Activity of Synthetic Peptide KP and Its Derivatives against Biofilm-Producing Escherichia coli Strains Resistant to Cephalosporins
Abstract
:1. Introduction
2. Results
2.1. Bactericidal Activity against Planktonic E. coli Cells
2.2. Inhibition of Biofilm Formation on Polystyrene Surfaces
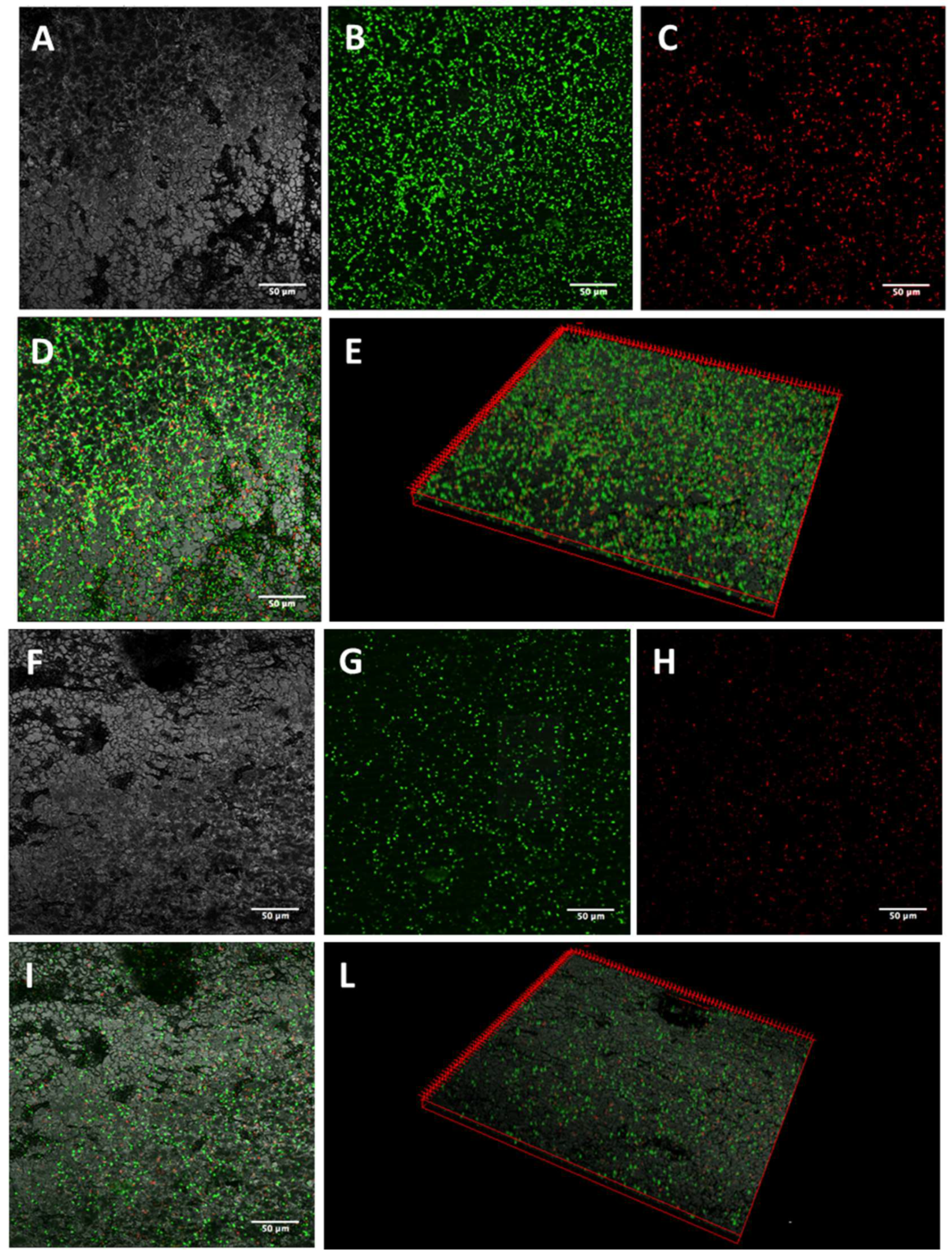
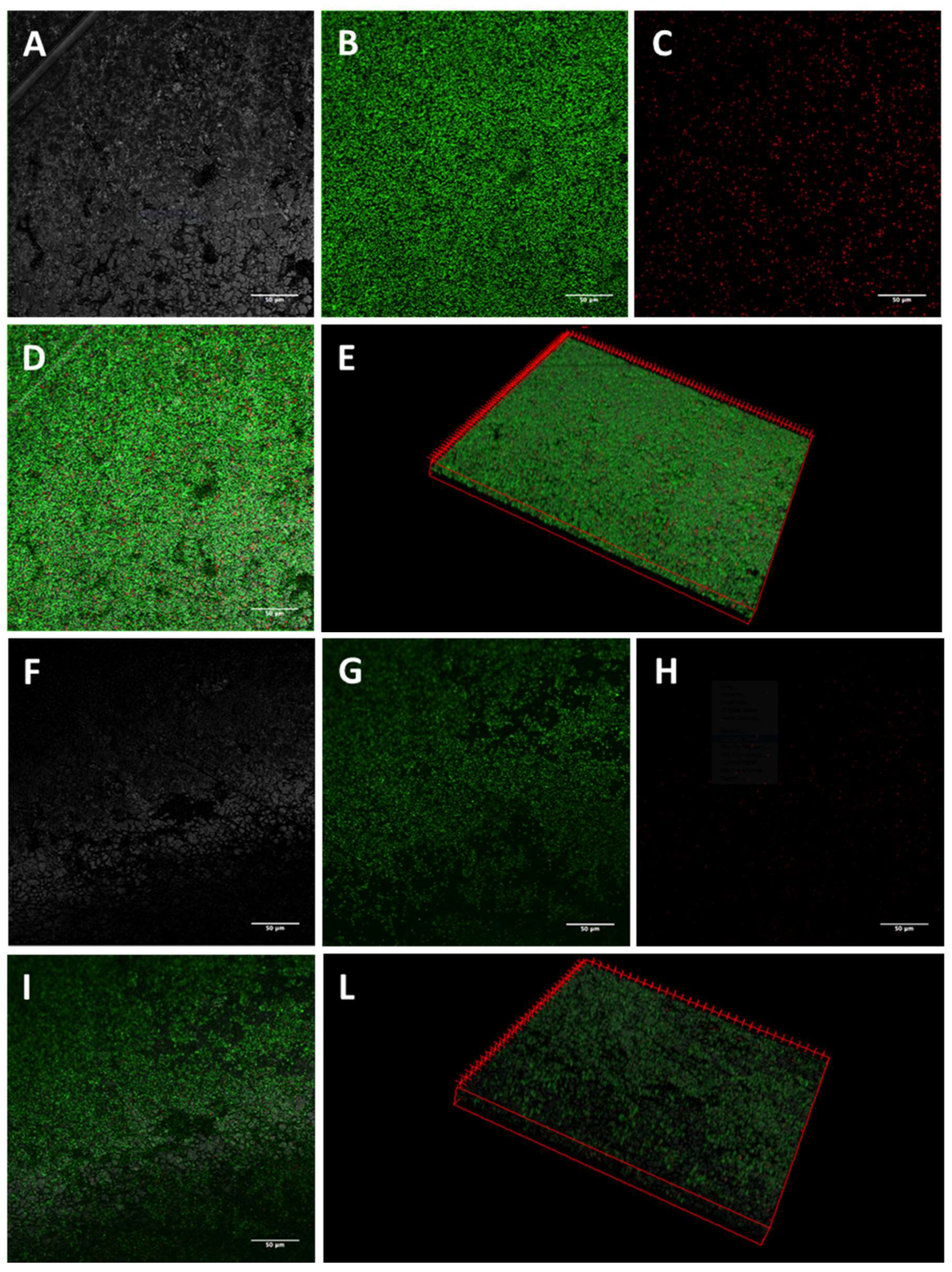
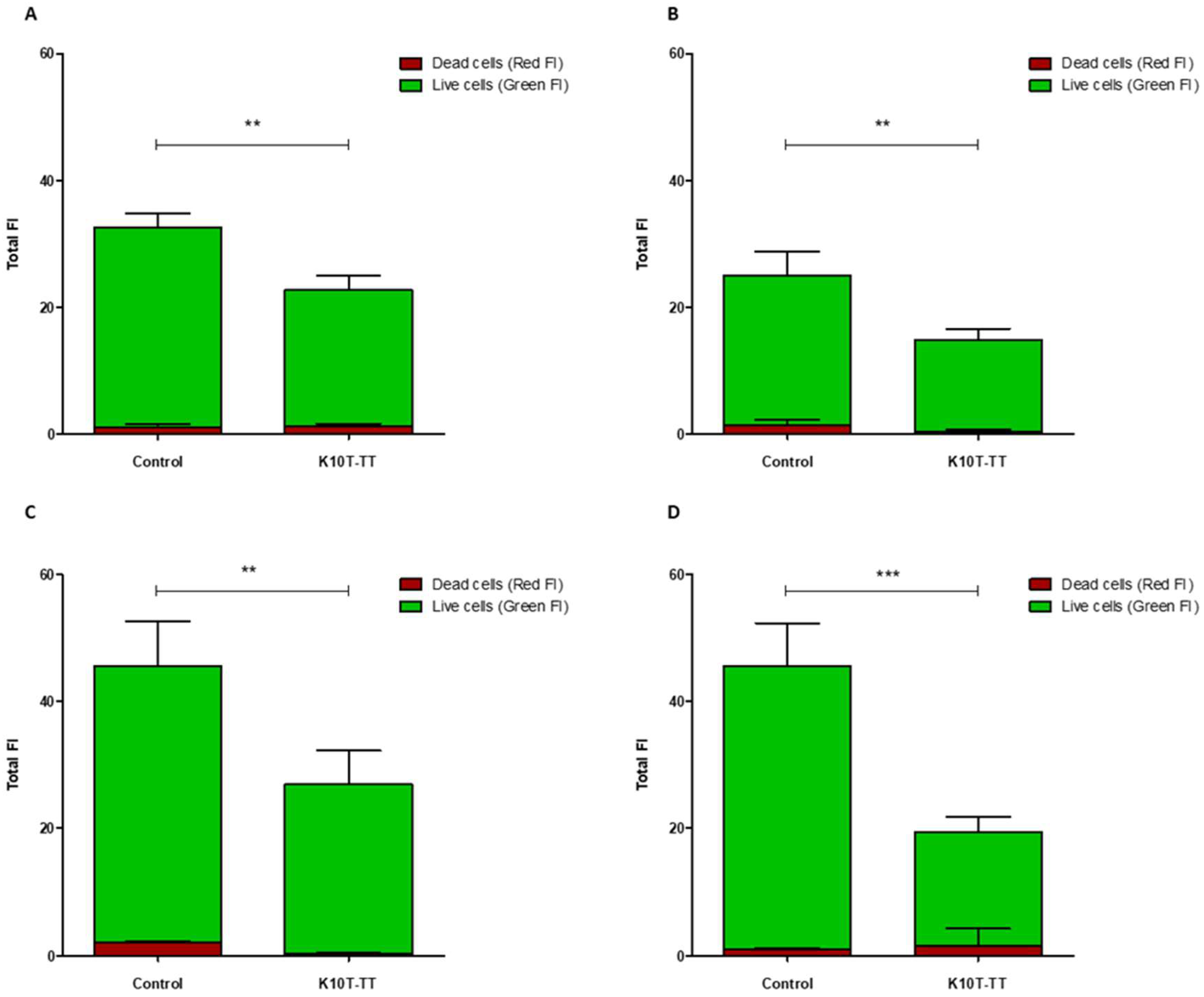
2.3. Activity of K10T-TT on Mature E. coli Biofilms Formed on Stainless Steel Surfaces Assessed by Confocal Microscopy
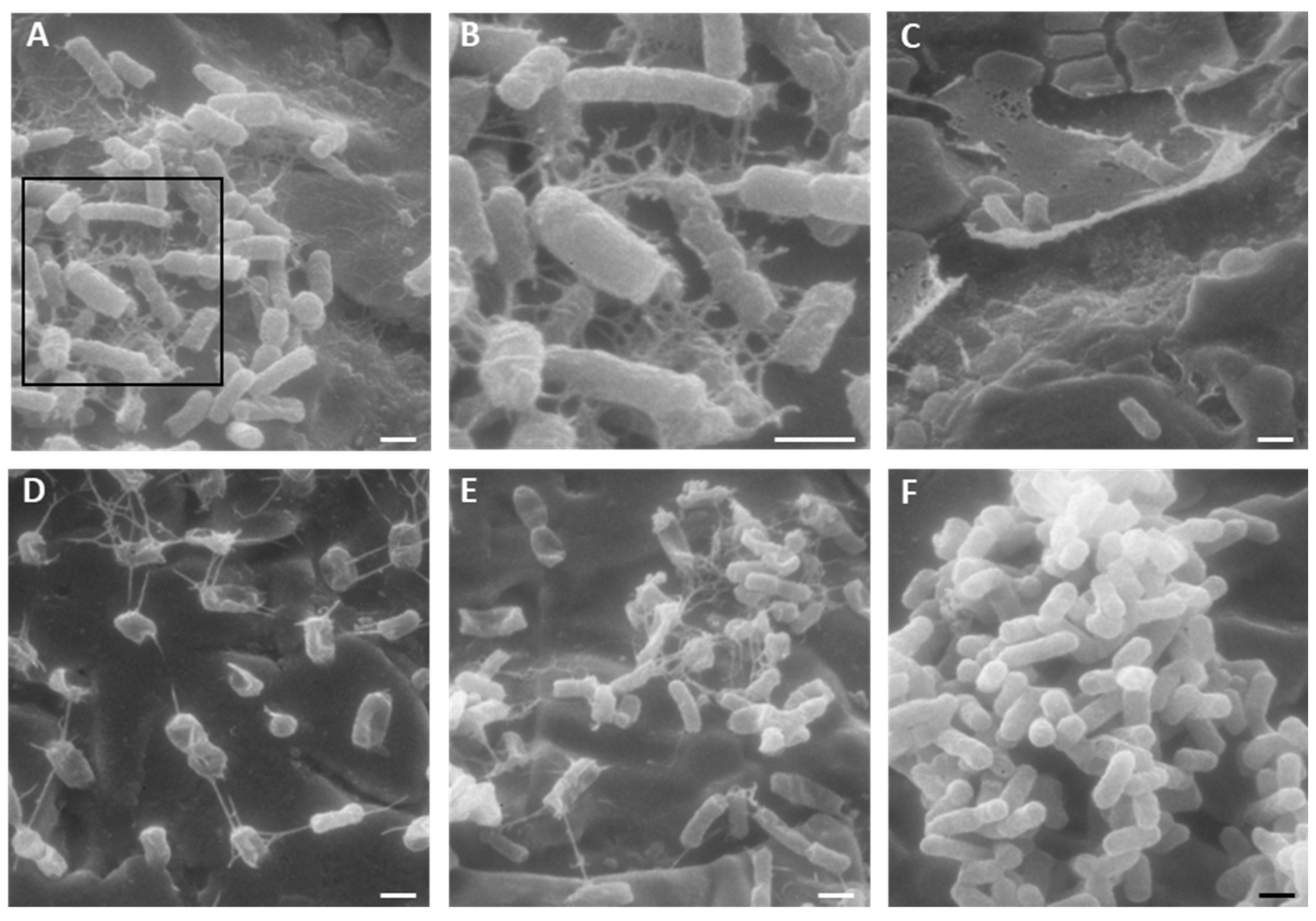
2.4. Activity of K10T-TT on Mature E. coli Biofilms Formed on Stainless Steel Surfaces Assessed by Scanning Electron Microscopy
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains
5.2. Antimicrobial Peptides
5.3. Bactericidal Activity against Planktonic Cells
5.4. Inhibition of Biofilm Formation on Polystyrene Surfaces
5.5. Inhibition of Mature E. coli Biofilms on Stainless Steel Surfaces
5.5.1. Confocal Laser Scanning Microscopy
5.5.2. Scanning Electron Microscopy
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2022; World Health Organization: Geneva, Switzerland, 2022.
