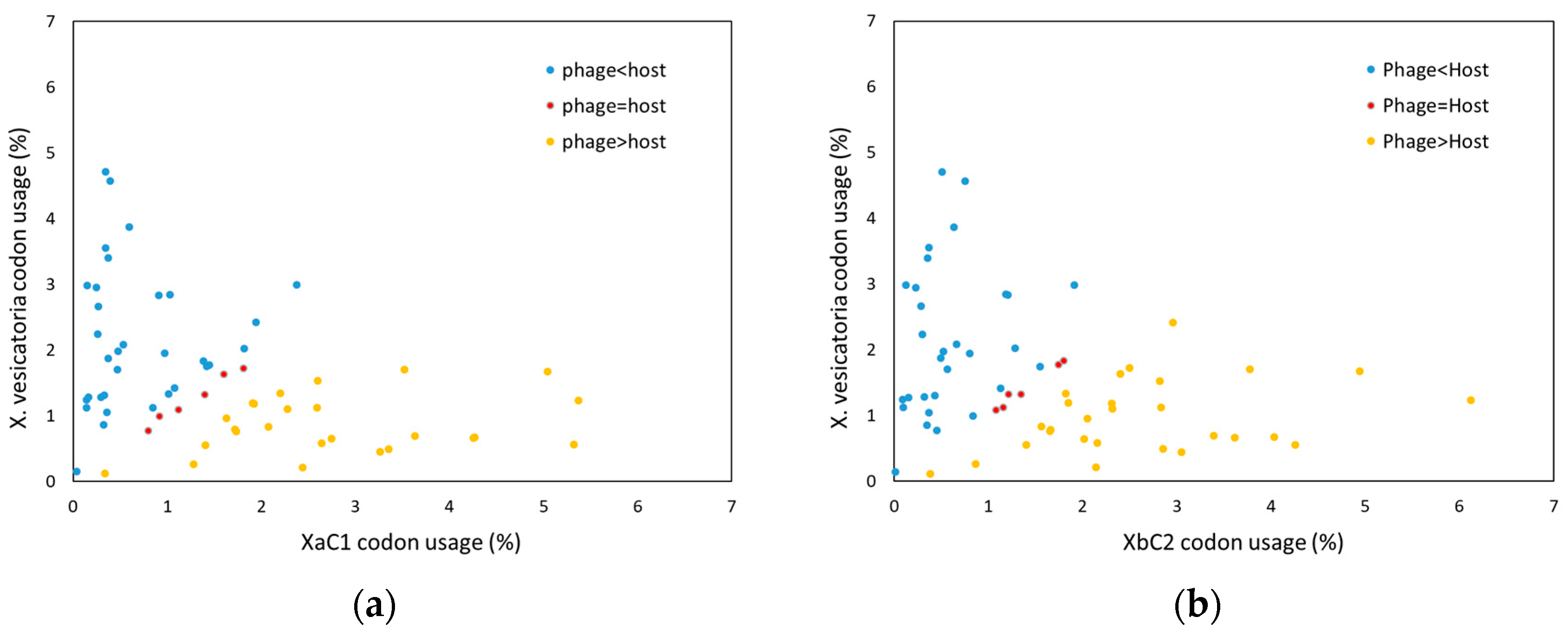
2.5. Detailed Analysis of Putative ORFs
2.5.1. Structural Genes, DNA Packaging and Lysis
2.5.2. DNA Metabolism, Replication, Repair and Recombination
2.5.3. DNA Cleavage and Modification
2.5.4. Transcription, Translation and tRNA Processing
2.5.5. Miscellaneous Functions
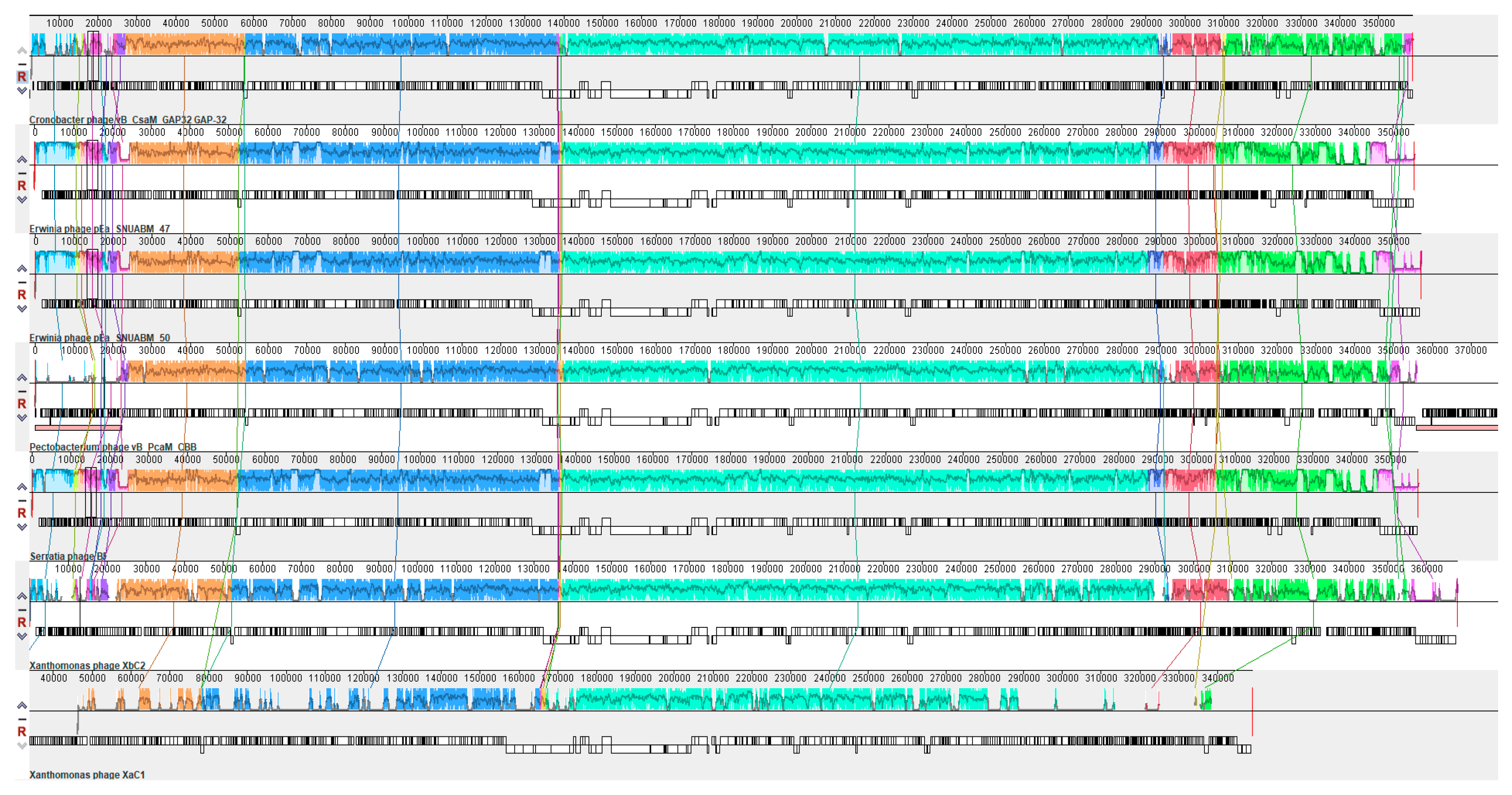
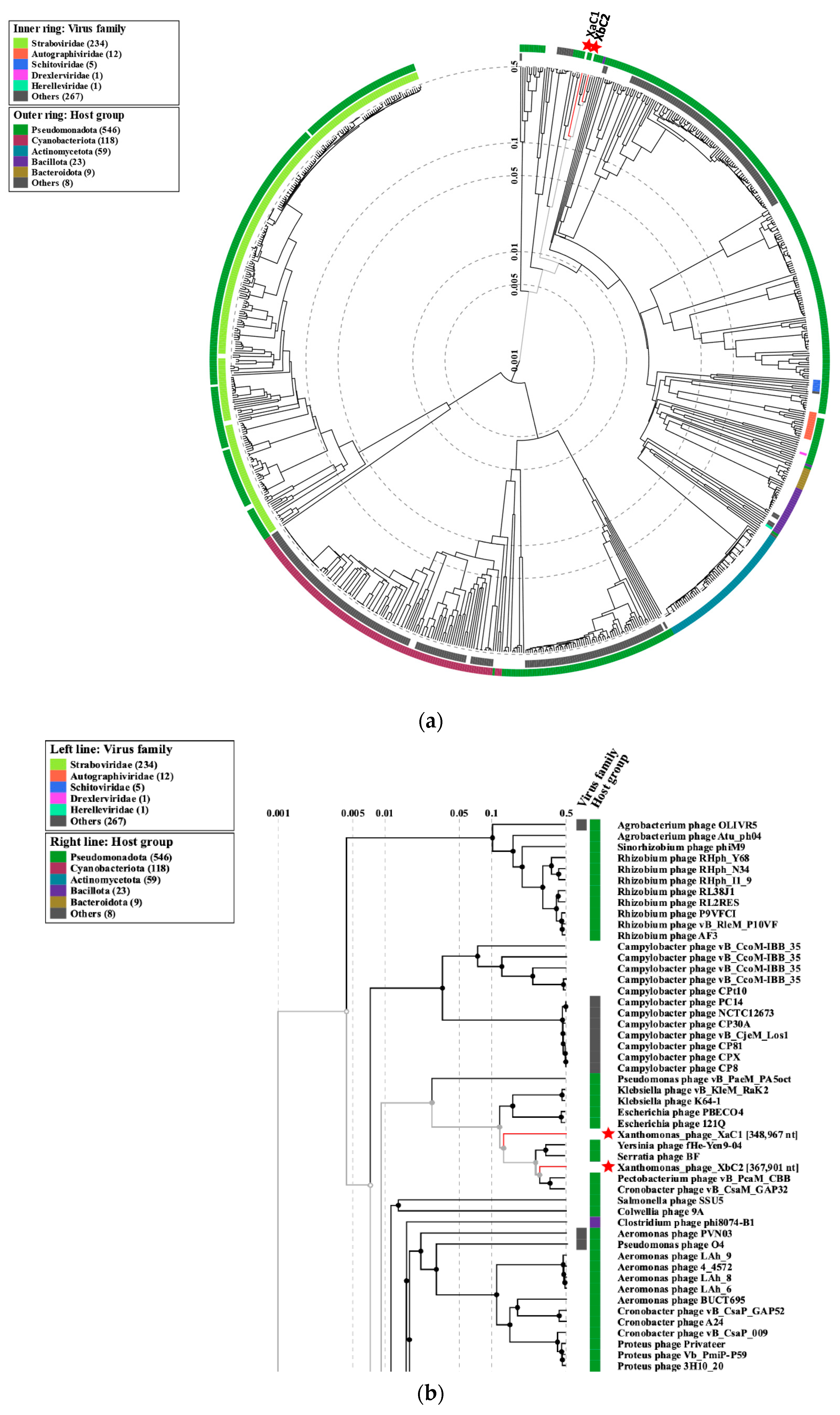
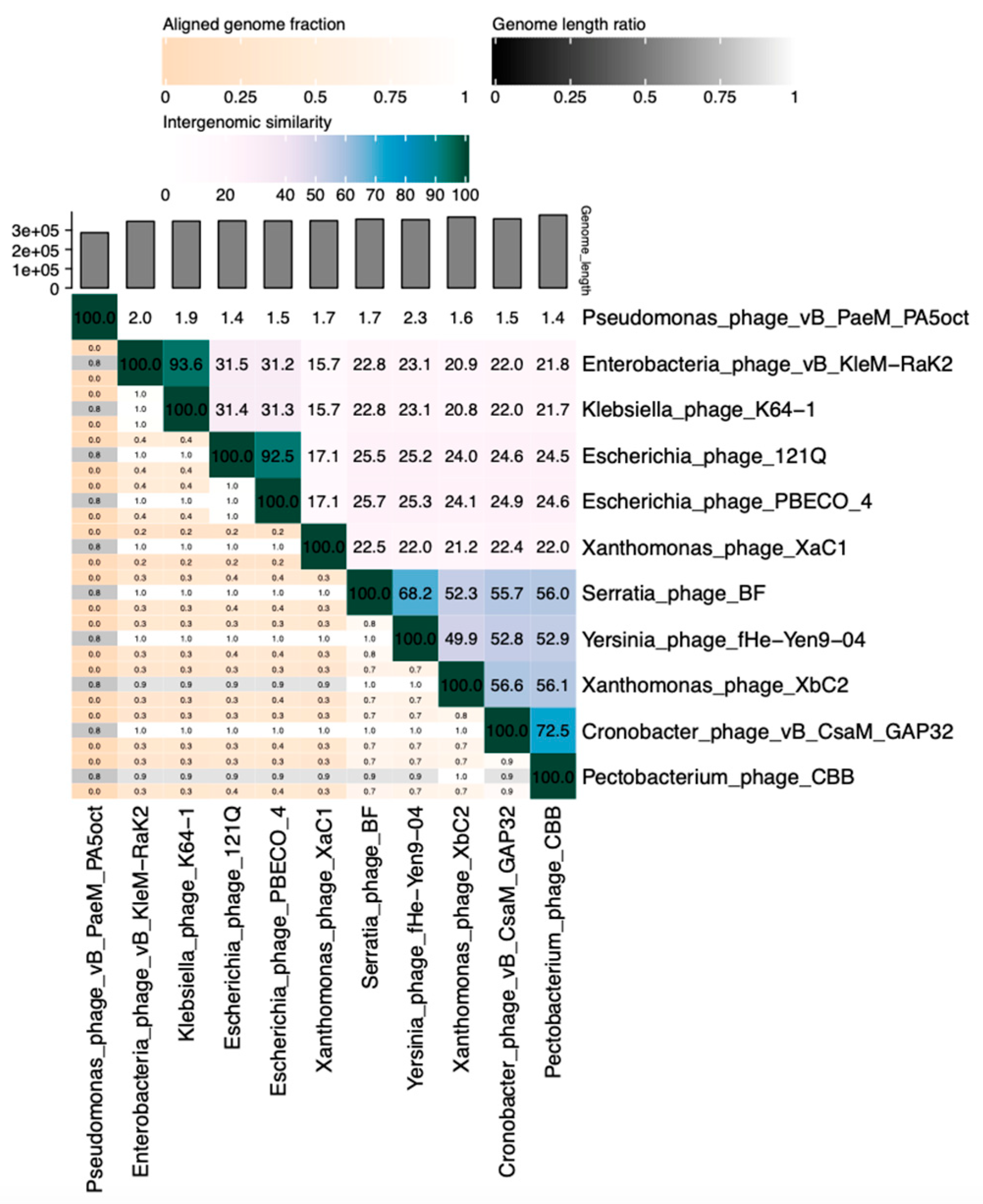
2.6. Phylogenetic and Comparative Analysis
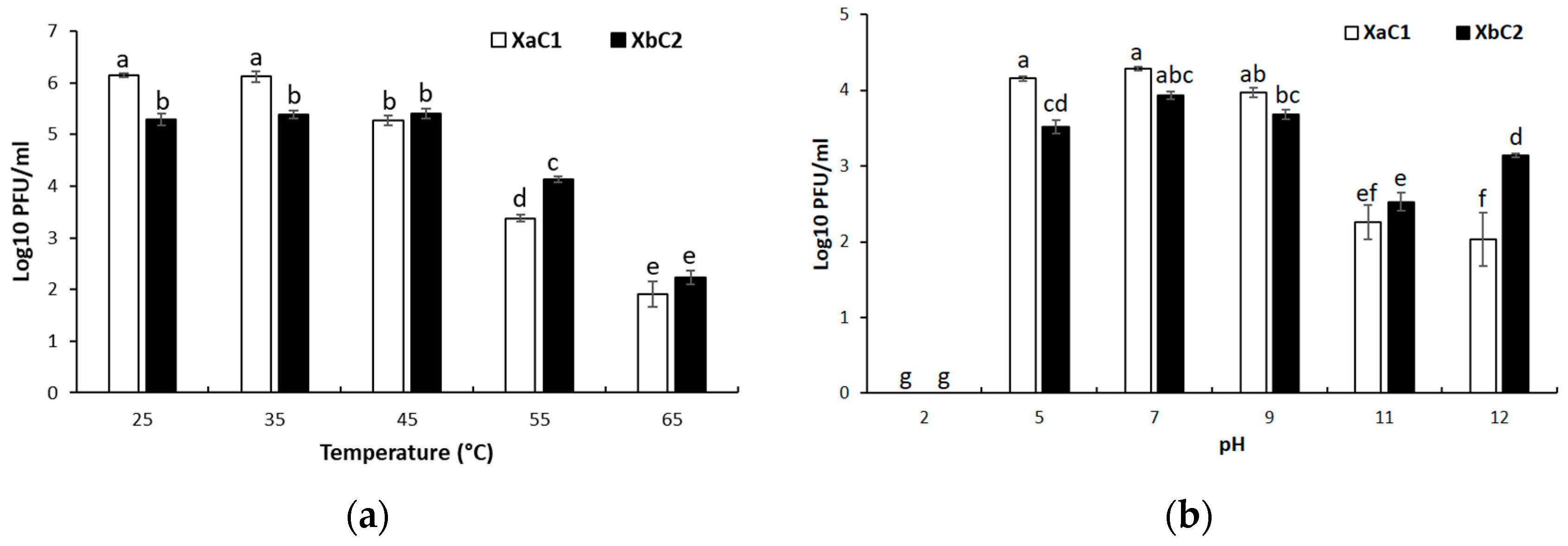
2.7. Thermal and pH Stability Assays
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Isolation and Purification of Phages
4.3. Propagation and Quantification of Phages
4.4. Transmission Electron Microscopy
4.5. Host Range
4.6. DNA Extraction and Restriction Analysis
4.7. Genome Sequencing, Assembly and Annotation
4.8. Phylogenetic and Comparative Genomics
4.9. Thermal and pH Stability Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nakayinga, R.; Makumi, A.; Tumuhaise, V.; Tinzaara, W. Xanthomonas bacteriophages: A review of their biology and biocontrol applications in agricultures. BMC Microbiol. 2021, 21, 291. [Google Scholar] [CrossRef]
- An, S.Q.; Potnis, N.; Dow, M.; Vorhölter, F.J.; He, Y.Q.; Becker, A.; Teper, D.; Li, Y.; Wang, N.; Bleris, L.; et al. Mechanistic insights into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas. FEMS Microbiol. Rev. 2020, 44, 1–32. [Google Scholar] [CrossRef]
- Constantin, E.C.; Cleenwerck, I.; Maes, M.; Baeyen, S.; Van Malderghem, C.; De Vos, P.; Cottyn, B. Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathol. 2016, 65, 792–806. [Google Scholar] [CrossRef]
- Morinière, L.; Burlet, A.; Rosenthal, E.R.; Nesme, X.; Portier, P.; Bull, C.T.; Lavire, C.; Fischer-Le Saux, M.; Bertolla, F. Clarifying the taxonomy of the causal agent of bacterial leaf spot of lettuce through a polyphasic approach reveals that Xanthomonas cynarae Trébaol et al. 2000 emend. Timilsina et al. 2019 is a later heterotypic synonym of Xanthomonas hortorum Vauterin et al. 1995. Syst. Appl. Microbiol. 2020, 43, 126087. [Google Scholar]
- Ritchie, D.F. Bacterial spot of pepper and tomato. Plant Health Instr. 2000. Available online: https://www.apsnet.org/edcenter/disandpath/prokaryote/pdlessons/Pages/Bacterialspot.aspx (accessed on 5 July 2024). [CrossRef]
