The pH-Insensitive Antimicrobial and Antibiofilm Activities of the Frog Skin Derived Peptide Esc(1-21): Promising Features for Novel Anti-Infective Drugs
Abstract
1. Introduction
2. Materials and Methods
2.1. Peptides
2.2. Materials
2.3. Bacterial Strains and Eukaryotic Cells
2.4. Antimicrobial Assays
2.5. Preparation of Large Unilamellar Vesicles (LUVs) and CyF Leakage Assay
2.6. Cell Viability Assay
3. Results and Discussion
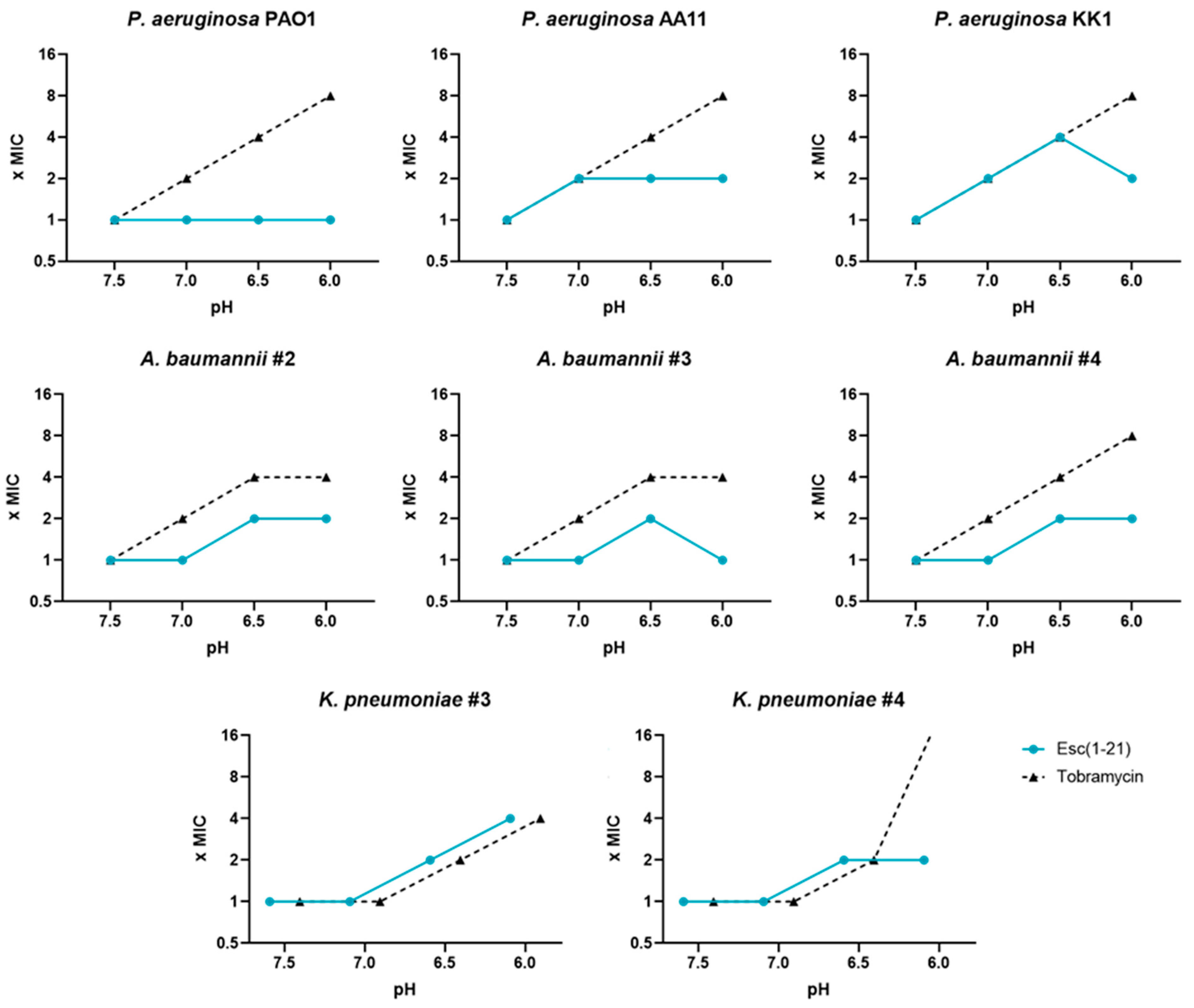
