Identifying Cell-Penetrating Peptides for Effectively Delivering Antimicrobial Molecules into Streptococcus suis
Abstract
:1. Introduction
2. Results
2.1. CPPs Penetration into S. suis
2.2. CPPs Uptake Efficiency Analysis by Flow Cytometry
2.3. Toxicity Analysis of HIV-1 TAT and (RXR)4XB
2.4. HIV-1 TAT-Coupled gyrA-Specific PNA Exhibits Bactericidal Activity
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. CPPs Synthesis and Fluorescent Labeling
4.3. SR-SIM Analysis
4.4. Flow Cytometry Analysis
4.5. Synthesis of PNAs and CPP-PNA Conjugates
4.6. MIC Determination
4.7. Determination of Bactericidal Effects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gori, A.; Lodigiani, G.; Colombarolli, S.G.; Bergamaschi, G.; Vitali, A. Cell Penetrating Peptides: Classification, Mechanisms, Methods of Study, and Applications. ChemMedChem 2023, 18, e202300236. [Google Scholar] [CrossRef] [PubMed]
- Zorko, M.; Langel, U. Cell-Penetrating Peptides. Methods Mol. Biol. 2022, 2383, 3–32. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef] [PubMed]
- Kotadiya, D.D.; Patel, P.; Patel, H.D. Cell-Penetrating Peptides: A Powerful Tool for Targeted Drug Delivery. Curr. Drug Deliv. 2024, 21, 368–388. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Y.; Zhang, C.; Yu, H.; Ma, Y.; Li, Z.; Shi, N. Recent Advances of Cell-Penetrating Peptides and Their Application as Vectors for Delivery of Peptide and Protein-Based Cargo Molecules. Pharmaceutics 2023, 15, 2093. [Google Scholar] [CrossRef] [PubMed]
- Tietz, O.; Cortezon-Tamarit, F.; Chalk, R.; Able, S.; Vallis, K.A. Tricyclic cell-penetrating peptides for efficient delivery of functional antibodies into cancer cells. Nat. Chem. 2022, 14, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Sauter, M.; Strieker, M.; Kleist, C.; Wischnjow, A.; Daniel, V.; Altmann, A.; Haberkorn, U.; Mier, W. Improving antibody-based therapies by chemical engineering of antibodies with multimeric cell-penetrating peptides for elevated intracellular delivery. J. Control. Release 2020, 322, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Pifer, R.; Greenberg, D.E. Antisense antibacterial compounds. Transl. Res. 2020, 223, 89–106. [Google Scholar] [CrossRef]
- Barkowsky, G.; Abt, C.; Pohner, I.; Bieda, A.; Hammerschmidt, S.; Jacob, A.; Kreikemeyer, B.; Patenge, N. Antimicrobial Activity of Peptide-Coupled Antisense Peptide Nucleic Acids in Streptococcus pneumoniae. Microbiol. Spectr. 2022, 10, e00497-22. [Google Scholar] [CrossRef]