- Djordjevic, S.P.; Jarocki, V.M.; Seemann, T.; Cummins, M.L.; Watt, A.E.; Drigo, B.; Wyrsch, E.R.; Reid, C.J.; Donner, E.; Howden, B.P. Genomic surveillance for antimicrobial resistance—A One Health perspective. Nat. Rev. Genet. 2024, 25, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Pokharel, P.; Dhakal, S.; Dozois, C.M. The Diversity of Escherichia coli Pathotypes and Vaccination Strategies against This Versatile Bacterial Pathogen. Microorganisms 2023, 11, 344. [Google Scholar] [CrossRef] [PubMed]
- Tokuda, M.; Shintani, M. Microbial evolution through horizontal gene transfer by mobile genetic elements. Microb. Biotechnol. 2024, 17, e14408. [Google Scholar] [CrossRef]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC-Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, L.F.; Nespolo, N.M.; Rossi, G.A.M.; Fairbrother, J.M. Exploring Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli in Food-Producing Animals and Animal-Derived Foods. Pathogens 2024, 13, 346. [Google Scholar] [CrossRef] [PubMed]
- Nasrollahian, S.; Graham, J.P.; Halaji, M. A review of the mechanisms that confer antibiotic resistance in pathotypes of E. coli. Front. Cell. Infect. Microbiol. 2024, 14, 1387497. [Google Scholar] [CrossRef] [PubMed]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Abdelhamid, A.G.; Yousef, A.E. Combating Bacterial Biofilms: Current and Emerging Antibiofilm Strategies for Treating Persistent Infections. Antibiotics 2023, 12, 1005. [Google Scholar] [CrossRef]
- Srivastava, A.; Verma, N.; Kumar, V.; Apoorva, P.; Agarwal, V. Biofilm inhibition/eradication: Exploring strategies and confronting challenges in combatting biofilm. Arch. Microbiol. 2024, 206, 212. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef]
- Luo, Y.; Song, Y. Mechanism of Antimicrobial Peptides: Antimicrobial, Anti-Inflammatory and Antibiofilm Activities. Int. J. Mol. Sci. 2021, 22, 11401. [Google Scholar] [CrossRef] [PubMed]
- Satchanska, G.; Davidova, S.; Gergova, A. Diversity and Mechanisms of Action of Plant, Animal, and Human Antimicrobial Peptides. Antibiotics 2024, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Mehraj, I.; Hamid, A.; Gani, U.; Iralu, N.; Manzoor, T.; Saleem Bhat, S. Combating Antimicrobial Resistance by Employing Antimicrobial Peptides: Immunomodulators and Therapeutic Agents against Infectious Diseases. ACS Appl. Bio Mater. 2024, 7, 2023–2035. [Google Scholar] [CrossRef] [PubMed]
- Batoni, G.; Maisetta, G.; Esin, S. Therapeutic Potential of Antimicrobial Peptides in Polymicrobial Biofilm-Associated Infections. Int. J. Mol. Sci. 2021, 22, 482. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Alford, M.A.; Haney, E.F. Antibiofilm activity of host defence peptides: Complexity provides opportunities. Nat. Rev. Microbiol. 2021, 19, 786–797. [Google Scholar] [CrossRef] [PubMed]
- Lopes, B.S.; Hanafiah, A.; Nachimuthu, R.; Muthupandian, S.; Md Nesran, Z.N.; Patil, S. The Role of Antimicrobial Peptides as Antimicrobial and Antibiofilm Agents in Tackling the Silent Pandemic of Antimicrobial Resistance. Molecules 2022, 27, 2995. [Google Scholar] [CrossRef] [PubMed]
- Anurag Anand, A.; Amod, A.; Anwar, S.; Sahoo, A.K.; Sethi, G.; Samanta, S.K. A comprehensive guide on screening and selection of a suitable AMP against biofilm-forming bacteria. Crit. Rev. Microbiol. 2023, 15, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Park, S.C.; Lee, M.Y.; Kim, J.Y.; Kim, H.; Jung, M.; Shin, M.K.; Lee, W.K.; Cheong, G.W.; Lee, J.R.; Jang, M.K. Anti-Biofilm Effects of Synthetic Antimicrobial Peptides against Drug-Resistant Pseudomonas aeruginosa and Staphylococcus aureus Planktonic Cells and Biofilm. Molecules 2019, 24, 4560. [Google Scholar] [CrossRef] [PubMed]