3.1. Antimicrobial Activity of AMPs and Antibiotics at Different pH Values, against Reference Gram-Negative Bacterial Strains
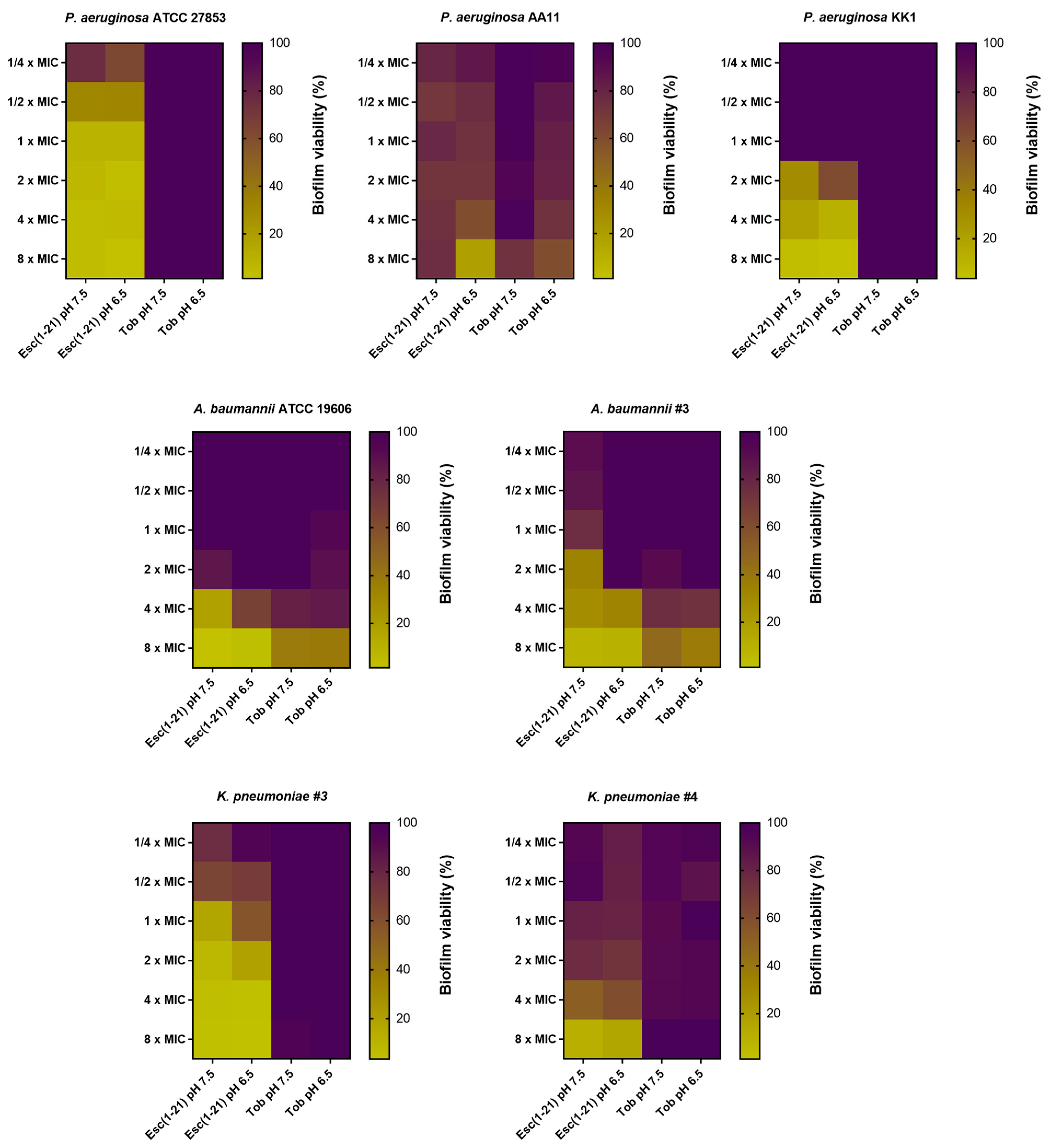
3.2. Antibiofilm Activity of Esc(1-21) Compared to Tobramycin against Preformed Biofilm in LB Broth
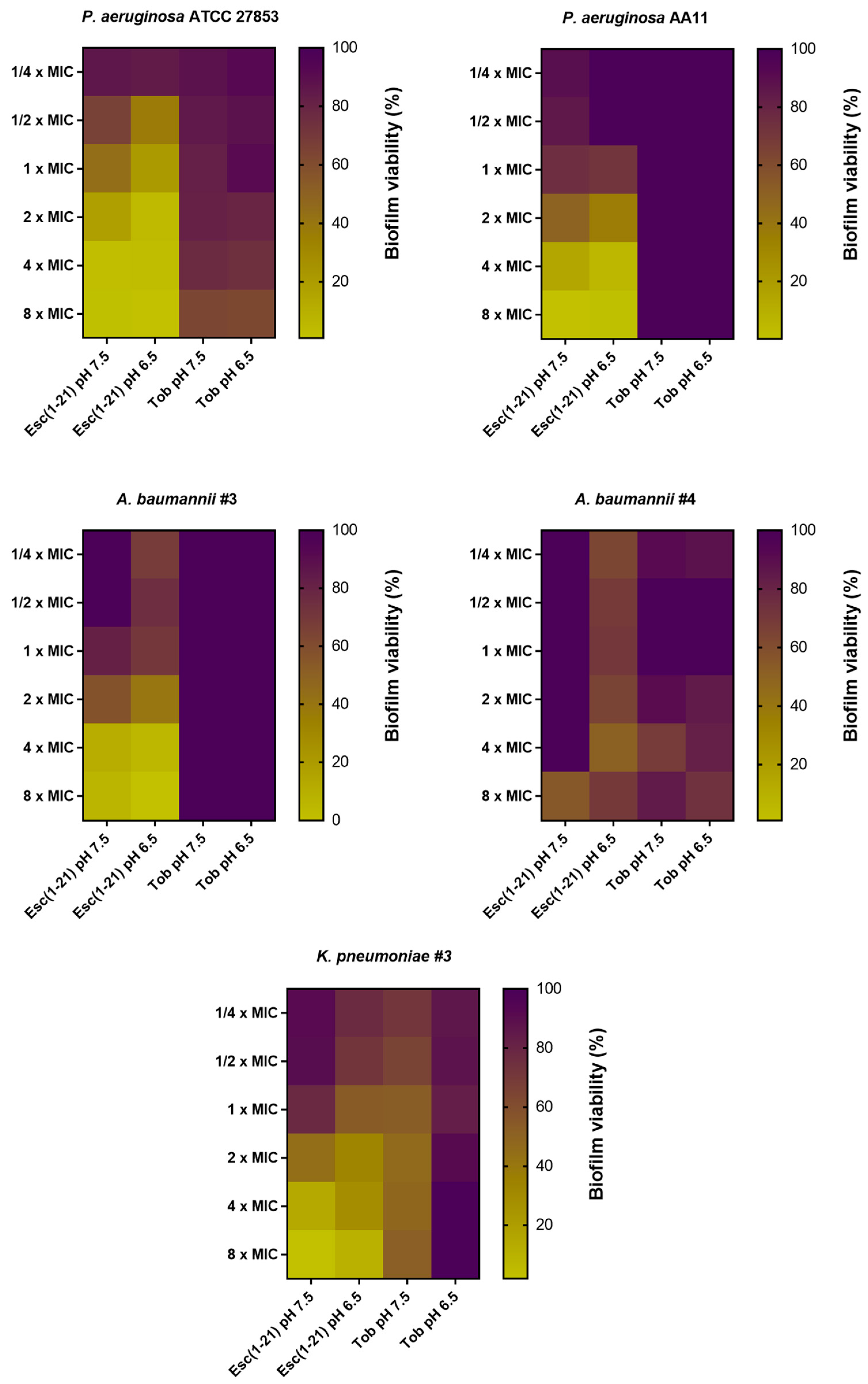
3.3. Antibiofilm Activity of Esc(1-21) Compared to Tobramycin against Preformed Biofilm Grown in m63 Medium at pH 7.5 and 6.5
3.4. Activity of Esc(1-21) at pHs 7.5 and 6.5 against Model Membranes of Gram-Negative Bacteria
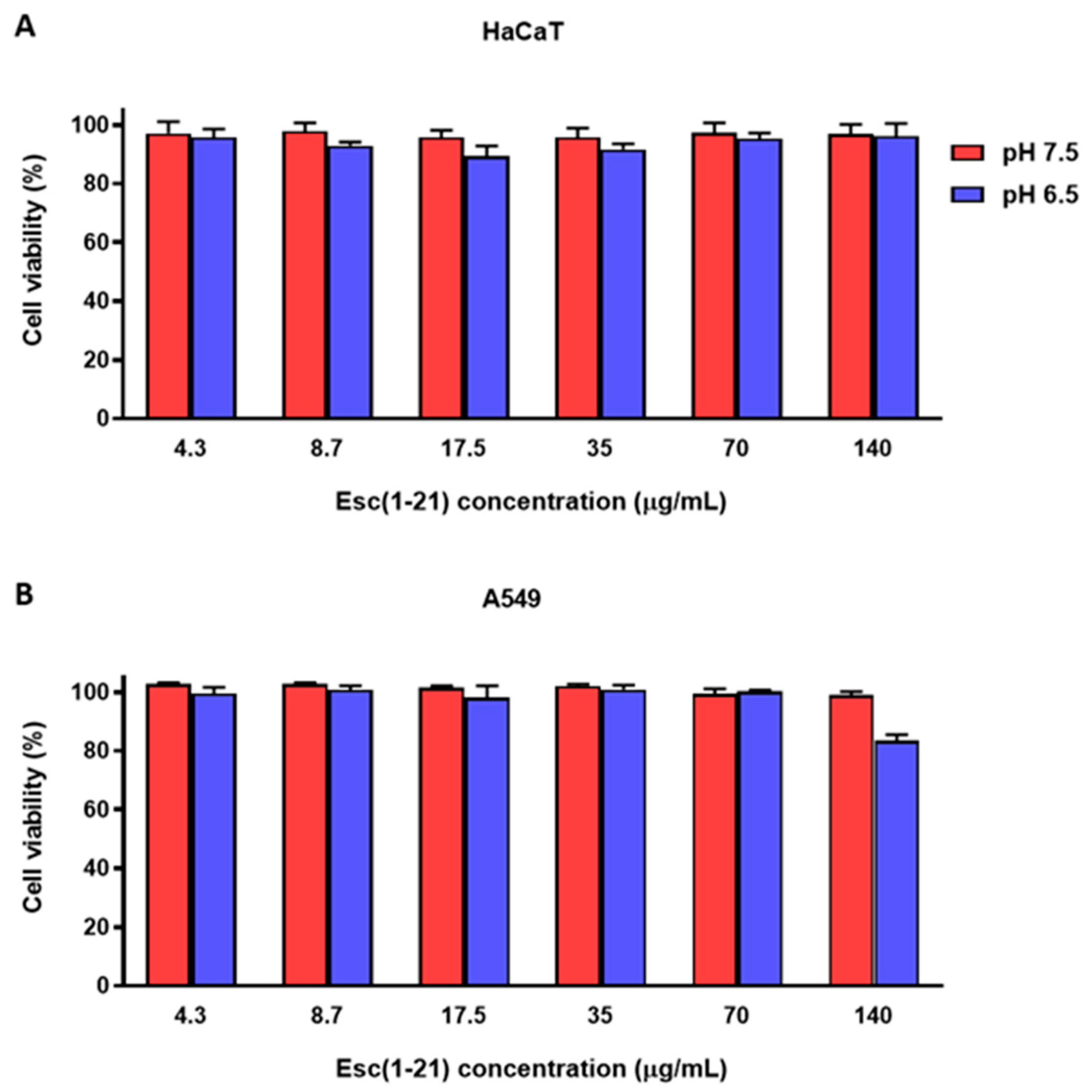
3.5. Cytotoxic Activity of Esc(1-21) at Different pHs against Mammalian Cell Lines
4. Summary and Conclusions
- (i)
- It slightly loses activity (from 2- to 4-fold) against the planktonic form of Gram-negative bacteria. This is in contrast with what happens with traditional antibiotics that lose their activity up to 32-fold;
- (ii)
- It retains antibiofilm activity against the sessile form of several Gram-negative bacteria grown in media with neutral pH; this does not happen for tobramycin, which is used as antibiotic control.