- Moustafa, D.A.; Wu, A.W.; Zamora, D.; Daly, S.M.; Sturge, C.R.; Pybus, C.; Geller, B.L.; Goldberg, J.B.; Greenberg, D.E. Peptide-Conjugated Phosphorodiamidate Morpholino Oligomers Retain Activity against Multidrug-Resistant Pseudomonas aeruginosa In Vitro and In Vivo. mBio 2021, 12, e02411-20. [Google Scholar] [CrossRef]
- Geller, B.L.; Marshall-Batty, K.; Schnell, F.J.; McKnight, M.M.; Iversen, P.L.; Greenberg, D.E. Gene-silencing antisense oligomers inhibit acinetobacter growth in vitro and in vivo. J. Infect. Dis. 2013, 208, 1553–1560. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J. An RNA biology perspective on species-specific programmable RNA antibiotics. Mol. Microbiol. 2020, 113, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Sully, E.K.; Geller, B.L. Antisense antimicrobial therapeutics. Curr. Opin. Microbiol. 2016, 33, 47–55. [Google Scholar] [CrossRef] [PubMed]
- El-Fateh, M.; Chatterjee, A.; Zhao, X. A systematic review of peptide nucleic acids (PNAs) with antibacterial activities: Efficacy, potential and challenges. Int. J. Antimicrob. Agents 2024, 63, 107083. [Google Scholar] [CrossRef] [PubMed]
- Eller, K.A.; Aunins, T.R.; Courtney, C.M.; Campos, J.K.; Otoupal, P.B.; Erickson, K.E.; Madinger, N.E.; Chatterjee, A. Facile accelerated specific therapeutic (FAST) platform develops antisense therapies to counter multidrug-resistant bacteria. Commun. Biol. 2021, 4, 331. [Google Scholar] [CrossRef]
- Good, L.; Nielsen, P.E. Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat. Biotechnol. 1998, 16, 355–358. [Google Scholar] [CrossRef]
- Geller, B.L.; Li, L.; Martinez, F.; Sully, E.; Sturge, C.R.; Daly, S.M.; Pybus, C.; Greenberg, D.E. Morpholino oligomers tested in vitro, in biofilm and in vivo against multidrug-resistant Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018, 73, 1611–1619. [Google Scholar] [CrossRef] [PubMed]
- Barkowsky, G.; Lemster, A.L.; Pappesch, R.; Jacob, A.; Kruger, S.; Schroder, A.; Kreikemeyer, B.; Patenge, N. Influence of Different Cell-Penetrating Peptides on the Antimicrobial Efficiency of PNAs in Streptococcus pyogenes. Mol. Ther. Nucleic Acids 2019, 18, 444–454. [Google Scholar] [CrossRef]
- Patenge, N.; Pappesch, R.; Krawack, F.; Walda, C.; Mraheil, M.A.; Jacob, A.; Hain, T.; Kreikemeyer, B. Inhibition of Growth and Gene Expression by PNA-peptide Conjugates in Streptococcus pyogenes. Mol. Ther. Nucleic Acids 2013, 2, e132. [Google Scholar] [CrossRef]
- Popella, L.; Jung, J.; Popova, K.; Ethurica-Mitic, S.; Barquist, L.; Vogel, J. Global RNA profiles show target selectivity and physiological effects of peptide-delivered antisense antibiotics. Nucleic Acids Res. 2021, 49, 4705–4724. [Google Scholar] [CrossRef]
