Abstract
The asymptomatic gastrointestinal colonization of multidrug-resistant (MDR) bacteria can lead to difficult-to-treat infections. We investigated the role of host factors influencing colonization in an orogastrical murine infection model using a CTX-M-15- and OXA-162-producing Klebsiella pneumoniae ST15 (MDR-KP) strain, as well as Escherichia coli J53 (EC) and E. coli transconjugants with an IncFII(K) plasmid carrying CTX-M-15 (EC-CTXM), and with an IncL plasmid carrying OXA-162 (EC-OXA) genes. The fecal bacterial count in colony-forming unit/gram stool (CFU/g) was determined by cultivation, IgA and defensin levels by ELISA, and gut microbiota by 16S rRNA analysis. The CFU was the lowest in EC, followed by EC-OXA and EC-CTXM, and the highest in the MDR-KP group. The IgA level in feces increased in MDR-KP, EC-CTXM, and EC-OXA, and did not change in EC. The beta-defensin 3 level markedly increased in all groups, with the highest values in MDR-KP and EC-CTXM. Alpha-defensin-5 increased in all groups especially in EC. In microbiota, the Bacteroidota phylum was dominant in MDR-KP, EC-CTXM, and EC-OXA, whereas Proteobacteria was dominant in EC. The Muribaculaceae family was significantly more common in the MDR-KP and EC-OXA groups, while the Lachnospiraceae family was dominant in the EC group. While fecal IgA levels positively correlated with colonizing bacterial CFU, the alpha-defensin 5 levels inversely correlated with CFUs and IgA levels. The presence of the IncFII(K) plasmid induced beta-defensin 3 production. The amounts of the Muribaculaceae family members exhibited a correlation with the IncL plasmid. The detected amounts of the Lachnospiraceae family indicated the protective role against the high-risk clone and the resistance plasmids’ dissemination. Our results suggest that not only the MDR-KP clone itself but also the resistance plasmids play a primary role in the colonization rate in the gastrointestinal tract. Both the MDR-KP clone as well as the IncFII(K) and IncL resistance plasmids provide survival and colonization benefits in the gut.
Keywords:
colonization; gut; multidrug resistance; mouse model; defensins; microbiome; ESBL; CTX-M; OXA-carbapenemase 1. Introduction
The global spread and increasing prevalence of multidrug-resistant (MDR) Enterobacterales pose a significant threat to our healthcare systems. These Gram-negative pathogens often resist commonly used third-generation cephalosporins and carbapenems due to the production of extended-spectrum β-lactamases (ESBLs), such as CTX-M types, and carbapenemases, including KPC, metallo-beta-lactamase, and OXA-type carbapanemase [1]. Making antibiotic-resistance dissemination more widespread, the genes encoding these ESBLs and carbapenemases are located on mobile genetic elements [2]. Infections caused by ESBL- or carbapenemase-producing Enterobacterales are associated with increased morbidity and mortality rates compared to infections caused by less resistant organisms [3]. In response to this growing threat, the World Health Organization in 2018 designated both ESBL-producing and/or carbapenemase-producing Enterobacterales as critical priority pathogens for the research and development of new therapeutic strategies and rapid diagnostics [4].
Since Enterobacterales are common commensal bacteria of the intestinal microbiota, infections caused by MDR-Enterobacterales—particularly, Escherichia coli and Klebsiella pneumoniae—often originate from prior asymptomatic gut colonization. The relationship between the intestinal microbiota and IgA production, as well as defensin production, has been investigated in connection with several primarily gastrointestinal diseases [5,6,7,8]. Defensins help to maintain the balance of microbiota by controlling the growth of pathogens and promoting the survival of beneficial bacteria [9]. Generally, IgA production in the gut plays a crucial role in the immune response. K. pneumoniae, as a common gut pathobiont, can induce intestinal inflammation [10]. However, the pathogenicity of Klebsiella is sensitive to the colonization status of gut microbiota [5].
High-risk K. pneumoniae clones are detected worldwide in hospital settings, and these are capable of acquiring diverse antibiotic-resistance mechanisms that enable them to survive in the hospital environment. Furthermore, high-risk clones can asymptomatically colonize the gut, and these clones are responsible for a high number of difficult-to-treat infections, because these exhibit multidrug resistance; therefore, limited number of effective antibiotics are available for treatment [11,12].
K. pneumoniae ST15 is an internationally disseminated high-risk clone that has been identified globally and appears to be resistant to multiple antibiotics, including cephalosporins, carbapenems, and fluoroquinolones. The high prevalence and virulence of K. pneumoniae ST15 strains make them a significant clinical and public health concern, particularly in hospital settings where they can spread rapidly and induce outbreaks [13,14,15].
CTX-M-15-producing K. pneumoniae ST15 is a widely disseminated clone that has been identified globally, particularly in Europe. The clone has been found to be highly transferable and has undergone multiclonal spread, contributing to its widespread presence in different parts of Europe. In the context of hospital settings, the presence of IncFII(K) plasmids carrying CTX-M-15 can contribute to the spread of multidrug-resistant bacteria, strongly limiting treatment options. IncL plasmids are often associated with antimicrobial-resistance genes, such as OXA-type carbapenemase [2,16].
The purpose of the current study was to determine different host factors influencing the gastrointestinal colonization of multidrug-resistant Enterobacterales strains. Our goal was to examine separately the role of resistance plasmids during colonization. We aimed to assess the effects of intestinal colonization with a CTX-M-15 ESBL- and OXA-162 carbapenemase-producing K. pneumoniae ST15 high-risk clone; and a sensitive, laboratory E. coli J53 strain and its transconjugants—either with a CTX-M-15-harboring IncFII(K) plasmid or with an OXA-162-harboring IncL plasmid—in a murine model to quantify the effects of the K. pneumoniae ST15 high-risk clone itself and the resistance plasmids on the establishment and elimination of intestinal colonization [17].
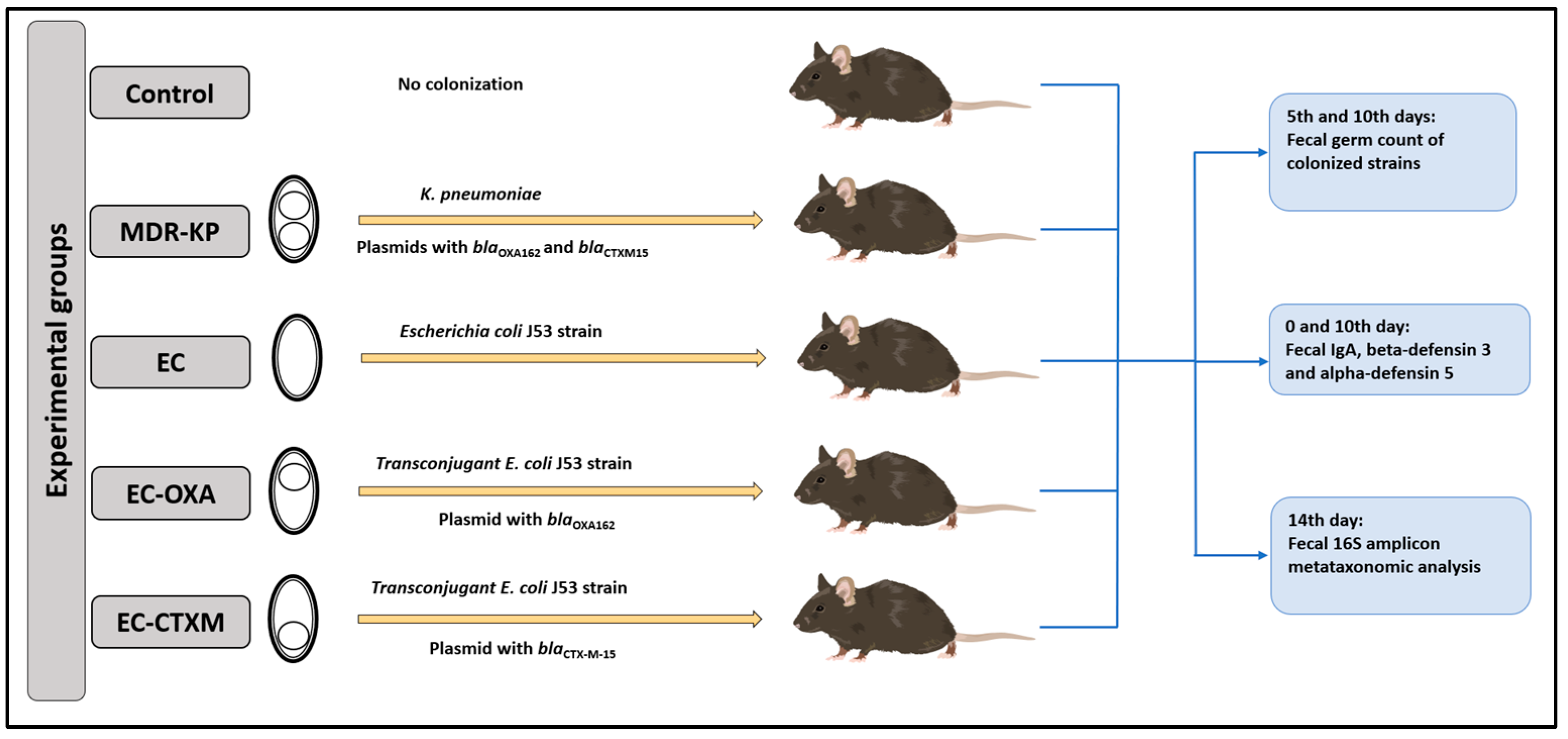
One particular aim of our study was to perform gastrointestinal colonization in oral-ampicillin-pretreated mice (C57BL/6) with a clinical CTX-M-15 ESBL- and OXA-162 carbapenemase-producing K. pneumoniae ST15 strain (MDR-KP), as well as its E. coli J53 transconjugants with an IncFII(K) plasmid containing the blaCTX-M-15 resistance gene (EC-CTXM), E. coli J53 transconjugants with an IncL plasmid containing the blaOXA-162 resistance gene within a Tn1991.2 genetic element (EC-OXA), and E. coli J53 strain itself (EC) [17]. After the orogastric colonization of the mice with the strains listed above, the bacterial count of the feces for the colonizing bacteria, the content of IgA, and the levels of beta-defensin-3 and alpha-defensin-5 were determined at different time points (Figure 1).