- (iii)
- It shows a similar or higher antibiofilm activity against the sessile form of bacteria grown in acidic media as that found in infectious site microenvironments, including CF lung.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McDonald, E.F.; Meiler, J.; Plate, L. CFTR Folding: From Structure and Proteostasis to Cystic Fibrosis Personalized Medicine. ACS Chem. Biol. 2023, 18, 2128–2143. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.D.; White, R.; Tobin, P. Keep them breathing: Cystic fibrosis pathophysiology, diagnosis, and treatment. JAAPA 2017, 30, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Pezzulo, A.A.; Tang, X.X.; Hoegger, M.J.; Abou Alaiwa, M.H.; Ramachandran, S.; Moninger, T.O.; Karp, P.H.; Wohlford-Lenane, C.L.; Haagsman, H.P.; van Eijk, M.; et al. Reduced airway surface pH impairs bacterial killing in the porcine cystic fibrosis lung. Nature 2012, 487, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Simonin, J.; Bille, E.; Crambert, G.; Noel, S.; Dreano, E.; Edwards, A.; Hatton, A.; Pranke, I.; Villeret, B.; Cottart, C.H.; et al. Airway surface liquid acidification initiates host defense abnormalities in Cystic Fibrosis. Sci. Rep. 2019, 9, 6516. [Google Scholar] [CrossRef]
- McShane, D.; Davies, J.C.; Davies, M.G.; Bush, A.; Geddes, D.M.; Alton, E.W. Airway surface pH in subjects with cystic fibrosis. Eur. Respir. J. 2003, 21, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Massip-Copiz, M.M.; Santa-Coloma, T.A. Extracellular pH and lung infections in cystic fibrosis. Eur. J. Cell Biol. 2018, 97, 402–410. [Google Scholar] [CrossRef]
- Shah, V.S.; Meyerholz, D.K.; Tang, X.X.; Reznikov, L.; Abou Alaiwa, M.; Ernst, S.E.; Karp, P.H.; Wohlford-Lenane, C.L.; Heilmann, K.P.; Leidinger, M.R.; et al. Airway acidification initiates host defense abnormalities in cystic fibrosis mice. Science 2016, 351, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Coakley, R.D.; Grubb, B.R.; Paradiso, A.M.; Gatzy, J.T.; Johnson, L.G.; Kreda, S.M.; O’Neal, W.K.; Boucher, R.C. Abnormal surface liquid pH regulation by cultured cystic fibrosis bronchial epithelium. Proc. Natl. Acad. Sci. USA 2003, 100, 16083–16088. [Google Scholar] [CrossRef]
- Wu, Z.; Pope, S.D.; Ahmed, N.S.; Leung, D.L.; Hajjar, S.; Yue, Q.; Anand, D.M.; Kopp, E.B.; Okin, D.; Ma, W.; et al. Control of Inflammatory Response by Tissue Microenvironment. bioRxiv 2024. [Google Scholar] [CrossRef]
- Li, Q. Bacterial infection and microbiota in carcinogenesis and tumor development. Front. Cell Infect. Microbiol. 2023, 13, 1294082. [Google Scholar] [CrossRef]
- Jablonska-Trypuc, A. The Role of the Microbiome in Inflammation and Carcinogenesis. Front. Biosci. (Elite Ed.) 2023, 15, 28. [Google Scholar] [CrossRef]
- Rocha, G.A.; Lima, D.F.; Rodrigues, E.R.; Leao, R.S.; Folescu, T.W.; Firmida, M.C.; Cohen, R.W.F.; Albano, R.M.; Marques, E.A. Species distribution, sequence types and antimicrobial resistance of Acinetobacter spp. from cystic fibrosis patients. Epidemiol. Infect. 2018, 146, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Phang, S.; Greysson-Wong, J.; Somayaji, R.; Storey, D.G.; Rabin, H.R.; Surette, M.G.; Parkins, M. Incidence, impact and natural history of Klebsiella species infections in cystic fibrosis: A longitudinal single center study. Can. J. Resp. Crit. Care 2019, 3, 148–154. [Google Scholar] [CrossRef]
- Sarkar, P.K.; Akand, N.; Tahura, S.; Kamruzzaman, M.; Akter, J.; Zaman, K.A.; Farhana, T.; Rima, S.S.; Alam, M.J.; Hassan, M.K.; et al. Antimicrobial sensitivity pattern of children with cystic fibrosis in Bangladesh: A lesson from a specialized Sishu (Children) Hospital. Egypt. Pediatr. Assoc. Gaz. 2022, 70, 33. [Google Scholar] [CrossRef]
- Azimi, L.; Ghanaiee, R.M.; Shirdust, M.; Karimi, A.M.; Armin, S.; Tabatabaei, S.R.; Shiva, F.; Fallah, F.; Rezaei, M.; Fahimzad, S.A.; et al. Molecular Identification of Gram-Negative Bacteria in Respiratory Samples of Cystic Fibrosis Patients from a Children Referral Hospital in Tehran. Arch. Pediatr. Infect. 2019, 7, e64834. [Google Scholar] [CrossRef]
- Jenkinson, A.C.; Mulligan, M.; White, R.; Power, N.; Lynch, C.; O’Connell, N.; Dunne, C.P.; Linnane, B. Acinetobacter baumannii Infection in a Child with Cystic Fibrosis. J. Paediatr. Child. Health 2021, 57, 1148–1149. [Google Scholar] [CrossRef]