- Tripathy, S.; Sahu, S.K. FtsZ inhibitors as a new genera of antibacterial agents. Bioorg. Chem. 2019, 91, 103169. [Google Scholar] [CrossRef]
- Abushahba, M.F.; Mohammad, H.; Thangamani, S.; Hussein, A.A.; Seleem, M.N. Impact of different cell penetrating peptides on the efficacy of antisense therapeutics for targeting intracellular pathogens. Sci. Rep. 2016, 6, 20832. [Google Scholar] [CrossRef]
- Popella, L.; Jung, J.; Do, P.T.; Hayward, R.J.; Barquist, L.; Vogel, J. Comprehensive analysis of PNA-based antisense antibiotics targeting various essential genes in uropathogenic Escherichia coli. Nucleic Acids Res. 2022, 50, 6435–6452. [Google Scholar] [CrossRef]
- Inoue, G.; Toyohara, D.; Mori, T.; Muraoka, T. Critical Side Chain Effects of Cell-Penetrating Peptides for Transporting Oligo Peptide Nucleic Acids in Bacteria. ACS Appl. Bio Mater. 2021, 4, 3462–3468. [Google Scholar] [CrossRef]
- Murray, G.G.R.; Hossain, A.; Miller, E.L.; Bruchmann, S.; Balmer, A.J.; Matuszewska, M.; Herbert, J.; Hadjirin, N.F.; Mugabi, R.; Li, G.; et al. The emergence and diversification of a zoonotic pathogen from within the microbiota of intensively farmed pigs. Proc. Natl. Acad. Sci. USA 2023, 120, e2307773120. [Google Scholar] [CrossRef] [PubMed]
- Goyette-Desjardins, G.; Auger, J.P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014, 3, e45. [Google Scholar] [CrossRef]
- Segura, M.; Aragon, V.; Brockmeier, S.L.; Gebhart, C.; Greeff, A.; Kerdsin, A.; O’Dea, M.A.; Okura, M.; Salery, M.; Schultsz, C.; et al. Update on Streptococcus suis Research and Prevention in the Era of Antimicrobial Restriction: 4th International Workshop on S. suis. Pathogens 2020, 9, 374. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, J.; Kang, W.; Zhang, X.; Kerdsin, A.; Yao, H.; Zheng, H.; Wu, Z. Streptococcus suis serotype 4: A population with the potential pathogenicity in humans and pigs. Emerg. Microbes Infect. 2024, 13, 2352435. [Google Scholar] [CrossRef] [PubMed]
- Hatrongjit, R.; Fittipaldi, N.; Jenjaroenpun, P.; Wongsurawat, T.; Visetnan, S.; Zheng, H.; Gottschalk, M.; Kerdsin, A. Genomic comparison of two Streptococcus suis serotype 1 strains recovered from porcine and human disease cases. Sci. Rep. 2023, 13, 5380. [Google Scholar] [CrossRef]
- Liang, P.; Wang, M.; Gottschalk, M.; Vela, A.I.; Estrada, A.A.; Wang, J.; Du, P.; Luo, M.; Zheng, H.; Wu, Z. Genomic and pathogenic investigations of Streptococcus suis serotype 7 population derived from a human patient and pigs. Emerg. Microbes Infect. 2021, 10, 1960–1974. [Google Scholar] [CrossRef]