- Pontes, J.T.C.; Toledo Borges, A.B.; Roque-Borda, C.A.; Pavan, F.R. Antimicrobial Peptides as an Alternative for the Eradication of Bacterial Biofilms of Multi-Drug Resistant Bacteria. Pharmaceutics 2022, 14, 642. [Google Scholar] [CrossRef]
- Lee, J.K.; Park, S.; Kim, Y.M.; Guk, T.; Lee, M.Y.; Park, S.C.; Lee, J.R.; Jang, M.K. Candidacidal and Antibiofilm Activity of PS1-3 Peptide against Drug-Resistant Candida albicans on Contact Lenses. Pharmaceutics 2022, 14, 1602. [Google Scholar] [CrossRef]
- Cresti, L.; Cappello, G.; Pini, A. Antimicrobial Peptides towards Clinical Application-A Long History to Be Concluded. Int. J. Mol. Sci. 2024, 25, 4870. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef] [PubMed]
- Magliani, W.; Conti, S.; Ciociola, T.; Giovati, L.; Zanello, P.P.; Pertinhez, T.; Spisni, A.; Polonelli, L. Killer peptide: A novel paradigm of antimicrobial, antiviral and immunomodulatory auto-delivering drugs. Future Med. Chem. 2011, 3, 1209–1231. [Google Scholar] [CrossRef] [PubMed]
- Ciociola, T.; Pertinhez, T.A.; De Simone, T.; Magliani, W.; Ferrari, E.; Belletti, S.; D’Adda, T.; Conti, S.; Giovati, L. In Vitro and In Vivo Anti-Candida Activity and Structural Analysis of Killer Peptide (KP)-Derivatives. J. Fungi 2021, 7, 129. [Google Scholar] [CrossRef] [PubMed]
- Ciociola, T.; Magliani, W.; De Simone, T.; Pertinhez, T.A.; Conti, S.; Cozza, G.; Marin, O.; Giovati, L. In Silico Predicted Antifungal Peptides: In Vitro and In Vivo Anti-Candida Activity. J. Fungi 2021, 7, 439. [Google Scholar] [CrossRef] [PubMed]
- Barilli, E.; Vismarra, A.; Villa, Z.; Bonilauri, P.; Bacci, C. ESβL E. coli isolated in pig’s chain: Genetic analysis associated to the phenotype and biofilm synthesis evaluation. Int. J. Food Microbiol. 2019, 289, 162–167. [Google Scholar] [CrossRef]
- Mulchandani, R.; Wang, Y.; Gilbert, M.; Van Boeckel, T.P. Global trends in antimicrobial use in food-producing animals: 2020 to 2030. PLOS Glob. Public Health 2023, 3, e0001305. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Thomsen, L.E.; Olsen, J.E. Antimicrobial-induced horizontal transfer of antimicrobial resistance genes in bacteria: A mini-review. J. Antimicrob. Chemother. 2022, 77, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Pillay, S.; Calderón-Franco, D.; Urhan, A.; Abeel, T. Metagenomic-based surveillance systems for antibiotic resistance in non-clinical settings. Front. Microbiol. 2022, 13, 1066995. [Google Scholar] [CrossRef]
- Sundaramoorthy, N.S.; Shankaran, P.; Gopalan, V.; Nagarajan, S. New tools to mitigate drug resistance in Enterobacteriaceae—Escherichia coli and Klebsiella pneumoniae. Crit. Rev. Microbiol. 2023, 49, 435–454. [Google Scholar] [CrossRef]
- Lewnard, J.A.; Charani, E.; Gleason, A.; Hsu, L.Y.; Khan, W.A.; Karkey, A.; Chandler, C.I.R.; Mashe, T.; Khan, E.A.; Bulabula, A.N.H.; et al. Burden of bacterial antimicrobial resistance in low-income and middle-income countries avertible by existing interventions: An evidence review and modelling analysis. Lancet 2024, 403, 2439–2454. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, L.M.C.; Consol, P.; Chen, Y. Drug Discovery in the Field of β-Lactams: An Academic Perspective. Antibiotics 2024, 13, 59. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2024.