- Strayer-Scherer, A.; Liao, Y.Y.; Abrahamian, P.; Timilsina, S.; Paret, M.; Momol, T.; Jones, J.; Vallad, G. Integrated Management of Bacterial Spot on Tomato in Florida; Plant Pathology Department, UF/IFAS Extension: Gainesville, FL, USA, 2019; pp. 1–8. [Google Scholar]
- Soto-Caro, A.; Vallad, G.E.; Xavier, K.V.; Abrahamian, P.; Wu, F.; Guan, Z. Managing Bacterial Spot of Tomato: Do Chemical Controls Pay Off? Agronomy 2023, 13, 972. [Google Scholar] [CrossRef]
- Adhikari, P.; Adhikari, T.B.; Louws, F.J.; Panthee, D.R. Advances and Challenges in Bacterial Spot Resistance Breeding in Tomato (Solanum lycopersicum L.). Int. J. Mol. Sci. 2020, 21, 1734. [Google Scholar] [CrossRef]
- Rodriguez, H.; Aguilar, L.; Lao, M. Variations in Xanthan production by antibiotic resistant mutants of Xanthomonas campestris. App. Microbiol. Biotech. 1997, 48, 626–629. [Google Scholar] [CrossRef]
- Herbert, A.; Hancock, C.N.; Cox, B.; Schnabel, G.; Moreno, D.; Carvalho, R.; Jones, J.; Paret, M.; Geng, X.; Wang, H. Oxytetracycline and Streptomycin Resistance Genes in Xanthomonas arboricola pv. pruni, the Causal Agent of Bacterial Spot in Peach. Front. Microbiol. 2022, 13, 821808. [Google Scholar] [CrossRef] [PubMed]
- Brives, C.; Pourraz, J. Phage therapy as a potential solution in the fight against AMR: Obstacles and possible futures. Palgrave Commun. 2020, 6, 100. [Google Scholar] [CrossRef]
- Villalpando-Aguilar, J.L.; Matos-Pech, G.; López-Rosas, I.; Castelán-Sánchez, H.G.; Alatorre-Cobos, F. Phage Therapy for Crops: Concepts, Experimental and Bioinformatics Approaches to Direct Its Application. Int. J. Mol. Sci. 2022, 24, 325. [Google Scholar] [CrossRef]
- Stefani, E.; Obradović, A.; Gašić, K.; Altin, I.; Nagy, I.K.; Kovács, T. Bacteriophage-Mediated Control of Phytopathogenic Xanthomonads: A Promising Green Solution for the Future. Microorganisms 2021, 9, 1056. [Google Scholar] [CrossRef]
- Gašić, K.; Kuzmanović, N.; Ivanović, M.; Prokić, A.; Šević, M.; Obradović, A. Complete Genome of the Xanthomonas euvesicatoria Specific Bacteriophage KΦ1, Its Survival and Potential in Control of Pepper Bacterial Spot. Front. Microbiol. 2018, 9, 2021. [Google Scholar] [CrossRef]
- Kizheva, Y.; Urshev, Z.; Dimitrova, M.; Bogatzevska, N.; Moncheva, P.; Hristova, P. Phenotypic and Genotypic Characterization of Newly Isolated Xanthomonas euvesicatoria-Specific Bacteriophages and Evaluation of Their Biocontrol Potential. Plants 2023, 12, 947. [Google Scholar] [CrossRef]