- Petrocchi-Rilo, M.; Martinez-Martinez, S.; Aguaron-Turrientes, A.; Roca-Martinez, E.; Garcia-Iglesias, M.J.; Perez-Fernandez, E.; Gonzalez-Fernandez, A.; Herencia-Lagunar, E.; Gutierrez-Martin, C.B. Anatomical Site, Typing, Virulence Gene Profiling, Antimicrobial Susceptibility and Resistance Genes of Streptococcus suis Isolates Recovered from Pigs in Spain. Antibiotics 2021, 10, 707. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.F.; Tan, J.; Zeng, Y.B.; Li, H.Q.; Yang, Q.; Zhou, R. Antimicrobial resistance phenotypes and genotypes of Streptococcus suis isolated from clinically healthy pigs from 2017 to 2019 in Jiangxi Province, China. J. Appl. Microbiol. 2021, 130, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Matajira, C.E.C.; Moreno, L.Z.; Poor, A.P.; Gomes, V.T.M.; Dalmutt, A.C.; Parra, B.M.; Oliveira, C.H.; Barbosa, M.R.F.; Sato, M.I.Z.; Calderaro, F.F.; et al. Streptococcus suis in Brazil: Genotypic, Virulence, and Resistance Profiling of Strains Isolated from Pigs between 2001 and 2016. Pathogens 2019, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Yongkiettrakul, S.; Maneerat, K.; Arechanajan, B.; Malila, Y.; Srimanote, P.; Gottschalk, M.; Visessanguan, W. Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs, asymptomatic pigs, and human patients in Thailand. BMC Vet. Res. 2019, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Vaara, M.; Porro, M. Group of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob. Agents Chemother. 1996, 40, 1801–1805. [Google Scholar] [CrossRef] [PubMed]
- Vives, E.; Brodin, P.; Lebleu, B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 1997, 272, 16010–16017. [Google Scholar] [CrossRef] [PubMed]
- Abes, R.; Moulton, H.M.; Clair, P.; Yang, S.T.; Abes, S.; Melikov, K.; Prevot, P.; Youngblood, D.S.; Iversen, P.L.; Chernomordik, L.V.; et al. Delivery of steric block morpholino oligomers by (R-X-R)4 peptides: Structure-activity studies. Nucleic Acids Res. 2008, 36, 6343–6354. [Google Scholar] [CrossRef] [PubMed]
- MacNair, C.R.; Rutherford, S.T.; Tan, M.W. Alternative therapeutic strategies to treat antibiotic-resistant pathogens. Nat. Rev. Microbiol. 2024, 22, 262–275. [Google Scholar] [CrossRef]
- Demidov, V.V.; Potaman, V.N.; Frank-Kamenetskii, M.D.; Egholm, M.; Buchard, O.; Sonnichsen, S.H.; Nielsen, P.E. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol. 1994, 48, 1310–1313. [Google Scholar] [CrossRef]
- Good, L.; Awasthi, S.K.; Dryselius, R.; Larsson, O.; Nielsen, P.E. Bactericidal antisense effects of peptide-PNA conjugates. Nat. Biotechnol. 2001, 19, 360–364. [Google Scholar] [CrossRef]