- Ciofu, O.; Moser, C.; Jensen, P.; Høiby, N. Tolerance and resistance of microbial biofilms. Nat. Rev. Microbiol. 2022, 20, 621–635. [Google Scholar] [CrossRef] [PubMed]
- Bouhrour, N.; Nibbering, P.H.; Bendali, F. Medical Device-Associated Biofilm Infections and Multidrug-Resistant Pathogens. Pathogens 2024, 13, 393. [Google Scholar] [CrossRef] [PubMed]
- Almatroudi, A. Investigating Biofilms: Advanced Methods for Comprehending Microbial Behavior and Antibiotic Resistance. Front. Biosci. Landmark Ed. 2024, 29, 133. [Google Scholar] [CrossRef] [PubMed]
- Solheim, H.T.; Sekse, C.; Urdahl, A.M.; Wasteson, Y.; Nesse, L.L. Biofilm as an environment for dissemination of stx genes by transduction. Appl. Environ. Microbiol. 2013, 79, 896–900. [Google Scholar] [CrossRef]
- Flores-Vargas, G.; Bergsveinson, J.; Lawrence, J.R.; Korber, D.R. Environmental Biofilms as Reservoirs for Antimicrobial Resistance. Front. Microbiol. 2021, 12, 766242. [Google Scholar] [CrossRef]
- Castañeda-Barba, S.; Top, E.M.; Stalder, T. Plasmids, a molecular cornerstone of antimicrobial resistance in the One Health era. Nat. Rev. Microbiol. 2024, 22, 18–32. [Google Scholar] [CrossRef]
- Pitts, B.; Hamilton, M.A.; Zelver, N.; Stewart, P.S. A microtiter-plate screening method for biofilm disinfection and removal. J. Microbiol. Methods 2003, 54, 269–276. [Google Scholar] [CrossRef]
- Dieltjens, L.; Appermans, K.; Lissens, M.; Lories, B.; Kim, W.; Van der Eycken, E.V.; Foster, K.R.; Steenackers, H.P. Inhibiting bacterial cooperation is an evolutionarily robust anti-biofilm strategy. Nat. Commun. 2020, 11, 107. [Google Scholar] [CrossRef] [PubMed]
- Silveira, G.; Torres, M.D.T.; Ribeiro, C.F.A.; Meneguetti, B.T.; Carvalho, C.M.E.; de la Fuente-Nunez, C.; Franco, O.L.; Cardoso, M.H. Antibiofilm Peptides: Relevant Preclinical Animal Infection Models and Translational Potential. ACS Pharmacol. Transl. Sci. 2021, 4, 55–73. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Gao, X.; Li, M.; Liu, Y.; Ma, J.; Wang, X.; Yu, Z.; Cheng, W.; Zhang, W.; Sun, H.; et al. Relationship between biofilm formation and antibiotic resistance of Klebsiella pneumoniae and updates on antibiofilm therapeutic strategies. Front. Cell. Infect. Microbiol. 2024, 14, 1324895. [Google Scholar] [CrossRef] [PubMed]
- Mountcastle, S.E.; Vyas, N.; Villapun, V.M.; Cox, S.C.; Jabbari, S.; Sammons, R.L.; Shelton, R.M.; Walmsley, A.D.; Kuehne, S.A. Biofilm viability checker: An open-source tool for automated biofilm viability analysis from confocal microscopy images. NPJ Biofilms Microbiomes 2021, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Mhade, S.; Kaushik, K.S. Tools of the Trade: Image Analysis Programs for Confocal Laser-Scanning Microscopy Studies of Biofilms and Considerations for Their Use by Experimental Researchers. ACS Omega 2023, 8, 20163–20177. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; Willcox, M.D.P.; Dutta, D. Action of Antimicrobial Peptides against Bacterial Biofilms. Materials 2018, 11, 2468. [Google Scholar] [CrossRef] [PubMed]
- Di Somma, A.; Moretta, A.; Canè, C.; Cirillo, A.; Duilio, A. Antimicrobial and Antibiofilm Peptides. Biomolecules 2020, 10, 652. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Wang, J.; Song, Y.; Liu, S.; Lu, Z.; Luo, H.; Tang, X. Antibacterial Potential Analysis of Novel α-Helix Peptides in the Chinese Wolf Spider Lycosa sinensis. Pharmaceutics 2022, 14, 2540. [Google Scholar] [CrossRef] [PubMed]
- Overhage, J.; Campisano, A.; Bains, M.; Torfs Ellen, C.W.; Rehm Bernd, H.A.; Hancock Robert, E.W. Human Host Defense Peptide LL-37 Prevents Bacterial Biofilm Formation. Infect. Immun. 2008, 76, 4176–4182. [Google Scholar] [CrossRef]