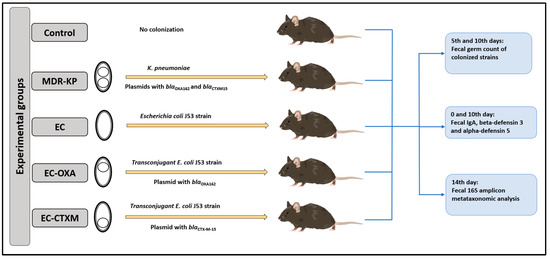
Figure 1.
Experimental design.
2. Results
2.1. Bacterial Loads in Feces During Gastrointestinal Colonization
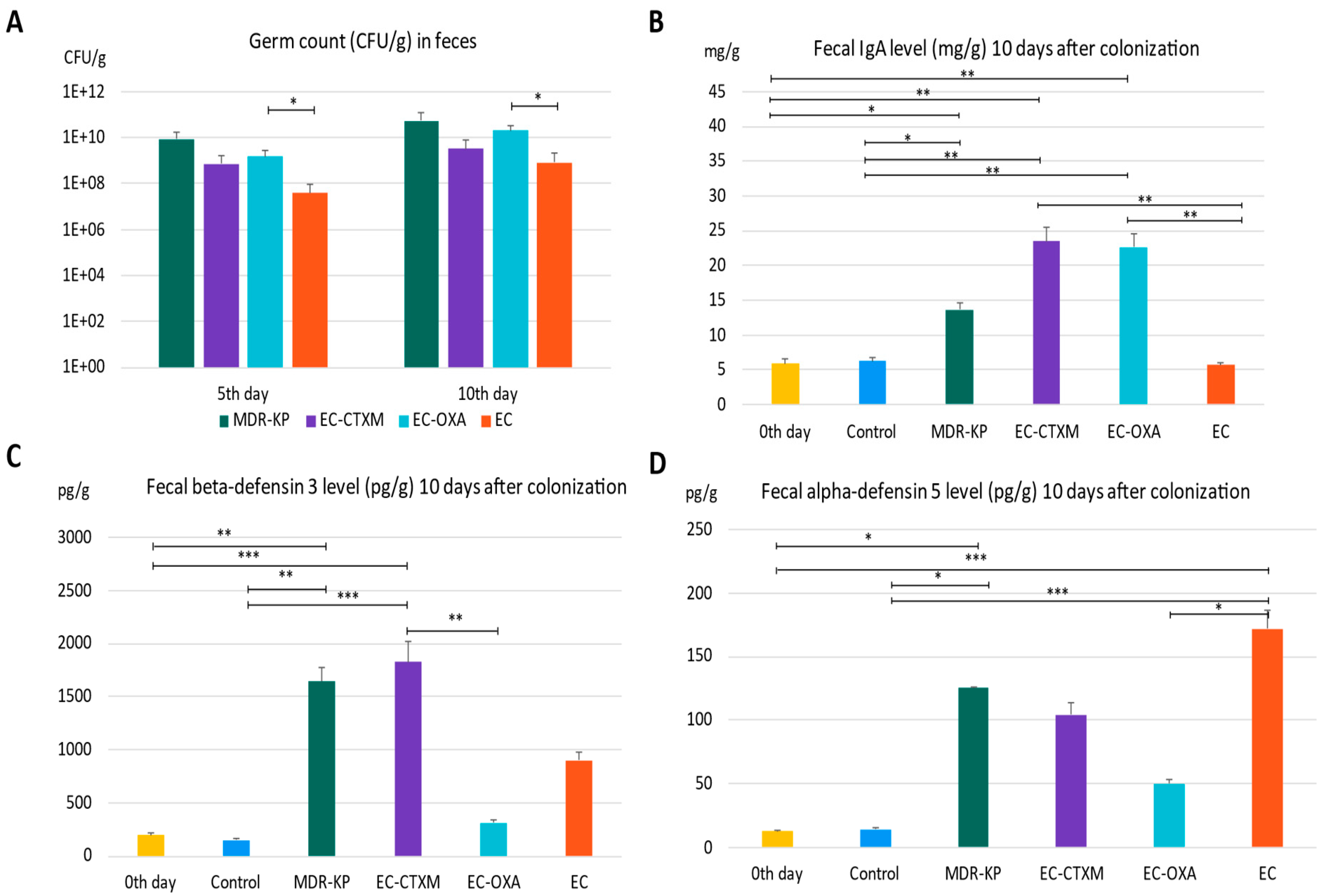
The mice were colonized orogastrically with MDR-KP (CTX-M-15- and OXA-162-producing K. pneumoniae ST15 strain), EC (E. coli J53), EC-CTXM (E. coli J53 transconjugant with an IncFII(K) plasmid containing the blaCTX-M-15 resistance gene), and EC-OXA (E. coli J53 transconjugant with an IncL plasmid containing the blaOXA-162 resistance gene within a Tn1991.2 genetic element). Fecal samples were collected on the fifth and tenth day of the colonization in order to determine the colonizing bacteria amount in the feces. On the fifth day of colonization, the CFU was the lowest at 3.77 × 107 CFU/g in the EC group, indicating the low colonization capability of laboratory-sensitive strains. The colonization rate was the highest in the MDR-KP group with a mean value of 8.64 × 109 CFU/g, and was also high in the EC-CTXM group at 7.06 × 108 CFU/g and in the EC-OXA group at 1.54 × 109 CFU/g, indicating the elevated gastrointestinal colonization ability of the transconjugant E. coli strains. The determination of germ counts from stool samples taken on the tenth day after colonization exhibited similar trends. On the tenth day, the fecal mean bacterial load was in the MDR-KP group at 5.43 × 1010 CFU/g, in the EC group at 8.43 × 108 CFU/g, in the EC-CTXM group at 3.18 × 109 CFU/g, and in the EC-OXA group at 2.01 × 1010 CFU/g (Figure 2A).
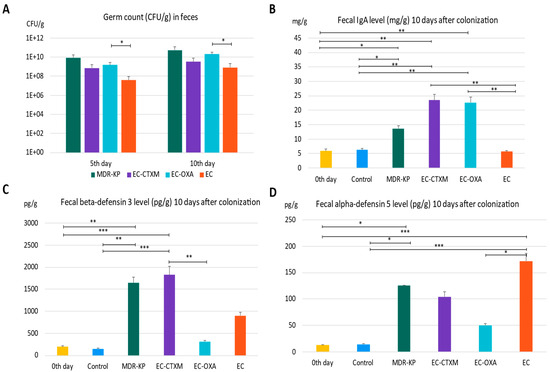
Figure 2.