- Perikleous, E.P.; Gkentzi, D.; Bertzouanis, A.; Paraskakis, E.; Sovtic, A.; Fouzas, S. Antibiotic Resistance in Patients with Cystic Fibrosis: Past, Present, and Future. Antibiotics 2023, 12, 217. [Google Scholar] [CrossRef]
- Vestby, L.K.; Gronseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Wan, N.; Wang, H.; Ng, C.K.; Mukherjee, M.; Ren, D.; Cao, B.; Tang, Y.J. Bacterial Metabolism During Biofilm Growth Investigated by (13)C Tracing. Front. Microbiol. 2018, 9, 2657. [Google Scholar] [CrossRef] [PubMed]
- Datta, M.; Rajeev, A.; Chattopadhyay, I. Application of antimicrobial peptides as next-generation therapeutics in the biomedical world. Biotechnol. Genet. Eng. Rev. 2023, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Cresti, L.; Cappello, G.; Pini, A. Antimicrobial Peptides towards Clinical Application-A Long History to Be Concluded. Int. J. Mol. Sci. 2024, 25, 4870. [Google Scholar] [CrossRef]
- Mehraj, I.; Hamid, A.; Gani, U.; Iralu, N.; Manzoor, T.; Saleem Bhat, S. Combating Antimicrobial Resistance by Employing Antimicrobial Peptides: Immunomodulators and Therapeutic Agents against Infectious Diseases. ACS Appl. Bio Mater. 2024, 7, 2023–2035. [Google Scholar] [CrossRef]
- Garcia, F.A.; Fuentes, T.F.; Alonso, I.P.; Bosch, R.A.; Brunetti, A.E.; Lopes, N.P. A Comprehensive Review of Patented Antimicrobial Peptides from Amphibian Anurans. J. Nat. Prod. 2024, 87, 600–616. [Google Scholar] [CrossRef]
- Mulukutla, A.; Shreshtha, R.; Kumar Deb, V.; Chatterjee, P.; Jain, U.; Chauhan, N. Recent advances in antimicrobial peptide-based therapy. Bioorg. Chem. 2024, 145, 107151. [Google Scholar] [CrossRef]
- Xuan, J.; Feng, W.; Wang, J.; Wang, R.; Zhang, B.; Bo, L.; Chen, Z.S.; Yang, H.; Sun, L. Antimicrobial peptides for combating drug-resistant bacterial infections. Drug Resist. Updat. 2023, 68, 100954. [Google Scholar] [CrossRef]
- Ahmad, A.; Khan, J.M.; Bandy, A. A Systematic Review of the Design and Applications of Antimicrobial Peptides in Wound Healing. Cureus 2024, 16, e58178. [Google Scholar] [CrossRef]
- Lin, Q.; Pilewski, J.M.; Di, Y.P. Acidic Microenvironment Determines Antibiotic Susceptibility and Biofilm Formation of Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 747834. [Google Scholar] [CrossRef]
- Kincses, A.; Racz, B.; Baaity, Z.; Vasarhelyi, O.; Kristof, E.; Somogyvari, F.; Spengler, G. The Relationship between Antibiotic Susceptibility and pH in the Case of Uropathogenic Bacteria. Antibiotics 2021, 10, 1431. [Google Scholar] [CrossRef]
- Dalhoff, A.; Schubert, S.; Ullmann, U. Effect of pH on the in vitro activity of and propensity for emergence of resistance to fluoroquinolones, macrolides, and a ketolide. Infection 2005, 33 (Suppl. S2), 36–43. [Google Scholar] [CrossRef]
- Luca, V.; Stringaro, A.; Colone, M.; Pini, A.; Mangoni, M.L. Esculentin(1-21), an amphibian skin membrane-active peptide with potent activity on both planktonic and biofilm cells of the bacterial pathogen Pseudomonas aeruginosa. Cell. Mol. Life Sci. 2013, 70, 2773–2786. [Google Scholar] [CrossRef]
- Loffredo, M.R.; Ghosh, A.; Harmouche, N.; Casciaro, B.; Luca, V.; Bortolotti, A.; Cappiello, F.; Stella, L.; Bhunia, A.; Bechinger, B.; et al. Membrane perturbing activities and structural properties of the frog-skin derived peptide Esculentin-1a(1-21)NH(2) and its Diastereomer Esc(1-21)-1c: Correlation with their antipseudomonal and cytotoxic activity. Biochim. Biophys. Acta Biomembr. 2017, 1859, 2327–2339. [Google Scholar] [CrossRef]
- Casciaro, B.; Dutta, D.; Loffredo, M.R.; Marcheggiani, S.; McDermott, A.M.; Willcox, M.D.; Mangoni, M.L. Esculentin-1a derived peptides kill Pseudomonas aeruginosa biofilm on soft contact lenses and retain antibacterial activity upon immobilization to the lens surface. Pept. Sci. 2018, 110, e23074. [Google Scholar] [CrossRef]
- Casciaro, B.; Loffredo, M.R.; Luca, V.; Verrusio, W.; Cacciafesta, M.; Mangoni, M.L. Esculentin-1a Derived Antipseudomonal Peptides: Limited Induction of Resistance and Synergy with Aztreonam. Protein Pept. Lett. 2018, 25, 1155–1162. [Google Scholar] [CrossRef]