- Balogh, B.; Nga, N.T.T.; Jones, J.B. Relative Level of Bacteriophage Multiplication in vitro or in Phyllosphere May Not Predict in planta Efficacy for Controlling Bacterial Leaf Spot on Tomato Caused by Xanthomonas perforans. Front. Microbiol. 2018, 9, 2176. [Google Scholar]
- de Sousa, D.M.; Janssen, L.; Rosa, R.B.; Belmok, A.; Yamada, J.K.; Corrêa, R.F.T.; de Souza Andrade, M.; Inoue-Nagata, A.K.; Ribeiro, B.M.; de Carvalho-Pontes, N. Isolation, characterization, and evaluation of putative new bacteriophages for controlling bacterial spot on tomato in Brazil. Arch. Virol. 2023, 168, 222. [Google Scholar] [CrossRef]
- Solís-Sánchez, G.A.; Quiñones-Aguilar, E.E.; Fraire-Velázquez, S.; Vega-Arreguín, J.; Rincón-Enríquez, G. Complete Genome Sequence of XaF13, a Novel Bacteriophage of Xanthomonas vesicatoria from Mexico. Microbiol. Resour. Announc. 2020, 9, e01371-19. [Google Scholar] [CrossRef]
- Ríos-Sandoval, M.; Quiñones-Aguilar, E.E.; Solís-Sánchez, G.A.; Enríquez-Vara, J.N.; Rincón-Enríquez, G. Complete Genome Sequence of Xanthomonas vesicatoria Bacteriophage ΦXaF18, a Contribution to the Biocontrol of Bacterial Spot of Pepper in Mexico. Microbiol. Resour. Announc. 2020, 9, e00213-20. [Google Scholar] [CrossRef]
- Korniienko, N.; Kharina, A.; Zrelovs, N.; Jindřichová, B.; Moravec, T.; Budzanivska, I.; Burketová, L.; Kalachova, T. Isolation and Characterization of Two Lytic Phages Efficient Against Phytopathogenic Bacteria from Pseudomonas and Xanthomonas Genera. Front. Microbiol. 2022, 13, 853593. [Google Scholar] [CrossRef]
- Yoshikawa, G.; Askora, A.; Blanc-Mathieu, R.; Kawasaki, T.; Li, Y.; Nakano, M.; Ogata, H.; Yamada, T. Xanthomonas citri jumbo phage XacN1 exhibits a wide host range and high complement of tRNA genes. Sci. Rep. 2018, 8, 4486. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Wang, S.C.; Yu, Y.J.; Fung, K.M.; Yang, M.T.; Tseng, Y.H.; Tsai, S.F.; Sun, H.S.; Lyu, P.C.; Chou, S.H. Complete Genome Sequence of Xanthomonas campestris pv. campestris Strain 17 from Taiwan. Genome Announc. 2015, 3, e01466-15. [Google Scholar] [CrossRef] [PubMed]
- Flodman, K.; Tsai, R.; Xu, M.Y.; Corrêa, I.R., Jr.; Copelas, A.; Lee, Y.J.; Xu, M.Q.; Weigele, P.; Xu, S.Y. Type II Restriction of Bacteriophage DNA With 5hmdU-Derived Base Modifications. Front. Microbiol. 2019, 10, 584. [Google Scholar] [CrossRef] [PubMed]