(A) The gastrointestinal colonization rate characterized by germ count (colony-forming units: CFU) in the feces by different bacteria—MDR-KP, EC, EC-OXA, and EC-CTX-M—on the fifth and tenth day of colonization. (B) The IgA level in feces of mice colonized by different bacteria MDR-KP, EC, EC-OXA, and EC-CTXM. (C) The beta-defensin-3 level in feces of mice colonized by different bacteria MDR-KP, EC, EC-OXA, and EC-CTXM. (D) The alfa-defensin-5 in feces of mice colonized by different bacteria MDR-KP, EC, EC-OXA, and EC-CTXM. Statistical differences are marked with * p < 0.05; ** p < 0.01; and *** p < 0.001.
2.2. Fecal IgA Levels During Gastrointestinal Colonization
The IgA level was measured in feces just before the colonization (mean of 5.89 mg/g) and it was increased in the MDR-KP group (mean of 13.67 mg/g) and did not change in the EC group (mean of 5.66 mg/g). However, it markedly increased in the group EC-CTXM (mean of 22.68 mg/g) and in the group EC-OXA (mean of 23.52 mg/g) by Day 10 (Figure 2B).
2.3. Fecal Beta-Defensin 3 Levels During Gastrointestinal Colonization
In the feces, the baseline mean beta-defensin 3 level was a mean of 196.63 pg/g. The beta-defensin-3 level was increased to a mean of 1640 pg/g in the MDR-KP group, to a mean of 898 pg/g in the EC group, to a mean of 309 pg/g in the EC-OXA group, and to a mean of 1825 pg/g in the EC-CTXM group, indicating the dominant effect of the blaCTX-M-15-containing IncFII(K) plasmid on beta-defensin 3 production (Figure 2C).
2.4. Fecal Alpha-Defensin Levels During Gastrointestinal Colonization
The level of alpha-defensin 5 increased to a mean of 125 pg/g in the MDR-KP group, to a mean of 172 pg/g in the EC group, to a mean of 104 pg/g in the EC-CTXM group, and to a mean of 50 pg/g in the EC-OXA group, and it was increased in the EC groups. Based on these results, the fecal alpha-defensin 5 levels were inversely correlated with CFUs and IgA levels (Figure 2D).
2.5. Fecal Microbiota Composition During Gastrointestinal Colonization
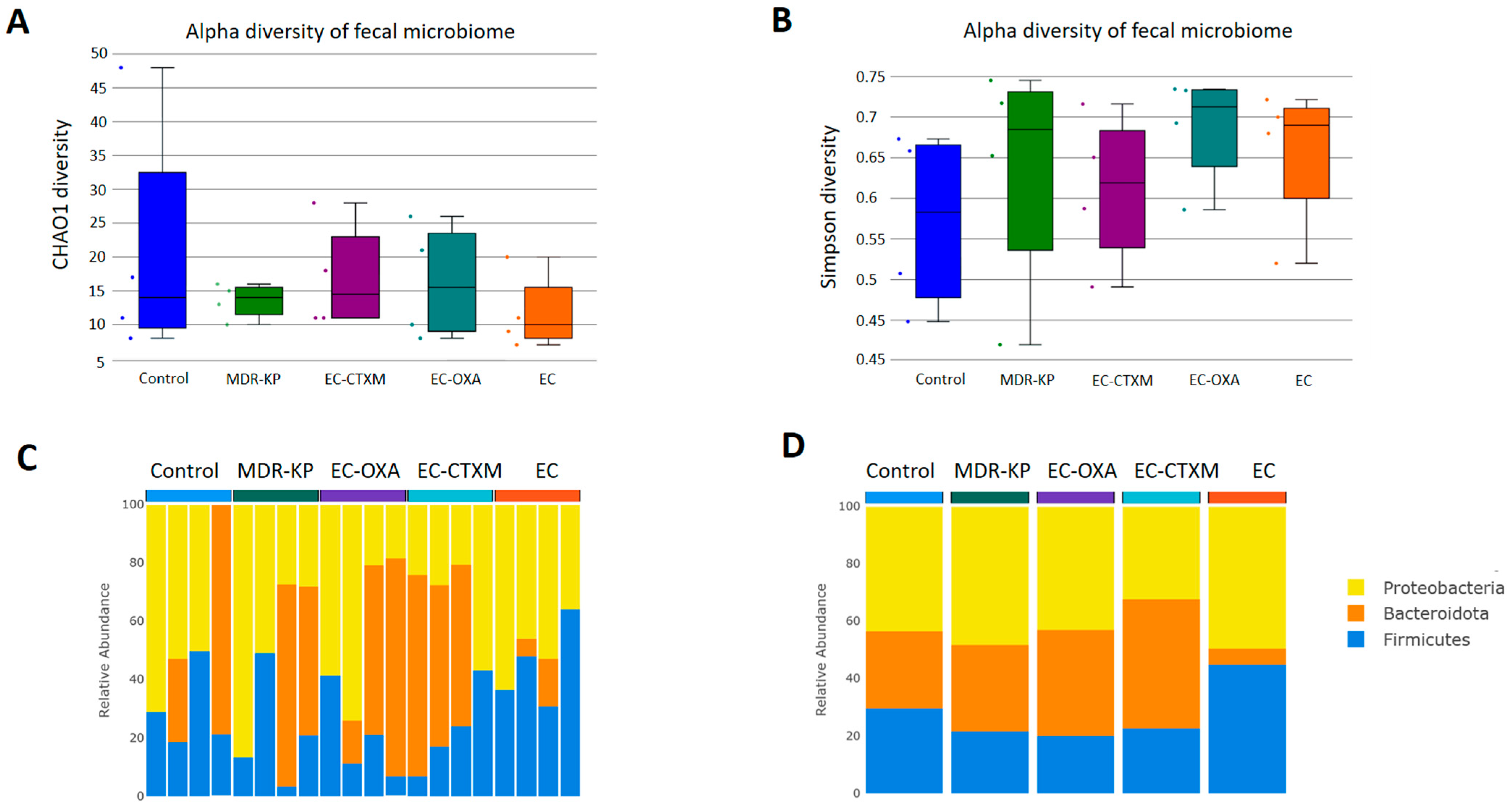
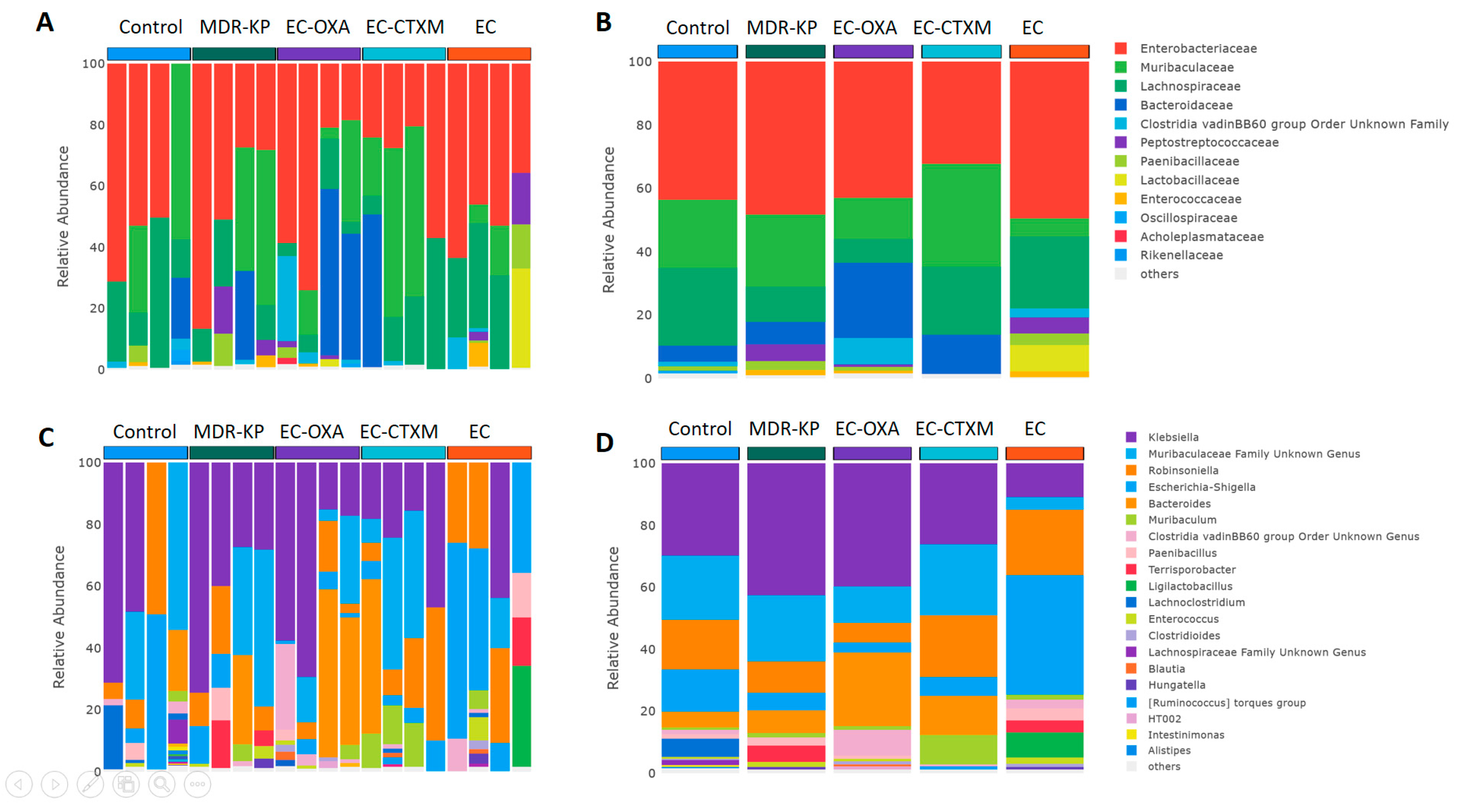
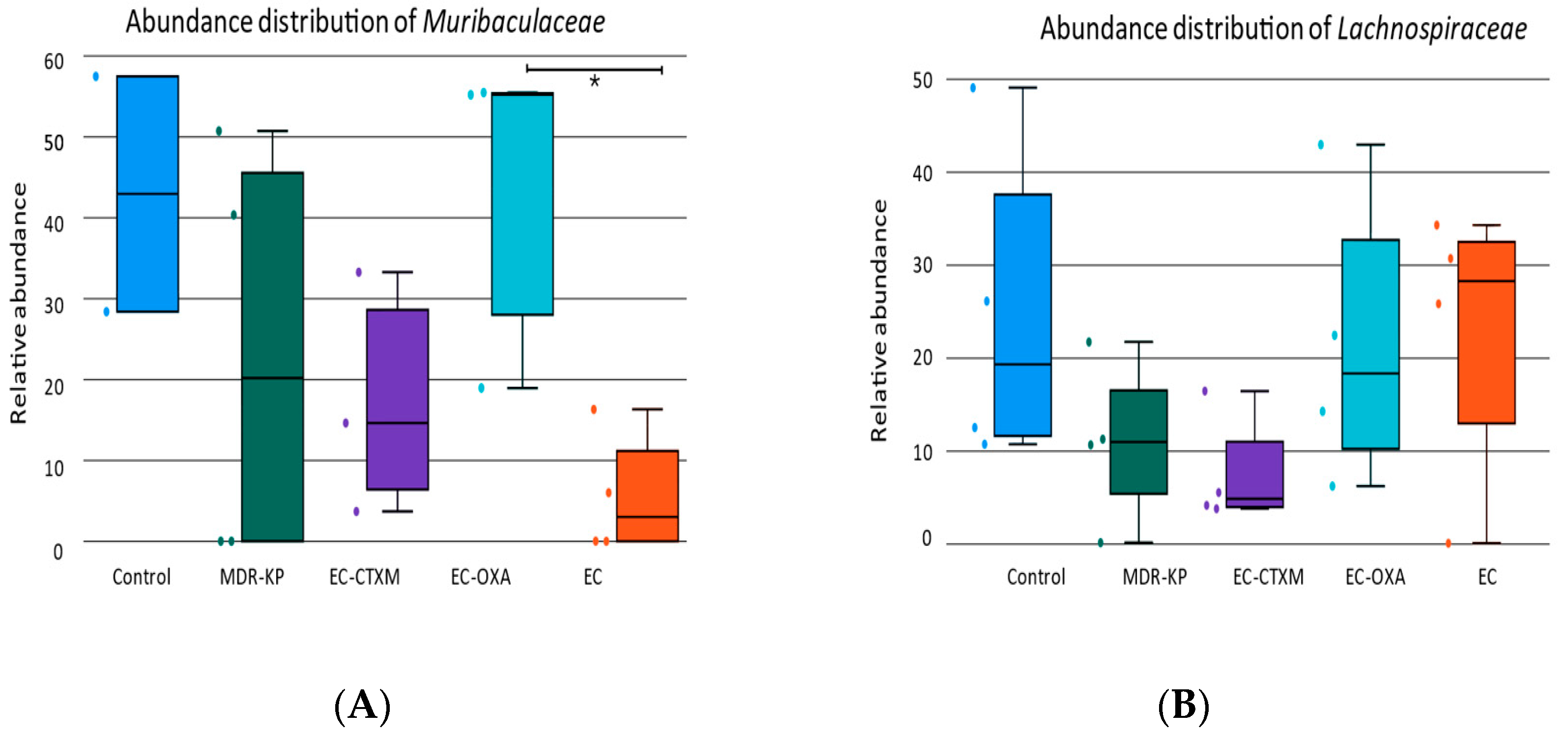