- Cappiello, F.; Carnicelli, V.; Casciaro, B.; Mangoni, M.L. Antipseudomonal and Immunomodulatory Properties of Esc Peptides: Promising Features for Treatment of Chronic Infectious Diseases and Inflammation. Int. J. Mol. Sci. 2021, 22, 557. [Google Scholar] [CrossRef]
- Sacco, F.; Bitossi, C.; Casciaro, B.; Loffredo, M.R.; Fabiano, G.; Torrini, L.; Raponi, F.; Raponi, G.; Mangoni, M.L. The Antimicrobial Peptide Esc(1-21) Synergizes with Colistin in Inhibiting the Growth and in Killing Multidrug Resistant Acinetobacter baumannii Strains. Antibiotics 2022, 11, 234. [Google Scholar] [CrossRef]
- Scotti, R.; Casciaro, B.; Stringaro, A.; Morgia, F.; Mangoni, M.L.; Gabbianelli, R. Derivatives of Esculentin-1 Peptides as Promising Candidates for Fighting Infections from Escherichia coli O157:H7. Antibiotics 2022, 11, 656. [Google Scholar] [CrossRef]
- Cappiello, F.; Casciaro, B.; Loffredo, M.R.; Puglisi, E.; Lin, Q.; Yang, D.; Conte, G.; d’Angelo, I.; Ungaro, F.; Ferrera, L.; et al. Pulmonary Safety Profile of Esc Peptides and Esc-Peptide-Loaded Poly(lactide-co-glycolide) Nanoparticles: A Promising Therapeutic Approach for Local Treatment of Lung Infectious Diseases. Pharmaceutics 2022, 14, 2297. [Google Scholar] [CrossRef]
- Casciaro, B.; Cappiello, F.; Loffredo, M.R.; Ghirga, F.; Mangoni, M.L. The Potential of Frog Skin Peptides for Anti-Infective Therapies: The Case of Esculentin-1a(1-21)NH2. Curr. Med. Chem. 2020, 27, 1405–1419. [Google Scholar] [CrossRef]
- Casciaro, B.; Loffredo, M.R.; Cappiello, F.; Verrusio, W.; Corleto, V.D.; Mangoni, M.L. Frog Skin-Derived Peptides Against Corynebacterium jeikeium: Correlation between Antibacterial and Cytotoxic Activities. Antibiotics 2020, 9, 448. [Google Scholar] [CrossRef]
- Bragonzi, A.; Paroni, M.; Nonis, A.; Cramer, N.; Montanari, S.; Rejman, J.; Di Serio, C.; Doring, G.; Tummler, B. Pseudomonas aeruginosa microevolution during cystic fibrosis lung infection establishes clones with adapted virulence. Am. J. Respir. Crit. Care Med. 2009, 180, 138–145. [Google Scholar] [CrossRef]
- Khosravi, M.; Lin, R.L.; Maskey, A.P.; Pandey, S.; Lin, A.H.; Lee, L.Y. A Distinct Difference between Air and Mucosal Temperatures in Human Respiratory Tract. Front. Med. 2021, 8, 650637. [Google Scholar] [CrossRef]
- Di Grazia, A.; Cappiello, F.; Cohen, H.; Casciaro, B.; Luca, V.; Pini, A.; Di, Y.P.; Shai, Y.; Mangoni, M.L. D-Amino acids incorporation in the frog skin-derived peptide esculentin-1a(1-21)NH2 is beneficial for its multiple functions. Amino Acids 2015, 47, 2505–2519. [Google Scholar] [CrossRef]
- Stewart, J.C. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal. Biochem. 1980, 104, 10–14. [Google Scholar] [CrossRef]
- Jafarinejad, S.; Martin, W.H.C.; Ras, B.A.; Isreb, M.; Jacob, B.; Aziz, A.; Adoul, Z.; Lagnado, R.; Bowen, R.D.; Najafzadeh, M. The anticancer/cytotoxic effect of a novel gallic acid derivative in non-small cell lung carcinoma A549 cells and peripheral blood mononuclear cells from healthy individuals and lung cancer patients. Biofactors 2024, 50, 201–213. [Google Scholar] [CrossRef]
- Mi, B.; Liu, J.; Liu, G.; Zhou, W.; Liu, Y.; Hu, L.; Xiong, L.; Ye, S.; Wu, Y. Icariin promotes wound healing by enhancing the migration and proliferation of keratinocytes via the AKT and ERK signaling pathway. Int. J. Mol. Med. 2018, 42, 831–838. [Google Scholar] [CrossRef]
- Nussbaumer-Proll, A.K.; Eberl, S.; Schober, C.; Zeitlinger, M. Impact of pH on the activity of novel cephalosporin cefiderocol in human urine. J. Antimicrob. Chemother. 2024, 79, 166–171. [Google Scholar] [CrossRef]
- Abou Alaiwa, M.H.; Reznikov, L.R.; Gansemer, N.D.; Sheets, K.A.; Horswill, A.R.; Stoltz, D.A.; Zabner, J.; Welsh, M.J. pH modulates the activity and synergism of the airway surface liquid antimicrobials beta-defensin-3 and LL-37. Proc. Natl. Acad. Sci. USA 2014, 111, 18703–18708. [Google Scholar] [CrossRef]
- Holdbrook, D.A.; Singh, S.; Choong, Y.K.; Petrlova, J.; Malmsten, M.; Bond, P.J.; Verma, N.K.; Schmidtchen, A.; Saravanan, R. Influence of pH on the activity of thrombin-derived antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2018, 1860, 2374–2384. [Google Scholar] [CrossRef]