- Dean, S.N.; Bishop, B.M.; Van Hoek, M.L. Susceptibility of Pseudomonas aeruginosa Biofilm to Alpha-Helical Peptides: D-enantiomer of LL-37. Front. Microbiol. 2011, 2, 128. [Google Scholar] [CrossRef]
- Ma, Z.; Zhang, R.; Hai, D.; Lu, Z.; Lv, F.; Zhao, H.; Zhang, C.; McAllister, T.A.; Stanford, K.; Bie, X. Antibiofilm activity and modes of action of a novel β-sheet peptide against multidrug-resistant Salmonella enterica. Food Res. Int. 2019, 125, 108520. [Google Scholar] [CrossRef] [PubMed]
- Ciociola, T.; Giovati, L.; Giovannelli, A.; Conti, S.; Castagnola, M.; Vitali, A. The activity of a mammalian proline-rich peptide against Gram-negative bacteria, including drug-resistant strains, relies on a nonmembranolytic mode of action. Infect. Drug Resist. 2018, 11, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Mergoni, G.; Manfredi, M.; Bertani, P.; Ciociola, T.; Conti, S.; Giovati, L. Activity of Two Antimicrobial Peptides against Enterococcus faecalis in a Model of Biofilm-Mediated Endodontic Infection. Antibiotics 2021, 10, 1220. [Google Scholar] [CrossRef] [PubMed]


| Peptide | Sequence | MM (Da) | pI | Charge | AI | GRAVY | Ref. |
|---|---|---|---|---|---|---|---|
| KP (A10S) | AKVTMTCSAS | 998.18 | 8.27 | 1+ | 49 | 0.53 | [24] |
| K10S | KKVTMTCSAS | 1055.27 | 9.31 | 2+ | 39 | −0.04 | [25] |
| K10T-TT | KKVTMTCTAT | 1083.33 | 9.31 | 2+ | 39 | −0.02 | [26] |
| K10S-SS | KKVSMSCSAS | 1027.22 | 9.31 | 2+ | 39 | −0.06 | [26] |
| E. coli Strain | EC50 1 (95% Confidence Intervals) | |||
|---|---|---|---|---|
| KP | K10S | K10T-TT | K10S-SS | |
| ATCC 25922 | 0.309 (0.233–0.375) | 0.347 (0.282–0.427) | 0.394 (0.312–0.497) | 0.195 (0.183–0.209) |
| FS3 | 0.848 (0.827–0.870) | 0.139 (0.133–0.146) | 0.385 (0.367–0.405) | 0.587 (0.540–0.639) |
| FS63 | 0.751 (0.732–0.770) | 0.051 (0.033–0.079) | 0.475 (0.427–0.529) | 0.203 (0.201–0.205) |
| CS160 | 1.099 (1.059–1.142) | 0.998 (0.724–1.377) | 0.607 (0.488–0.756) | 0.149 (0.137–0.162) |
| E. coli Strain | EC50 1 (95% Confidence Intervals) | |
|---|---|---|
| K10T-TT | K10S-SS | |
| ATCC 25922 | 49.569 (31.491–78.039) | 89.442 (39.212–204.00) |
| FS3 | 13.792 (4.863–39.093) | 26.187 (5.615–122.17) |
| FS63 | 44.109 (19.288–100.87) | 55.569 (9.346–330.45) |
| CS160 | 56.42 (54.566–58.335) | 69.062 (51.205–93.135) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Artesani, L.; Ciociola, T.; Vismarra, A.; Bacci, C.; Conti, S.; Giovati, L. Activity of Synthetic Peptide KP and Its Derivatives against Biofilm-Producing Escherichia coli Strains Resistant to Cephalosporins. Antibiotics 2024, 13, 683. https://doi.org/10.3390/antibiotics13080683
Artesani L, Ciociola T, Vismarra A, Bacci C, Conti S, Giovati L. Activity of Synthetic Peptide KP and Its Derivatives against Biofilm-Producing Escherichia coli Strains Resistant to Cephalosporins. Antibiotics. 2024; 13(8):683. https://doi.org/10.3390/antibiotics13080683
Chicago/Turabian StyleArtesani, Lorenza, Tecla Ciociola, Alice Vismarra, Cristina Bacci, Stefania Conti, and Laura Giovati. 2024. "Activity of Synthetic Peptide KP and Its Derivatives against Biofilm-Producing Escherichia coli Strains Resistant to Cephalosporins" Antibiotics 13, no. 8: 683. https://doi.org/10.3390/antibiotics13080683
APA StyleArtesani, L., Ciociola, T., Vismarra, A., Bacci, C., Conti, S., & Giovati, L. (2024). Activity of Synthetic Peptide KP and Its Derivatives against Biofilm-Producing Escherichia coli Strains Resistant to Cephalosporins. Antibiotics, 13(8), 683. https://doi.org/10.3390/antibiotics13080683