- Topka-Bielecka, G.; Dydecka, A.; Necel, A.; Bloch, S.; Nejman-Faleńczyk, B.; Węgrzyn, G.; Węgrzyn, A. Bacteriophage-Derived Depolymerases against Bacterial Biofilm. Antibiotics 2021, 10, 175. [Google Scholar] [CrossRef] [PubMed]
- Awile, O.; Krisko, A.; Sbalzarini, I.F.; Zagrovic, B. Intrinsically disordered regions may lower the hydration free energy in proteins: A case study of nudix hydrolase in the bacterium Deinococcus radiodurans. PLoS Comput. Biol. 2010, 6, e1000854. [Google Scholar] [CrossRef] [PubMed]
- Pellicer, M.T.; Nuñez, M.F.; Aguilar, J.; Badia, J.; Baldoma, L. Role of 2-phosphoglycolate phosphatase of Escherichia coli in metabolism of the 2-phosphoglycolate formed in DNA repair. J. Bacteriol. 2003, 185, 5815–5821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, D.; Huang, Y.; Zhu, X.; Rui, M.; Wan, T.; Zheng, X.; Shen, Y.; Chen, X.; Ma, K.; et al. Structural and functional characterization of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease. Sci. Rep. 2017, 7, 42542. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.; Klumpp, J.; Mahony, J.; O’Connell-Motherway, M.; Nauta, A.; van Sinderen, D. Methyltransferases acquired by lactococcal 936-type phage provide protection against restriction endonuclease activity. BMC Genom. 2014, 15, 831. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Bu, W.; Sheppard, K.; Kitabatake, M.; Kwon, S.T.; Söll, D.; Smith, J.L. Insights into tRNA-dependent amidotransferase evolution and catalysis from the structure of the Aquifex aeolicus enzyme. J. Mol. Biol. 2009, 391, 703–716. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, J.; Yu, Z.; Jin, J.; Liu, X.; Wang, G. Novel groups and unique distribution of phage phoH genes in paddy waters in northeast China. Sci. Rep. 2016, 6, 38428. [Google Scholar] [CrossRef]
- Thabet, M.A.; Penadés, J.R.; Haag, A.F. The ClpX protease is essential for inactivating the CI master repressor and completing prophage induction in Staphylococcus aureus. Nat. Commun. 2023, 14, 6599. [Google Scholar] [CrossRef]
- Farias, P.; Francisco, R.; Morais, P.V. Potential of tellurite resistance in heterotrophic bacteria from mining environments. iScience 2022, 25, 104566. [Google Scholar] [CrossRef]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef]