- Hitchner, M.A.; Santiago-Ortiz, L.E.; Necelis, M.R.; Shirley, D.J.; Palmer, T.J.; Tarnawsky, K.E.; Vaden, T.D.; Caputo, G.A. Activity and characterization of a pH-sensitive antimicrobial peptide. Biochim. Biophys. Acta Biomembr. 2019, 1861, 182984. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Q.; Li, J.; Shang, L.; Li, J.; Chou, S.; Lyu, Y.; Shan, A. pH-Responsive Antimicrobial Peptide with Selective Killing Activity for Bacterial Abscess Therapy. J. Med. Chem. 2022, 65, 5355–5373. [Google Scholar] [CrossRef]
- Mohamed, M.F.; Brezden, A.; Mohammad, H.; Chmielewski, J.; Seleem, M.N. A short D-enantiomeric antimicrobial peptide with potent immunomodulatory and antibiofilm activity against multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii. Sci. Rep. 2017, 7, 6953. [Google Scholar] [CrossRef]
- Moriarty, T.F.; Elborn, J.S.; Tunney, M.M. Effect of pH on the antimicrobial susceptibility of planktonic and biofilm-grown clinical Pseudomonas aeruginosa isolates. Br. J. Biomed. Sci. 2007, 64, 101–104. [Google Scholar] [CrossRef]
- Becker, K.; Cao, S.; Nilsson, A.; Erlandsson, M.; Hotop, S.K.; Kuka, J.; Hansen, J.; Haldimann, K.; Grinberga, S.; Berruga-Fernandez, T.; et al. Antibacterial activity of apramycin at acidic pH warrants wide therapeutic window in the treatment of complicated urinary tract infections and acute pyelonephritis. EBioMedicine 2021, 73, 103652. [Google Scholar] [CrossRef]
- Scoffone, V.C.; Barbieri, G.; Irudal, S.; Trespidi, G.; Buroni, S. New Antimicrobial Strategies to Treat Multi-Drug Resistant Infections Caused by Gram-Negatives in Cystic Fibrosis. Antibiotics 2024, 13, 71. [Google Scholar] [CrossRef]
- Batoni, G.; Maisetta, G.; Esin, S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim. Biophys. Acta 2016, 1858, 1044–1060. [Google Scholar] [CrossRef]
- Ruhal, R.; Kataria, R. Biofilm patterns in gram-positive and gram-negative bacteria. Microbiol. Res. 2021, 251, 126829. [Google Scholar] [CrossRef]
- Wu, K.C.; Hua, K.F.; Yu, Y.H.; Cheng, Y.H.; Cheng, T.T.; Huang, Y.K.; Chang, H.W.; Chen, W.J. Antibacterial and Antibiofilm Activities of Novel Antimicrobial Peptides against Multidrug-Resistant Enterotoxigenic Escherichia Coli. Int. J. Mol. Sci. 2021, 22, 3926. [Google Scholar] [CrossRef]
- de la Fuente-Nunez, C.; Cardoso, M.H.; de Souza Candido, E.; Franco, O.L.; Hancock, R.E. Synthetic antibiofilm peptides. Biochim. Biophys. Acta 2016, 1858, 1061–1069. [Google Scholar] [CrossRef]
- Lund, P.A.; De Biase, D.; Liran, O.; Scheler, O.; Mira, N.P.; Cetecioglu, Z.; Fernandez, E.N.; Bover-Cid, S.; Hall, R.; Sauer, M.; et al. Understanding How Microorganisms Respond to Acid pH Is Central to Their Control and Successful Exploitation. Front. Microbiol. 2020, 11, 556140. [Google Scholar] [CrossRef]
- Guan, N.; Liu, L. Microbial response to acid stress: Mechanisms and applications. Appl. Microbiol. Biotechnol. 2020, 104, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Pennacchietti, E.; D’Alonzo, C.; Freddi, L.; Occhialini, A.; De Biase, D. The Glutaminase-Dependent Acid Resistance System: Qualitative and Quantitative Assays and Analysis of Its Distribution in Enteric Bacteria. Front. Microbiol. 2018, 9, 2869. [Google Scholar] [CrossRef] [PubMed]
- Krulwich, T.A.; Sachs, G.; Padan, E. Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 2011, 9, 330–343. [Google Scholar] [CrossRef]
- Russell, J.B. The energy spilling reactions of bacteria and other organisms. J. Mol. Microbiol. Biotechnol. 2007, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mozaheb, N.; Rasouli, P.; Kaur, M.; Van Der Smissen, P.; Larrouy-Maumus, G.; Mingeot-Leclercq, M.P. A Mildly Acidic Environment Alters Pseudomonas aeruginosa Virulence and Causes Remodeling of the Bacterial Surface. Microbiol. Spectr. 2023, 11, e0483222. [Google Scholar] [CrossRef]
- Hostacka, A.; Ciznar, I.; Stefkovicova, M. Temperature and pH affect the production of bacterial biofilm. Folia Microbiol. 2010, 55, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Ignatova, N.; Abidullina, A.; Streltsova, O.; Elagin, V.; Kamensky, V. Effect of pH, Norepinephrine and Glucose on Metabolic and Biofilm Activity of Uropathogenic Microorganisms. Microorganisms 2023, 11, 862. [Google Scholar] [CrossRef] [PubMed]