A 16S rRNA taxonomic analysis was performed on the feces samples on Day 14. There were no significant differences in the alpha-diversity by the Chao1 and Simpson tests and in the beta-diversity among the groups. The Bacteroidota phylum was the most dominant phyla in the MDR-KP, EC-CTXM, and EC-OXA-162 groups, whereas, in the EC group, the Proteobacteria phylum was dominant (Figure 3). At the family level, the Muribaculaceae family was significantly (p < 0.05) more common in the EC-OXA-162 groups than in the EC group, showing a correlation with the presence of the OXA-162 plasmid. The Lachnospiraceae family was dominant in the EC group, indicating the protective effect of the Lachnospiraceae family against the high-risk Klebsiella clone and the CTX-M15- and OXA-162-containing resistance plasmid dissemination (Figure 4).
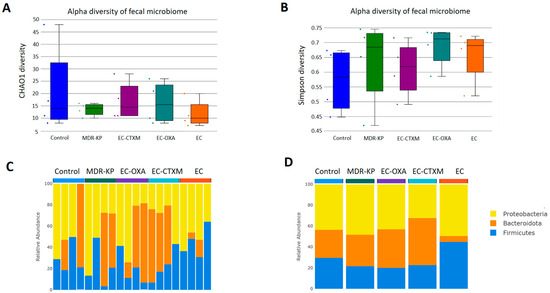
Figure 3.
(A) Relative abundances of abundant taxonomic phylum in each mouse. Elements are shown if they have at least 2% relative abundance in at least one of the averaged samples. (B) Average values of relative abundances at the phylum level were calculated for samples from the same treatment groups. Elements are shown if they have at least 2% relative abundance in at least one of the averaged samples. (C) Chao1 alpha-diversity of fecal samples in the different groups (Control, MDR-KP, EC, EC-CTXM, and EC-OXA). Box plots show the distribution of diversities in each group. (D) Simpson alpha-diversity of fecal samples in the different groups (Control, MDR-KP, EC, EC-CTXM, and EC-OXA). Box plots show the distribution of diversities in each group.
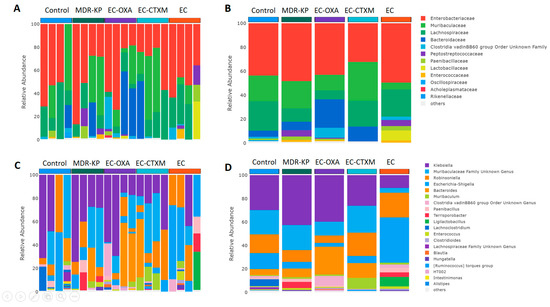
Figure 4.
(A) Relative abundances of most abundant taxonomic families in each mouse. Elements are shown if they have at least 2% relative abundance. (B) Average values of relative abundances at the family level were calculated for samples from the same treatment groups. Elements are shown if they have at least 2% relative abundance in at least one of the averaged samples. (C) Relative abundances of most abundant taxonomic genera in each mouse. Elements are shown if they have at least 2% relative abundance. (D) Average values of relative abundances at the genus level were calculated for samples from the same treatment groups. Elements are shown if they have at least 2% relative abundance in at least one of the averaged samples.
On one hand, the abundance of the Lachnospiraceae family showed an inverse relationship with the gastrointestinal carriage of the MDR-KP strain and harboring of the resistance plasmid in the EC-CTXM and EC-OXA-162 groups. On the other, the Enterobacteriaceae family showed a correlational relationship with the high-risk Klebsiella clone and the CTX-M15- and OXA-162-containing resistance plasmids (Figure 5).