- Nekhotiaeva, N.; Awasthi, S.K.; Nielsen, P.E.; Good, L. Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol. Ther. 2004, 10, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Tavakoli, M.; Hashemi, A.; Vaezjalali, M.; Mohammadzadeh, M.; Goudarzi, H. Inhibition of growth and gene expression in Staphylococcus aureus by anti-gyrA peptide nucleic acid. Future Microbiol. 2019, 14, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; He, Y.; Xia, Y.; Wang, L.; Liang, S. Inhibition of gene expression and growth of multidrug-resistant Acinetobacter baumannii by antisense peptide nucleic acids. Mol. Biol. Rep. 2014, 41, 7535–7541. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, D.J.; Kim, D.T.; Steinman, L.; Fathman, C.G.; Rothbard, J.B. Polyarginine enters cells more efficiently than other polycationic homopolymers. J. Pept. Res. 2000, 56, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Howard, J.J.; Sturge, C.R.; Moustafa, D.A.; Daly, S.M.; Marshall-Batty, K.R.; Felder, C.F.; Zamora, D.; Yabe-Gill, M.; Labandeira-Rey, M.; Bailey, S.M.; et al. Inhibition of Pseudomonas aeruginosa by Peptide-Conjugated Phosphorodiamidate Morpholino Oligomers. Antimicrob. Agents Chemother. 2017, 61, e01938-16. [Google Scholar] [CrossRef] [PubMed]
- Daly, S.M.; Sturge, C.R.; Felder-Scott, C.F.; Geller, B.L.; Greenberg, D.E. MCR-1 Inhibition with Peptide-Conjugated Phosphorodiamidate Morpholino Oligomers Restores Sensitivity to Polymyxin in Escherichia coli. mBio 2017, 8, e01315-17. [Google Scholar] [CrossRef]
- Aunins, T.R.; Erickson, K.E.; Chatterjee, A. Transcriptome-based design of antisense inhibitors potentiates carbapenem efficacy in CRE Escherichia coli. Proc. Natl. Acad. Sci. USA 2020, 117, 30699–30709. [Google Scholar] [CrossRef]
- Oh, E.; Zhang, Q.; Jeon, B. Target optimization for peptide nucleic acid (PNA)-mediated antisense inhibition of the CmeABC multidrug efflux pump in Campylobacter jejuni. J. Antimicrob. Chemother. 2014, 69, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Goh, S.; Loeffler, A.; Lloyd, D.H.; Nair, S.P.; Good, L. Oxacillin sensitization of methicillin-resistant Staphylococcus aureus and methicillin-resistant Staphylococcus pseudintermedius by antisense peptide nucleic acids in vitro. BMC Microbiol. 2015, 15, 262. [Google Scholar] [CrossRef]
- Fleitas Martinez, O.; Cardoso, M.H.; Ribeiro, S.M.; Franco, O.L. Recent Advances in Anti-virulence Therapeutic Strategies with a Focus on Dismantling Bacterial Membrane Microdomains, Toxin Neutralization, Quorum-Sensing Interference and Biofilm Inhibition. Front. Cell Infect. Microbiol. 2019, 9, 74. [Google Scholar] [CrossRef]