- Van den Bossche, S.; De Broe, E.; Coenye, T.; Van Braeckel, E.; Crabbe, A. The cystic fibrosis lung microenvironment alters antibiotic activity: Causes and effects. Eur. Respir. Rev. 2021, 30, 210055. [Google Scholar] [CrossRef]
- Malik, E.; Dennison, S.R.; Harris, F.; Phoenix, D.A. pH Dependent Antimicrobial Peptides and Proteins, Their Mechanisms of Action and Potential as Therapeutic Agents. Pharmaceuticals 2016, 9, 67. [Google Scholar] [CrossRef]
- Biondi, B.; Casciaro, B.; Di Grazia, A.; Cappiello, F.; Luca, V.; Crisma, M.; Mangoni, M.L. Effects of Aib residues insertion on the structural-functional properties of the frog skin-derived peptide esculentin-1a(1-21)NH(2). Amino Acids 2017, 49, 139–150. [Google Scholar] [CrossRef]
- Walsh, T.R.; Gales, A.C.; Laxminarayan, R.; Dodd, P.C. Antimicrobial Resistance: Addressing a Global Threat to Humanity. PLoS Med. 2023, 20, e1004264. [Google Scholar] [CrossRef]
- Guillaume, O.; Butnarasu, C.; Visentin, S.; Reimhult, E. Interplay between biofilm microenvironment and pathogenicity of Pseudomonas aeruginosa in cystic fibrosis lung chronic infection. Biofilm 2022, 4, 100089. [Google Scholar] [CrossRef]
- Thomas, J.; Linton, S.; Corum, L.; Slone, W.; Okel, T.; Percival, S.L. The affect of pH and bacterial phenotypic state on antibiotic efficacy. Int. Wound J. 2012, 9, 428–435. [Google Scholar] [CrossRef]
- Yang, L.; Wang, K.; Li, H.; Denstedt, J.D.; Cadieux, P.A. The influence of urinary pH on antibiotic efficacy against bacterial uropathogens. Urology 2014, 84, 731.e1–731.e7. [Google Scholar] [CrossRef]



| Strain | Resistance Profile |
|---|---|
| K. pneumoniae #3 | AMI–CEFE–CEFO–CEFT–CIP–ERT–FOS–PIP/TAZ |
| K. pneumoniae #4 | AMI–AMO/CLA–CEFE–CEFO–CEFT–CIP–ERT–PIP/TAZ |
| A. baumannii #2 | AMI–AMO/CLA–CEFE–CEFO–CEFT–CIP–GEN–IMI–MER–PIP/TAZ–TRI/SUL |
| A. baumannii #3 | AMO/CLA–AMP–CEFO–CIP–COL–ERT–GEN–IMI–PIP/TAZ–TRI/SUL |
| A. baumannii #4 | AMO/CLA–AMP–CEFO–CIP–COL–ERT–GEN–IMI–PIP/TAZ–TRI/SUL |
| MICs (µg/mL) | |||
|---|---|---|---|
| Compound | A. baumannii ATCC 19606 | E. coli ATCC 25922 | P. aeruginosa ATCC 27853 |
| Esc(1-21) | 4.3 | 4.3 | 8.7 |
| Bombinin H2 | 15.3 | 61.3 | 245 |
| Temporin L | 3.25 | 6.5 | 6.5 |
| LL-37 | 9.0 | 9.0 | 4.5 |
| Colistin | 0.5 | 0.5 | 0.25 |
| Tobramycin | 2.0 | 1.0 | 0.125 |
| Ciprofloxacin | 0.25 | 0.25 | 0.125 |
| MICs (µg/mL) | ||
|---|---|---|
| Strain | Esc(1-21) | Tobramycin |
| P. aeruginosa PAO1 | 8.7 | 0.25 |
| P. aeruginosa AA11 | 2.2 | 0.25 |
| P. aeruginosa KK1 | 17.5 | 0.125 |
| A. baumannii #2 | 2.2 | 16 |
| A. baumannii #3 | 2.2 | 16 |
| A. baumannii #4 | 2.2 | 16 |
| K. pneumoniae #3 | 8.7 | 32 |
| K. pneumoniae #4 | 8.7 | 64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loffredo, M.R.; Cappiello, F.; Cappella, G.; Capuozzo, E.; Torrini, L.; Diaco, F.; Di, Y.P.; Mangoni, M.L.; Casciaro, B. The pH-Insensitive Antimicrobial and Antibiofilm Activities of the Frog Skin Derived Peptide Esc(1-21): Promising Features for Novel Anti-Infective Drugs. Antibiotics 2024, 13, 701. https://doi.org/10.3390/antibiotics13080701
Loffredo MR, Cappiello F, Cappella G, Capuozzo E, Torrini L, Diaco F, Di YP, Mangoni ML, Casciaro B. The pH-Insensitive Antimicrobial and Antibiofilm Activities of the Frog Skin Derived Peptide Esc(1-21): Promising Features for Novel Anti-Infective Drugs. Antibiotics. 2024; 13(8):701. https://doi.org/10.3390/antibiotics13080701
Chicago/Turabian StyleLoffredo, Maria Rosa, Floriana Cappiello, Giacomo Cappella, Elisabetta Capuozzo, Luisa Torrini, Fabiana Diaco, Yuanpu Peter Di, Maria Luisa Mangoni, and Bruno Casciaro. 2024. "The pH-Insensitive Antimicrobial and Antibiofilm Activities of the Frog Skin Derived Peptide Esc(1-21): Promising Features for Novel Anti-Infective Drugs" Antibiotics 13, no. 8: 701. https://doi.org/10.3390/antibiotics13080701
APA StyleLoffredo, M. R., Cappiello, F., Cappella, G., Capuozzo, E., Torrini, L., Diaco, F., Di, Y. P., Mangoni, M. L., & Casciaro, B. (2024). The pH-Insensitive Antimicrobial and Antibiofilm Activities of the Frog Skin Derived Peptide Esc(1-21): Promising Features for Novel Anti-Infective Drugs. Antibiotics, 13(8), 701. https://doi.org/10.3390/antibiotics13080701