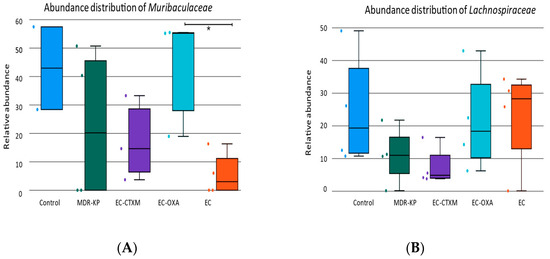
Figure 5.
(A) The relative abundance of Muribaculaceae family in each group (Control, MDR-KP, EC, EC-CTXM, and EC-OXA). (B) The relative abundance of Lachnospiraceae family in each group (Control, MDR-KP, EC, EC-CTXM, and EC-OXA). Statistical difference is marked with * p < 0.05.
3. Discussion
Gastrointestinal colonization by multidrug-resistant strains of Enterobacterales has been the focus of attention worldwide because these multidrug-resistant strains (e.g., ESBL- and/or carbapenemase-producing K. pneumoniae, and E. coli) can spread easily from person to person between healthy individuals as well as among hospitalized patients. The gut environment provides optimal conditions (e.g., a high bacterial density, optimal temperature, and a source of nutrients for bacteria) for horizontal gene transfer, that enhances the further dissemination of resistance genes among intestinal bacteria, and multidrug-resistant strains can evolve [18,19].
Several studies have investigated the risk factors of colonization with multidrug-resistant ESBL- and carbapenemase-producing Enterobacterales, such as earlier antibiotic treatment, previous hospitalization, intensive care unit treatment, travel abroad, etc. [20,21,22,23,24,25,26,27,28,29,30,31]. The screening for intestinal colonization with ESBL- and carbapenemase-producing Enterobacterales strains has been implemented in healthcare settings in several countries. Its importance is well-described among patients (e.g., newborns, and patients who are transferred between hospitals); however, healthy people after travel can be also screened [20,21,22,23,24,25].
Different decolonization strategies have been investigated in order to diminish the intestinal colonization of ESBL- and carbapenemase-producing Enterobacterales strains, because the prior gastrointestinal colonization can induce systemic infections and can initiate community-acquired infections. Furthermore, intestinally carried MDR-Enterobacterales strains can induce several outbreaks in hospitals as well [32].
In our animal study, after the colonization assay, the amount of MDR-Klebsiella and the E. coli strains harboring the CTX-M or the OXA-162 plasmids markedly increased in feces. Interestingly, the E. coli strain without the plasmid was not able to colonize the gastrointestinal tract; however, K. pneumoniae and E. coli harboring different resistance plasmids were able to successfully colonize.
It seems that the presence of the IncFII(K) plasmid with blaCTX-M-15 and the IncL plasmid with blaOXA-162 in E. coli changed the colonization properties of the original all-sensitive E. coli strain. Several underlying mechanisms can explain the reason, that the carriage of plasmids was accompanied by an increase in the bacterial cell count in stool samples.
In recent years, the relationship between the intestinal microbiome composition and ESBL-producing E. coli has been investigated. Davies et al. conducted a point-prevalence metagenomics study on fecal samples from international travelers before and after travel, observed changes in the microbiome composition during travel, and found that these changes were primarily associated with the development of travelers’ diarrhea rather than the acquisition of ESBL-producing E. coli [33]. In another study, no differences were found in the diversity parameters or relative abundance of bacterial species in the gut microbiome between healthy individuals, who were colonized or not colonized with ESBL-producing E. coli [34]. Recently, Ducarmon et al. analyzed the potential role of the gut microbiome in controlling the colonization of ESBL-producing E. coli, and no differences in the diversity parameters or in the relative abundance were observed between ESBL-producing E. coli and the negative groups [35]. Our obtained results are in good correlation with the previous literature data that describe human results, and we also found no difference in the alpha-diversity between the control group that does not carry ESBL or carbapenemase genes, and the group of ESBL- and carbapenemase-carrying strains [36,37,38]. Based on our findings, the abundance of Bacteroidota phylum was correlated with multidrug-resistance features. It was clearly dominant in the colonization with Klebsiella or with E. coli containing either the ESBL or OXA-162 plasmids. The gut microbiota exhibits remarkable alteration after colonization with the carpanemase-producing K. pneumoniae in animal studies with a specific dysbiosis characterized by a consistently marked decrease in Muribaculaceae, Rikenellaceae, and Lachnospiraceae_NK4A136_group [36]. However, we could not detect a significant difference in the abundance of the Lachnospiraceae and Muribaculaceae groups. Having said that, a significant difference could be observed in favor of ES and EC-OXA-162 in terms of Muribaculaceae, and a small difference could be detected in the EC group in terms of Lachnospiraceae in our study. The presence of the OXA-162 plasmids either in the K. pneuomoniae or E. coli strain led to changes in the gut microbiota composition, with a Muribaculaceae dominance, showing a correlation with the presence of the plasmid. The Muribaculaceae family specializes in the fermentation of complex polysaccharides. A genomic analysis has also suggested that the capacity for propionate production is widespread in the family [39,40].
Apart from the composition of the microbiome behind the gastrointestinal colonization with multidrug-resistant Klebsiella or E. coli, we are unaware of other mechanisms investigated in human or animal studies. The role of IgA and defensins in the gastrointestinal tract of mice colonized with a multidrug-resistant clone and its long-term colonization remains to be studied in detail. In order to identify the mechanisms involved in the gastrointestinal colonization of multidrug-resistant strains, we quantified the IgA and beta-defensin levels in mouse feces.
IgA binds to commensal bacteria and pathobionts like Klebsiella, which, in turn, can inhibit their growth and penetration of the mucus layer [40,41]. Interestingly, Klebsiella itself can induce IgA production in the gut [5]. Persistent colonization by resistant E. coli induces the secretion of luminal IgA, while commensal E. coli strain does not [40,41]. However, our results demonstrate that host immunity selectively recognizes pathobiont E. coli with the specific resistance plasmids, and develop specific IgA. The induced IgA specific to resistant E. coli, in turn, contributes to preventing the resistant strains from accessing the epithelium. K. pneumoniae induces a targeted IgA response in the gut, which helps to control its own colonization levels [10]. However, Klebsiella pathogenicity depends on the overall composition of the gut microbiota, with a dysbiotic state favoring Klebsiella overgrowth and inflammation [10,36]. Our results indicate that the presence of resistance plasmids—the IncFII(K) plasmid with CTXM-15 and the IncL plasmid with OXA-162—play a primary role in the MDR colonization rate in the gastrointestinal tract.
K. pneumoniae colonization in the gut can induce the production of human beta-defensins, which are antimicrobial peptides that help to regulate the gut microbiome [42,43]. Specifically, Klebsiella infection leads to increased levels of human beta-defensin 2 and human beta-defensin 3 in the intestine [44]. However, the total number of specific intestinal microbiota like Klebsiella does not differ significantly based on the different beta-defensin levels, suggesting that, while Klebsiella induces beta-defensin production, the defensin levels alone do not determine the abundance of Klebsiella in the gut [43,44]. Experimentally altering the gut microbiome composition can lead to changes in the beta-defensin-3 secretion, indicating a complex interplay between the microbiome, Klebsiella colonization, and host antimicrobial peptide production in the intestine [45]. Our results indicate that blaCTX-M-15 plays a dominant effect in containing the IncFII(K) plasmid in beta-defensin 3 production.
E. coli is known to play a significant role in the production and regulation of defensins in the gut. E. coli can induce the production of defensins in response to various stimuli, including the presence of pathogens. Different E. coli strains can have distinct effects on defensin production and gut health. For example, the probiotic E. coli Nissle 1917 can induce human beta-defensin 2 production, while other strains may have different effects on the gut microbiota [46]. Based on our results, the blaCTX-M-15-containing IncFII(K) plasmid presence plays an important role in human beta-defensin 3 production. Surprisingly, the alpha-defensin 5 level was the highest in the case of colonization with the apathogen E. coli and lower during the colonization with the plasmid carrying strains in this study. These findings highlight the importance of E. coli in the production and regulation of defensins in the gut, a process that plays a crucial role in maintaining gut health and controlling the growth of pathogens.
To our knowledge, this is the first paper that studied not only the gut microbiome dynamics, but also the role of IgA production and defensin levels during colonization by a multidrug-resistant K. pneumoniae high-risk clone. We documented that IgA levels and human beta-defensin 3 production have a crucial role in colonization and plasmid dissemination. All these findings confirm and emphasize that plasmids carrying resistance genes play a significant role in the spread of high-risk clones worldwide, whose role goes beyond the spread of resistance. The further identification of plasmid-mediated factors involved in colonization requires additional studies.