- Yuan, Y.; Gao, M. Jumbo Bacteriophages: An Overview. Front. Microbiol. 2017, 8, 403. [Google Scholar] [CrossRef] [PubMed]
- Garneau, J.R.; Depardieu, F.; Fortier, L.C.; Bikard, D.; Monot, M. PhageTerm: A tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 2017, 7, 8292. [Google Scholar] [CrossRef]
- Batinovic, S.; Stanton, C.R.; Rice, D.T.F.; Rowe, B.; Beer, M.; Petrovski, S. Tyroviruses are a new group of temperate phages that infect Bacillus species in soil environments worldwide. BMC Genom. 2022, 23, 777. [Google Scholar] [CrossRef] [PubMed]
- Bailly-Bechet, M.; Vergassola, M.; Rocha, E. Causes for the intriguing presence of tRNAs in phages. Genome Res. 2007, 17, 1486–1495. [Google Scholar] [CrossRef]
- van den Berg, D.F.; van der Steen, B.A.; Costa, A.R.; Brouns, S.J.J. Phage tRNAs evade tRNA-targeting host defenses through anticodon loop mutations. eLife 2023, 12, e85183. [Google Scholar] [CrossRef] [PubMed]
- Azam, A.H.; Chihara, K.; Kondo, K.; Nakamura, T.; Ojima, S.; Tamura, A.; Yamashita, W.; Cui, L.; Takahashi, Y.; Watashi, K.; et al. Viruses encode tRNA and anti-retron to evade bacterial immunity. bioRxiv 2023. [Google Scholar] [CrossRef]
- Erdrich, S.H.; Sharma, V.; Schurr, U.; Arsova, B.; Frunzke, J. Isolation of Novel Xanthomonas Phages Infecting the Plant Pathogens X. translucens and X. campestris. Viruses 2022, 14, 1449. [Google Scholar] [CrossRef]
- Shiimori, M.; Garrett, S.C.; Graveley, B.R.; Terns, M.P. Cas4 Nucleases Define the PAM, Length, and Orientation of DNA Fragments Integrated at CRISPR Loci. Mol. Cell 2018, 70, 814–824. [Google Scholar] [CrossRef]
- Hooton, S.P.T.; Connerton, I. Campylobacter jejuni acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein. Front. Microbiol. 2015, 5, 744. [Google Scholar] [CrossRef] [PubMed]
- Garb, J.; Lopatina, A.; Bernheim, A.; Zaremba, M.; Siksnys, V.; Melamed, S.; Leavitt, A.; Millman, A.; Amitai, G.; Sorek, R. Multiple phage resistance systems inhibit infection via SIR2-dependent NAD+ depletion. Nat. Microbiol. 2022, 7, 1849–1856. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, R.; Xu, M.; Liu, Y.; Zhu, X.; Qiu, J.; Liu, Q.; He, P.; Li, Q. A Novel Polysaccharide Depolymerase Encoded by the Phage SH-KP152226 Confers Specific Activity Against Multidrug-Resistant Klebsiella pneumoniae via Biofilm Degradation. Front. Microbiol. 2019, 10, 2768. [Google Scholar] [CrossRef]