The limitations of the study are as follows: Further studies have to identify the direct roles of defensins on the pathogenic-resistance plasmid or indirect effects through the microbiota modification. Other MDR high-risk clones of K. pneumoniae and E. coli that carry different resistant plasmids and resistance genes should be tested in a colonization model.
4. Materials and Methods
4.1. Bacterial Strain and Conjugation Assay
For colonizing the experimental groups of mice, different bacterial strains were used that were generated previously and described in detail [17]. We provide here a summary of the relevant information related to these experiments. A multiresistant K. pneumoniae isolate ST15 (5825) (MDR-KP) was used as a conjugation donor. It harbors different resistance plasmids, among them an IncF(II)K-replicon-type plasmid with the gene blaCTX-M-15 and an IncL-replicon-type plasmid with the gene blaOXA-162. MDR-KP were also resistant to ciprofloxacin, that was used as selection agent during cultivation for germ count determination. E. coli J53 (EC) strain was used as an acceptor in the conjugation assay. It is resistant to sodium-azide, used as selection agent for acceptor and transconjugant strains. Two transconjugant strain were isolated, one isolate harboring IncL plasmid with blaOXA162 (EC-OXA) and one isolate harboring IncF(II)K plasmid with blaCTX-M-15 (EC-CTXM). These four different bacterial strains were propagated on Luria–Bertani (LB) agar (Biolab, Budapest, Hungary) (EC) and LB agar with 8 mg/L ampicillin (Sandoz, Schaftenau, Austria) (MDR-KP, EC-OXA, and EC-CTXM). A suspension in phosphate buffer saline (PBS, VWR, Debrecen, Hungary) was made containing 109 CFU/mL for gastrointestinal colonization of mice.
4.2. Animal Study
For gastrointestinal colonization, C57BL/6 male mice (Jackson Laboratory, Bar Harbor, ME, USA), aged 6–8 weeks, were used. The mice were individually housed in standard ventilated cages (IVC) under a 12 h light–dark cycle, with controlled temperature (20–22 °C). They had ad libitum access to sterile food and water, along with sterile bedding material. To prevent potential interference from natural gut bacteria, each mouse was housed individually throughout the experiment. Prior to the commencement of the experiments, the mice underwent a two-week acclimation period to minimize stress and ensure adaptation to the new environment.
Before colonization, the mice were treated with ampicillin (Sandoz) in their drinking water at a concentration of 0.5 mg/L for two weeks to facilitate the establishment of bacterial strains in the gut environment.
Colonization was then achieved via orogastric gavage. Each mouse received a dose of 108 colony-forming units (CFUs) in 100 microliters of PBS for each bacterial strain. The bacterial strains used for colonization were MDR-KP, EC, EC-OXA, and EC-CTXM. The control group was treated with sterile PBS following the same protocol. Each experimental groups contained six mice. After orogastric colonization, ampicillin treatment through the drinking water (0.5 mg/L) was maintained until the end of the experiment. Fresh fecal samples were collected and weighed for further investigation on Days 5, 10, and 14 after colonization. For the determination of the germ count, samples were used immediately after collection. For ELISA and DNA extraction, separated samples were stored at −80°C until they were investigated.
During the acclimation period and throughout the experiment, mice were handled gently to maintain their welfare. Experimental procedures were conducted in compliance with ethical guidelines and approved by the institutional animal care and use committee. Animals were maintained and handled in accordance with the recommendations of the Guidelines for the Care and Use of Laboratory Animals and the experiments were approved by the Animal Care Committee of Semmelweis University (Permission No. PE/EA/60-8/2018, PE/EA/964-5/2018).
4.3. Determination of the Fecal Germ Count of Mice
Fecal shedding of the colonized bacterial strains was quantified by determination of germ count in feces. Freshly collected fecal samples were weighed and immediately suspended by mechanical dissection of fecal pellet with sterile inoculation loop in 1 mL sterile PBS (VWR, Hungary) followed by thorough vortexing to gain a homogenous suspension. The suspension was serially diluted tenfold, and twenty microliters from each dilution were streaked onto selective chromogenic agar plates and incubated overnight. The following day, colonies were identified by appropriate color and counted. The germ count was calculated based on the colony numbers and dilution factors, expressed as CFUs per gram of fecal mass. For the multiresistant K. pneumoniae (MDR-KP), Orientation CHROMagar plates containing 0.5 mg/L ciprofloxacin (Fresenius Kabi, Bad Homburg vor der Höhe, Germany) were used to selectively support their growth. For E. coli J53 (EC) and transconjugant E. coli strains (EC-OXA and EC-CTXM), Enterobacteriaceae CHROMagar plates containing 100 mg/L sodium-azide (Merck, Darmstadt, Germany) were used. Results were statistically compared with two-tailed Student’s t-test.
4.4. Determination of Total IgA and Defensin Levels in Stool by ELISA
Total IgA, murine beta-defensin 3, and murine alpha-defensin 5 were determined from mouse feces by commercial ELISA kits (MyBiosource, San Diego, CA, USA, MBS7725462, MBS7725303, and MBS7725358). Collected frozen fecal samples were thawed and weighed before they were suspended in PBS and vortexed thoroughly for 1 h at 4 °C. Suspensions were centrifuged at 2500 rpm for 10 min and the supernatants were used in further studies. Sandwich ELISA measurements were made according to manufacturer’s instructions. After stopping the reaction, optical density was measured at 450 nm and 690 nm as reference wavelengths. Results were calculated with a calibration curve gained from included standards. Lastly, total IgA and defensin content were calculated (mg/g or pg/g feces). Results were statistically compared using Wilcoxon rank-sum test.
4.5. Microbiome Composition with 16S Metagenomic Analysis
To investigate the effect of different colonizing bacterial strains on the composition of gastrointestinal microbiota of mice, stool samples were collected on day 14 after colonization. DNA was extracted from ~80 mg of feces using the ZymoBIOMICS DNA Miniprep Kit (Zymo Research, Irvine, CA, USA, D4300) according to manufacturer’s instructions. The V3–V4 region of the bacterial 16S rRNA genes were amplified by PCR. Dual indices (barcodes) and Illumina sequencing adapters were added to the amplicons using the Nextera XT Index kit (Illumina, Inc., San Diego, CA, USA), followed by DNA purification (Agencourt AMPure XP, Beckman Coulter, Brea, CA, USA). Individual barcoded DNA samples were then quantified with Qubit dsDNA HS Assay kit with Qubit 2.0 (Thermo Fisher Scientific, Waltham, MA, USA), quantified with DNA 7500 kit with Agilent 2100 Bioanalyzer (Santa Clara, CA, USA), normalized, and pooled. Multiplexed libraries were diluted to 7 pM and denatured with NaOH prior to sequencing on the MiSeq system (Illumina) using the MiSeq reagent kit v3 600 cycles (2 × 300 bp; Illumina). Results of the sequencing were uploaded and analyzed with the CosmosID-HUB software v2.0 [47]. Paired-end reads of samples were analyzed by DADA2 algorithm, after primer removal data were quality-trimmed with a threshold of median Phred score 20 over the length of reads. Forward and reversed reads were trimmed to a uniform length based on quality of reads and merged if they have at least 12 base long overlap followed by the removing of chimeric sequences. Data were then processed to amplicon sequence variants (ASVs). The taxonomical annotation of clusters was made using DADA2’s naive Bayesian classifier and the Silva version 138 database. Microbial composition of samples was characterized by relative abundance of identified taxa, diversity indices (CHAO1, Simpson), and comparison of abundance distribution of specified taxa between experimental groups. Statistical comparison was made using Wilcoxon rank-sum test.
5. Conclusions
In our study, a mouse model demonstrated that intestinal colonization with the MDR CTX-M-15- and OXA-162-producing K. pneumoniae ST15 high-risk clone is multifactorial. Not only the MDR clone itself but also the resistance plasmids, namely, IncFII(K) and IncL, play a primary role in the colonization rate in the gastrointestinal tract. The levels of IgA, beta-defensin-3, and alpha-defensin-5, as well as the intestinal microbiota composition influence the colonization of the MDR K. pneumoniae high-risk clone.