- Nguyen, T.T.H.; Kikuchi, T.; Tokunaga, T.; Iyoda, S.; Iguchi, A. Diversity of the Tellurite Resistance Gene Operon in Escherichia coli. Front. Microbiol. 2021, 12, 681175. [Google Scholar] [CrossRef]
- Li, X.; Sun, Y.; Liu, J.; Yao, Q.; Wang, G. Survey of the bacteriophage phoH gene in wetland sediments in northeast China. Sci. Rep. 2019, 9, 911. [Google Scholar] [CrossRef]
- Gross, M.; Marianovsky, I.; Glaser, G. MazG—A regulator of programmed cell death in Escherichia coli. Mol. Microbiol. 2006, 59, 590–601. [Google Scholar] [CrossRef] [PubMed]
- Warwick-Dugdale, J.; Buchholz, H.H.; Allen, M.J.; Temperton, B. Host-hijacking and planktonic piracy: How phages command the microbial high seas. Virol. J. 2019, 16, 15. [Google Scholar] [CrossRef]
- Ho, P.; Chen, Y.; Biswas, S.; Canfield, E.; Abdolvahabi, A.; Feldman, D.E. Bacteriophage antidefense genes that neutralize TIR and STING immune responses. Cell Rep. 2023, 42, 112305. [Google Scholar] [CrossRef]
- Sifuentes-Ibarra, E.; Macías-Cervantes, J.; Mendoza-Pérez, C.; Preciado-Rangel, P.; Salinas-Verduzco, D.A. Effect of direct sowing on soil properties and irrrigation water use in maize in Sinaloa, Mexico. Rev. Mex. Cienc. Agric. 2018, 9, 4235–4243. [Google Scholar]
- Rangel-Peraza, J.; Padilla-Gasca, E.; López-Corrales, R.; Medina, J.; Bustos-Terrones, Y.; Amabilis-Sosa, L.; Rodríguez-Mata, A.; Osuna-Enciso, T. Robust Soil Quality Index for Tropical Soils Influenced by Agricultural Activities. JACEN 2017, 6, 199–221. [Google Scholar] [CrossRef][Green Version]
- Tabare, E.; Glonti, T.; Cochez, C.; Ngassam, C.; Pirnay, J.P.; Amighi, K.; Goole, J. A Design of Experiment Approach to Optimize Spray-Dried Powders Containing Pseudomonas aeruginosa Podoviridae and Myoviridae Bacteriophages. Viruses 2021, 13, 1926. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, H.; Ghosh, D. The Stabilizing Excipients in Dry State Therapeutic Phage Formulations. AAPS Pharm. Sci. Tech. 2020, 21, 133. [Google Scholar] [CrossRef] [PubMed]
- Rahimzadeh, G.; Saeedi, M.; Moosazadeh, M.; Hashemi, S.M.H.; Babaei, A.; Rezai, M.S.; Kamel, K.; Asare-Addo, K.; Nokhodchi, A. Encapsulation of bacteriophage cocktail into chitosan for the treatment of bacterial diarrhea. Sci. Rep. 2021, 11, 15603. [Google Scholar] [CrossRef] [PubMed]
- Tapia-de la Barrera, L.B.; Báez-Sañudo, M.A.; García-Estrada, R.S.; Tovar-Pedraza, J.M.; Carrillo-Fasio, J.A. Identification of species and physiological races of Xanthomonas isolated from tomato (Solanum lycopersicum) and chili pepper (Capsicum annuum) in Sinaloa, Mexico. Rev. Mex. Fitopatol. 2023, 41, 326–342. [Google Scholar] [CrossRef]
- Rombouts, S.; Volckaert, A.; Venneman, S.; Declercq, B.; Vandenheuvel, D.; Allonsius, C.N.; Van Malderghem, C.; Jang, H.B.; Briers, Y.; Noben, J.P.; et al. Characterization of Novel Bacteriophages for Biocontrol of Bacterial Blight in Leek Caused by Pseudomonas syringae pv. porri. Front. Microbiol. 2016, 7, 279. [Google Scholar] [CrossRef] [PubMed]
- Jamalludeen, N.; Johnson, R.P.; Friendship, R.; Kropinski, A.M.; Lingohr, E.J.; Gyles, C.L. Isolation and characterization of nine bacteriophages that lyse O149 enterotoxigenic Escherichia coli. Vet. Microbiol. 2007, 124, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 2009, 501, 69–76. [Google Scholar]
- Gasic, K.; Ivanovic, M.M.; Ignjatov, M.; Calic, A.; Obradovic, A. Isolation and characterization of Xanthomonas euvesicatoria bacteriophages. J. Plant Pathol. 2011, 93, 415–423. [Google Scholar]
- Sambrook, J.; Russell, D. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- McNair, K.; Bailey, B.A.; Edwards, R.A. PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 2012, 28, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Krumsiek, J.; Arnold, R.; Rattei, T. Gepard: A rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 2007, 23, 1026–1028. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC-A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef]
- Jeon, J.; D’Souza, R.; Pinto, N.; Ryu, C.M.; Park, J.; Yong, D.; Lee, K. Characterization and complete genome sequence analysis of two Myoviral bacteriophages infecting clinical carbapenem-resistant Acinetobacter baumannii isolates. J. Appl. Microbiol. 2016, 121, 68–77. [Google Scholar] [CrossRef]


| Strains | Crop 1 | XaC1 | XbC2 |
|---|---|---|---|
| X. vesicatoria | |||
| XvA | Tomato | + | + |
| XvB | Tomato | + | + |
| Xv-2 | Tomato | − | + |
| Xv-17 | Tomato | + | − |
| Xv-44 | Tomato | − | − |
| X. euvesicatoria pv. euvesicatoria a | |||
| Xe-1 | Tomato | − | − |
| Xe-4 | Tomato | − | − |
| Xe-5 | Tomato | − | − |
| Xe-6 | Tomato | − | − |