Author Contributions
B.S.: data curation, methodology, and writing—original draft, review, and editing; J.D.: data curation and methodology; Z.A.D.: project administration and resources; N.M.: project administration and resources; J.J.: software, visualization, and formal analysis; E.O.: supervision and validation; B.K.: funding acquisition, validation, and writing—review and editing; D.S.: conceptualization, funding acquisition, and writing—original draft, review, and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by HUN-REN-SU, Human Microbiota Study Group No “0272”, and by the European Union’s Horizon 2020 research and innovation program (952491-AmReSu). B.K. was supported by the János Bolyai Scholarship (BO/00286/22/5) of the Hungarian Academy of Sciences.
Institutional Review Board Statement
The animals were maintained and handled in accordance with the recommendations of the Guidelines for the Care and Use of Laboratory Animals, and the experiments were approved by the Animal Care Committee of Semmelweis University (permission no. PE/EA/60-8/2018, PE/EA/964-5/2018).
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article. The sequencing data generated during the current study are available through following the figshare identifier: https://doi.org/10.6084/m9.figshare.26124370.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hansen, G.T. Continuous Evolution: Perspective on the Epidemiology of Carbapenemase Resistance Among Enterobacterales and Other Gram-Negative Bacteria. Infect. Dis. Ther. 2021, 10, 75–92. [Google Scholar] [CrossRef] [PubMed]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Sun, X.; Ma, X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 18. [Google Scholar] [CrossRef] [PubMed]
- Govindaraj Vaithinathan, A.; Vanitha, A. WHO global priority pathogens list on antibiotic resistance: An urgent need for action to integrate One Health data. Perspect. Public Health 2018, 138, 87–88. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Ohno, H. IgA in human health and diseases: Potential regulator of commensal microbiota. Front. Immunol. 2022, 13, 1024330. [Google Scholar] [CrossRef] [PubMed]
- DuPont, H.L.; Jiang, Z.D.; Alexander, A.S.; DuPont, A.W.; Brown, E.L. Intestinal IgA-Coated Bacteria in Healthy- and Altered-Microbiomes (Dysbiosis) and Predictive Value in Successful Fecal Microbiota Transplantation. Microorganisms 2022, 11, 93. [Google Scholar] [CrossRef]
- Meade, K.G.; O’Farrelly, C. beta-Defensins: Farming the Microbiome for Homeostasis and Health. Front. Immunol. 2018, 9, 3072. [Google Scholar]
- Suzuki, K.; Nakamura, K.; Shimizu, Y.; Yokoi, Y.; Ohira, S.; Hagiwara, M.; Wang, Y.; Song, Y.; Aizawa, T.; Ayabe, T. Decrease of alpha-defensin impairs intestinal metabolite homeostasis via dysbiosis in mouse chronic social defeat stress model. Sci. Rep. 2021, 11, 9915. [Google Scholar] [CrossRef] [PubMed]
- Zong, X.; Fu, J.; Xu, B.; Wang, Y.; Jin, M. Interplay between gut microbiota and antimicrobial peptides. Anim. Nutr. 2020, 6, 389–396. [Google Scholar] [CrossRef]
- Pope, J.L.; Yang, Y.; Newsome, R.C.; Sun, W.; Sun, X.; Ukhanova, M.; Neu, J.; Issa, J.P.; Mai, V.; Jobin, C. Microbial Colonization Coordinates the Pathogenesis of a Klebsiella pneumoniae Infant Isolate. Sci. Rep. 2019, 9, 3380. [Google Scholar] [CrossRef] [PubMed]
- Budia-Silva, M.; Kostyanev, T.; Ayala-Montaño, S.; Bravo-Ferrer Acosta, J.; Garcia-Castillo, M.; Cantón, R.; Goossens, H.; Rodriguez-Baño, J.; Grundmann, H.; Reuter, S. International and regional spread of carbapenem-resistant Klebsiella pneumoniae in Europe. Nat. Commun. 2024, 15, 5092. [Google Scholar] [CrossRef]
- Erdem, F.; Díez-Aguilar, M.; Oksuz, L.; Kayacan, C.; Abulaila, A.; Oncul, O.; Morosini, M.I.; Cantón, R.; Aktas, Z. Time kill-assays of antibiotic combinations for multidrug resistant clinical isolates of OXA-48 carbapenemase producing Klebsiella pneumoniae. Acta Microbiol. Immunol. Hung. 2022, 69, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Gato, E.; Rodino-Janeiro, B.K.; Gude, M.J.; Fernandez-Cuenca, F.; Pascual, A.; Fernandez, A.; Perez, A.; Bou, G. Diagnostic tool for surveillance, detection and monitoring of the high-risk clone K. pneumoniae ST15. J. Hosp. Infect. 2023, 142, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Chatzidimitriou, M.; Kavvada, A.; Kavvadas, D.; Kyriazidi, M.A.; Eleftheriadis, K.; Varlamis, S.; Papaliagkas, V.; Mitka, S. Carbapenem-resistant Klebsiella pneumoniae in the Balkans: Clonal distribution and associated resistance determinants. Acta Microbiol. Immunol. Hung. 2024, 71, 10–24. [Google Scholar] [CrossRef]
- Mohajer, H.B.; Salimizand, H.; Gharanizadeh, D.; Hossainpanahi, A.; Ramazanzadeh, R. Investigation of NDM-1 and OXA-48 producing carbapenem resistant Klebsiella pneumoniae ST15 in Iran. Acta Microbiol. Immunol. Hung. 2023, 70, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; Pitout, J.D.D. Extended-Spectrum beta-Lactamase-Producing Enterobacteriaceae: Update on Molecular Epidemiology and Treatment Options. Drugs 2019, 79, 1529–1541. [Google Scholar] [CrossRef]
- Stercz, B.; Farkas, F.B.; Tóth, A.; Gajdács, M.; Domokos, J.; Horváth, V.; Ostorházi, E.; Makra, N.; Kocsis, B.; Juhász, J.; et al. The influence of antibiotics on transitory resistome during gut colonization with CTX-M-15 and OXA-162 producing K. pneumoniae ST15. Sci. Rep. 2021, 11, 6335. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Tseng, W.P.; Chen, Y.C.; Chen, S.Y.; Chen, S.Y.; Chang, S.C. Risk for subsequent infection and mortality after hospitalization among patients with multidrug-resistant gram-negative bacteria colonization or infection. Antimicrob. Resist. Infect. Control 2018, 7, 93. [Google Scholar] [CrossRef]
- Jorgensen, S.B.; Soraas, A.; Sundsfjord, A.; Liestol, K.; Leegaard, T.M.; Jenum, P.A. Fecal carriage of extended spectrum beta-lactamase producing Escherichia coli and Klebsiella pneumoniae after urinary tract infection—A three year prospective cohort study. PLoS ONE 2017, 12, e0173510. [Google Scholar] [CrossRef]
- Rodriguez-Revuelta, M.J.; Lopez-Cerero, L.; Serrano, L.; Luna-Lagares, S.; Pascual, A.; Rodriguez-Bano, J. Incidence and Risk Factors for Acquisition of Extended-Spectrum beta-Lactamase-Producing Enterobacteriaceae in Newborns in Seville, Spain: A Prospective Cohort Study. Int. J. Antimicrob. Agents 2018, 52, 835–841. [Google Scholar] [CrossRef]
- Rossi, M.; Chatenoud, L.; Gona, F.; Sala, I.; Nattino, G.; D’Antonio, A.; Castelli, D.; Itri, T.; Morelli, P.; Bigoni, S.; et al. Characteristics and Clinical Implications of Carbapenemase-Producing Klebsiella pneumoniae Colonization and Infection, Italy. Emerg. Infect. Dis. 2021, 27, 1416–1426. [Google Scholar] [CrossRef]
- Denkel, L.A.; Maechler, F.; Schwab, F.; Kola, A.; Weber, A.; Gastmeier, P.; Pfafflin, F.; Weber, S.; Werner, G.; Pfeifer, Y.; et al. Infections caused by extended-spectrum beta-lactamase-producing Enterobacterales after rectal colonization with ESBL-producing Escherichia coli or Klebsiella pneumoniae. Clin. Microbiol. Infect. 2020, 26, 1046–1051. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Rojas, V.; Villanueva-Garcia, D.; Miranda-Vega, A.L.; Aldana-Vergara, R.; Aguilar-Rodea, P.; Lopez-Marceliano, B.; Reyes-Lopez, A.; Alcantar-Curiel, M.D. Gut colonization and subsequent infection of neonates caused by extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2023, 13, 1322874. [Google Scholar] [CrossRef]