| Xe-7 | Tomato | − | − |
| Xe-8 | Tomato | − | − |
| Xe-11 | Tomato | − | − |
| Xe-13 | Tomato | − | − |
| Xe-15 | Tomato | − | − |
| Xe-16 | Tomato | − | − |
| Xe-18 | Tomato | − | − |
| Xe-19 | Tomato | − | − |
| Xe-24 | Tomato | − | − |
| Xe-25 | Tomato | − | − |
| Xe-26 | Tomato | − | − |
| Xe-27 | Bell pepper | − | − |
| Xe-30 | Tomato | − | − |
| Xe-32 | Bell pepper | − | − |
| Xe-34 | Bell pepper | − | − |
| Xe-42 | Tomato | − | − |
| Xe-46 | Tomato | − | − |
| X. hortorum pv. gardneri b | |||
| Xg-28 | Tomato | − | − |
| Xg-29 | Tomato | − | − |
| Feature | XaC1 | XbC2 |
|---|---|---|
| Genome size | 348,967 bp | 367,901 bp |
| % GC | 35.1 | 37.5 |
| ORFs | 550 | 557 |
| ORFs with homology | 124 | 141 |
| Hypothetical proteins | 426 | 416 |
| tRNAs 1 | 8 | 34 |
| Lifestyle 2 | Lytic | Lytic |
| Virulence factor 3 | No | No |
| Antibiotic resistance genes 4 | No | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villicaña, C.; Rubí-Rangel, L.M.; Amarillas, L.; Lightbourn-Rojas, L.A.; Carrillo-Fasio, J.A.; León-Félix, J. Isolation and Characterization of Two Novel Genera of Jumbo Bacteriophages Infecting Xanthomonas vesicatoria Isolated from Agricultural Regions in Mexico. Antibiotics 2024, 13, 651. https://doi.org/10.3390/antibiotics13070651
Villicaña C, Rubí-Rangel LM, Amarillas L, Lightbourn-Rojas LA, Carrillo-Fasio JA, León-Félix J. Isolation and Characterization of Two Novel Genera of Jumbo Bacteriophages Infecting Xanthomonas vesicatoria Isolated from Agricultural Regions in Mexico. Antibiotics. 2024; 13(7):651. https://doi.org/10.3390/antibiotics13070651
Chicago/Turabian StyleVillicaña, Claudia, Lucía M. Rubí-Rangel, Luis Amarillas, Luis Alberto Lightbourn-Rojas, José Armando Carrillo-Fasio, and Josefina León-Félix. 2024. "Isolation and Characterization of Two Novel Genera of Jumbo Bacteriophages Infecting Xanthomonas vesicatoria Isolated from Agricultural Regions in Mexico" Antibiotics 13, no. 7: 651. https://doi.org/10.3390/antibiotics13070651
APA StyleVillicaña, C., Rubí-Rangel, L. M., Amarillas, L., Lightbourn-Rojas, L. A., Carrillo-Fasio, J. A., & León-Félix, J. (2024). Isolation and Characterization of Two Novel Genera of Jumbo Bacteriophages Infecting Xanthomonas vesicatoria Isolated from Agricultural Regions in Mexico. Antibiotics, 13(7), 651. https://doi.org/10.3390/antibiotics13070651