- Seekatz, A.M.; Bassis, C.M.; Fogg, L.; Moore, N.M.; Rhee, Y.; Lolans, K.; Weinstein, R.A.; Lin, M.Y.; Young, V.B.; Hayden, M.K.; et al. Gut Microbiota and Clinical Features Distinguish Colonization with Klebsiella pneumoniae Carbapenemase-Producing Klebsiella pneumoniae at the Time of Admission to a Long-term Acute Care Hospital. Open Forum Infect. Dis. 2018, 5, ofy190. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Kim, D.; Hong, J.S.; Park, S.Y.; Cho, N.H.; Kim, M.N.; Lee, Y.J.; Wi, Y.; Lee, E.H.; Han, S.H.; et al. Prevalence of carbapenemase producing Enterobacterales colonization and risk factor of clinical infection. J. Infect. Public Health 2023, 16, 1860–1869. [Google Scholar] [CrossRef] [PubMed]
- Callejon Fernandez, M.; Madueno Alonso, A.; Abreu Rodriguez, R.; Aguirre-Jaime, A.; Castro Hernandez, M.B.; Ramos-Real, M.J.; Pedroso-Fernandez, Y.; Lecuona Fernandez, M. Risk factors for colonization by carbapenemase-producing bacteria in Spanish long-term care facilities: A multicentre point-prevalence study. Antimicrob. Resist. Infect. Control 2022, 11, 163. [Google Scholar] [CrossRef]
- Yan, L.; Sun, J.; Xu, X.; Huang, S. Epidemiology and risk factors of rectal colonization of carbapenemase-producing Enterobacteriaceae among high-risk patients from ICU and HSCT wards in a university hospital. Antimicrob. Resist. Infect. Control 2020, 9, 155. [Google Scholar] [CrossRef]
- Errico, G.; Gagliotti, C.; Monaco, M.; Masiero, L.; Gaibani, P.; Ambretti, S.; Landini, M.P.; D’Arezzo, S.; Di Caro, A.; Parisi, S.G.; et al. Colonization and infection due to carbapenemase-producing Enterobacteriaceae in liver and lung transplant recipients and donor-derived transmission: A prospective cohort study conducted in Italy. Clin. Microbiol. Infect. 2019, 25, 203–209. [Google Scholar] [CrossRef]
- Giannella, M.; Bartoletti, M.; Campoli, C.; Rinaldi, M.; Coladonato, S.; Pascale, R.; Tedeschi, S.; Ambretti, S.; Cristini, F.; Tumietto, F.; et al. The impact of carbapenemase-producing Enterobacteriaceae colonization on infection risk after liver transplantation: A prospective observational cohort study. Clin. Microbiol. Infect. 2019, 25, 1525–1531. [Google Scholar] [CrossRef]
- Madueno, A.; Gonzalez Garcia, J.; Ramos, M.J.; Pedroso, Y.; Diaz, Z.; Oteo, J.; Lecuona, M. Risk factors associated with carbapenemase-producing Klebsiella pneumoniae fecal carriage: A case-control study in a Spanish tertiary care hospital. Am. J. Infect. Control 2017, 45, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Mazzaferri, F.; de Smet, A.M.; Bragantini, D.; Eggimann, P.; Huttner, B.D.; Kuijper, E.J.; Lucet, J.C.; Mutters, N.T.; Sanguinetti, M.; et al. ESCMID-EUCIC clinical guidelines on decolonization of multidrug-resistant Gram-negative bacteria carriers. Clin. Microbiol. Infect. 2019, 25, 807–817. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.; Galazzo, G.; van Hattem, J.M.; Arcilla, M.S.; Melles, D.C.; de Jong, M.D.; Schultsz, C.; Wolffs, P.; McNally, A.; Schaik, W.V.; et al. Enterobacteriaceae and Bacteroidaceae provide resistance to travel-associated intestinal colonization by multi-drug resistant Escherichia coli. Gut Microbes 2022, 14, 2060676. [Google Scholar] [CrossRef] [PubMed]
- Boyd, A.; El Dani, M.; Ajrouche, R.; Demontant, V.; Cheval, J.; Lacombe, K.; Cosson, G.; Rodriguez, C.; Pawlotsky, J.M.; Woerther, P.L.; et al. Gut microbiome diversity and composition in individuals with and without extended-spectrum beta-lactamase-producing Enterobacterales carriage: A matched case-control study in infectious diseases department. Clin. Microbiol. Infect. 2024. [Google Scholar] [CrossRef] [PubMed]
- Ducarmon, Q.R.; Zwittink, R.D.; Hornung, B.V.H.; van Schaik, W.; Young, V.B.; Kuijper, E.J. Gut Microbiota and Colonization Resistance against Bacterial Enteric Infection. Microbiol. Mol. Biol. Rev. 2019, 83, e00007. [Google Scholar] [CrossRef] [PubMed]
- Le Guern, R.; Grandjean, T.; Stabler, S.; Bauduin, M.; Gosset, P.; Kipnis, E.; Dessein, R. Gut colonisation with multidrug-resistant Klebsiella pneumoniae worsens Pseudomonas aeruginosa lung infection. Nat. Commun. 2023, 14, 78. [Google Scholar] [CrossRef]
- Baek, M.S.; Kim, S.; Kim, W.Y.; Kweon, M.N.; Huh, J.W. Gut microbiota alterations in critically Ill patients with carbapenem-resistant Enterobacteriaceae colonization: A clinical analysis. Front. Microbiol. 2023, 14, 1140402. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.L.; Olsen, M.H.; Palleja, A.; Ebdrup, S.R.; Sorensen, N.; Lukjancenko, O.; Marvig, R.L.; Moller, K.; Frimodt-Moller, N.; Hertz, F.B. Microbiome Compositions and Resistome Levels after Antibiotic Treatment of Critically Ill Patients: An Observational Cohort Study. Microorganisms 2021, 9, 2542. [Google Scholar] [CrossRef]
- Ormerod, K.L.; Wood, D.L.; Lachner, N.; Gellatly, S.L.; Daly, J.N.; Parsons, J.D.; Dal’Molin, C.G.; Palfreyman, R.W.; Nielsen, L.K.; Cooper, M.A.; et al. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome 2016, 4, 36. [Google Scholar] [CrossRef]
- Lagkouvardos, I.; Lesker, T.R.; Hitch, T.C.A.; Galvez, E.J.C.; Smit, N.; Neuhaus, K.; Wang, J.; Baines, J.F.; Abt, B.; Stecher, B.; et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome 2019, 7, 28. [Google Scholar] [CrossRef]
- Sterlin, D.; Fadlallah, J.; Adams, O.; Fieschi, C.; Parizot, C.; Dorgham, K.; Rajkumar, A.; Autaa, G.; El-Kafsi, H.; Charuel, J.L.; et al. Human IgA binds a diverse array of commensal bacteria. J. Exp. Med. 2020, 217, e20181635. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Ren, C.; Han, X.; Huang, W.; You, Y.; Zhan, J. Role of IgA in the early-life establishment of the gut microbiota and immunity: Implications for constructing a healthy start. Gut Microbes 2021, 13, 1908101. [Google Scholar] [CrossRef] [PubMed]
- Corebima, B.; Rohsiswatmo, R.; Gayatri, P.; Patole, S. Fecal human beta-defensin-2 (hBD-2) levels and gut microbiota patterns in preterm neonates with different feeding patterns. Iran. J. Microbiol. 2019, 11, 151–159. [Google Scholar] [PubMed]
- Moranta, D.; Regueiro, V.; March, C.; Llobet, E.; Margareto, J.; Larrarte, E.; Garmendia, J.; Bengoechea, J.A. Klebsiella pneumoniae capsule polysaccharide impedes the expression of beta-defensins by airway epithelial cells. Infect. Immun. 2010, 78, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Saqib, Z.; De Palma, G.; Lu, J.; Surette, M.; Bercik, P.; Collins, S.M. Alterations in fecal beta-defensin-3 secretion as a marker of instability of the gut microbiota. Gut Microbes 2023, 15, 2233679. [Google Scholar] [CrossRef] [PubMed]
- Cobo, E.R.; Chadee, K. Antimicrobial Human beta-Defensins in the Colon and Their Role in Infectious and Non-Infectious Diseases. Pathogens 2013, 2, 177–192. [Google Scholar] [CrossRef]
- Yan, Q.; Wi, Y.M.; Thoendel, M.J.; Raval, Y.S.; Greenwood-Quaintance, K.E.; Abdel, M.P.; Jeraldo, P.R.; Chia, N.; Patel, R. Evaluation of the CosmosID Bioinformatics Platform for Prosthetic Joint-Associated Sonicate Fluid Shotgun Metagenomic Data Analysis. J. Clin. Microbiol. 2019, 57, e01182. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).