- Rasko, D.A.; Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Muhlen, S.; Dersch, P. Anti-virulence Strategies to Target Bacterial Infections. Curr. Top. Microbiol. Immunol. 2016, 398, 147–183. [Google Scholar] [CrossRef]
- Daly, S.M.; Elmore, B.O.; Kavanaugh, J.S.; Triplett, K.D.; Figueroa, M.; Raja, H.A.; El-Elimat, T.; Crosby, H.A.; Femling, J.K.; Cech, N.B.; et al. omega-Hydroxyemodin limits Staphylococcus aureus quorum sensing-mediated pathogenesis and inflammation. Antimicrob. Agents Chemother. 2015, 59, 2223–2235. [Google Scholar] [CrossRef] [PubMed]
- Sully, E.K.; Malachowa, N.; Elmore, B.O.; Alexander, S.M.; Femling, J.K.; Gray, B.M.; DeLeo, F.R.; Otto, M.; Cheung, A.L.; Edwards, B.S.; et al. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 2014, 10, e1004174. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Odahara, M.; Yoshizumi, T.; Oikawa, K.; Kimura, M.; Su’etsugu, M.; Numata, K. Cell-Penetrating Peptide-Mediated Transformation of Large Plasmid DNA into Escherichia coli. ACS Synth. Biol. 2019, 8, 1215–1218. [Google Scholar] [CrossRef] [PubMed]
- Numata, K.; Kaplan, D.L. Silk-based gene carriers with cell membrane destabilizing peptides. Biomacromolecules 2010, 11, 3189–3195. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Dong, W.; Ma, J.; Zhang, Y.; Pan, Z.; Yao, H. Utilization of the ComRS system for the rapid markerless deletion of chromosomal genes in Streptococcus suis. Future Microbiol. 2019, 14, 207–222. [Google Scholar] [CrossRef] [PubMed]
- Zaccaria, E.; van Baarlen, P.; de Greeff, A.; Morrison, D.A.; Smith, H.; Wells, J.M. Control of competence for DNA transformation in streptococcus suis by genetically transferable pherotypes. PLoS ONE 2014, 9, e99394. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wang, W.; Tang, M.; Shao, J.; Dai, C.; Zhang, W.; Fan, H.; Yao, H.; Zong, J.; Chen, D.; et al. Comparative genomic analysis shows that Streptococcus suis meningitis isolate SC070731 contains a unique 105K genomic island. Gene 2014, 535, 156–164. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, W.; Lu, C. Comparative proteome analysis of secreted proteins of Streptococcus suis serotype 9 isolates from diseased and healthy pigs. Microb. Pathog. 2008, 45, 159–166. [Google Scholar] [CrossRef]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of quinolone action and resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef] [PubMed]
- Goltermann, L.; Nielsen, P.E. PNA Antisense Targeting in Bacteria: Determination of Antibacterial Activity (MIC) of PNA-Peptide Conjugates. Methods Mol. Biol. 2020, 2105, 231–239. [Google Scholar] [CrossRef] [PubMed]
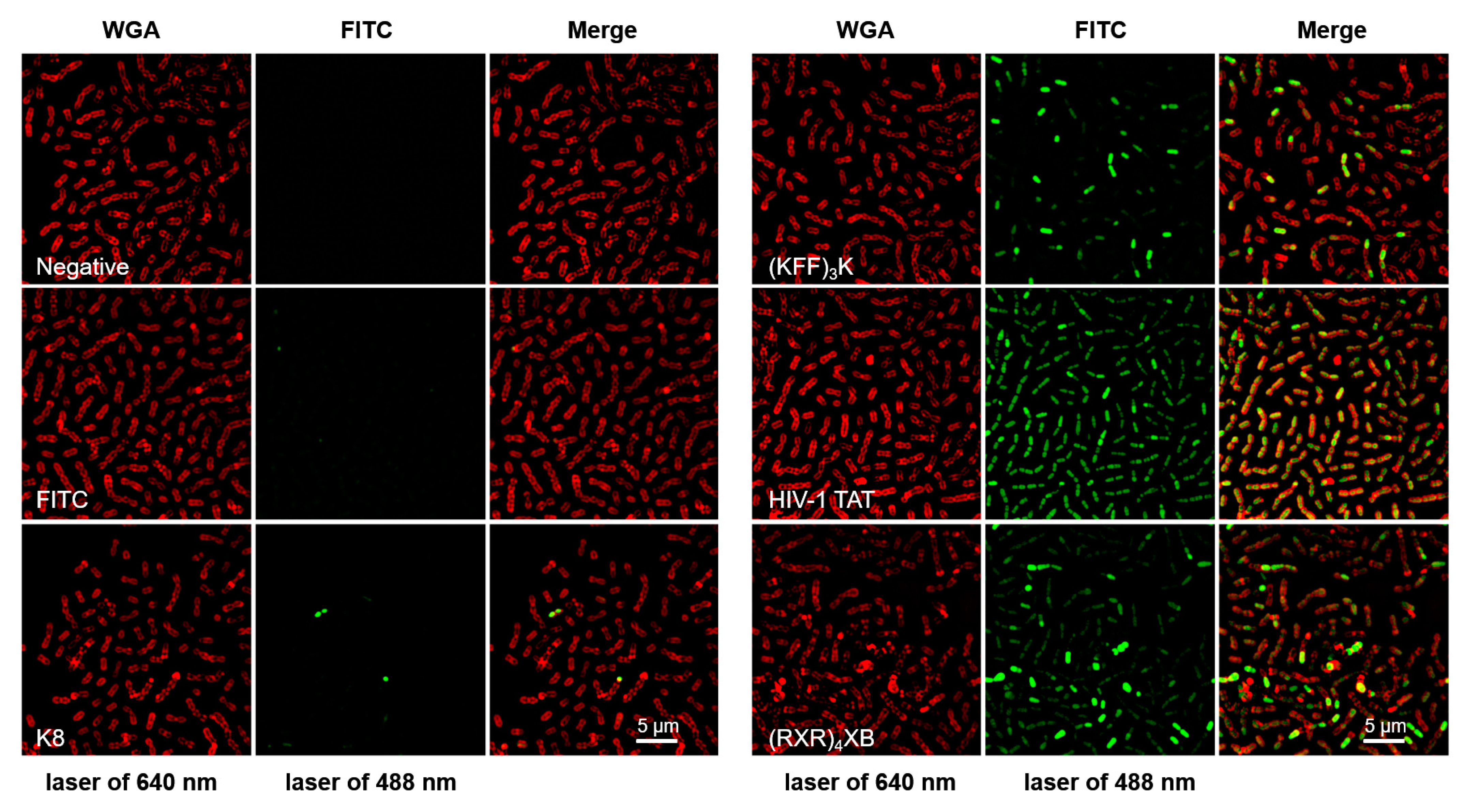
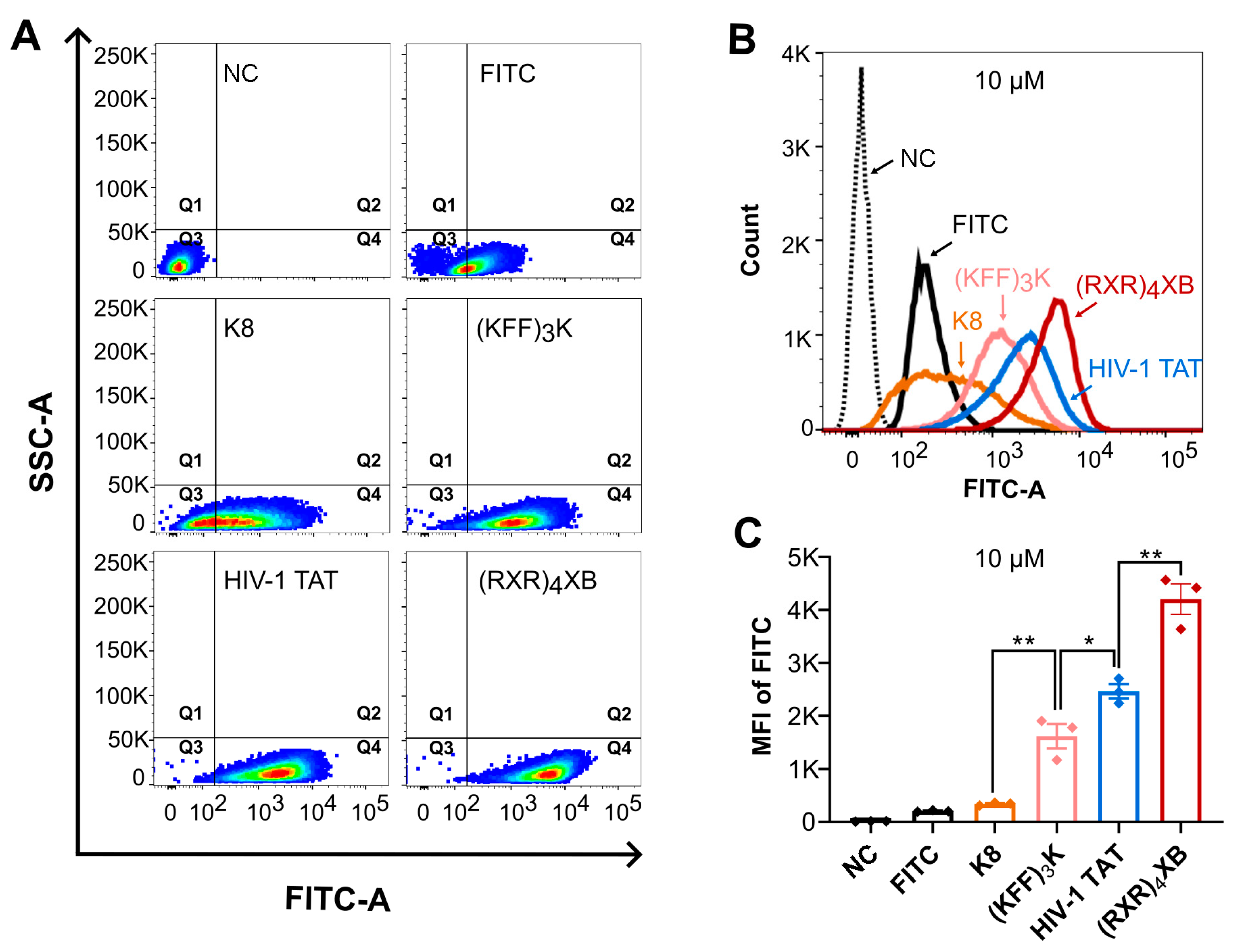

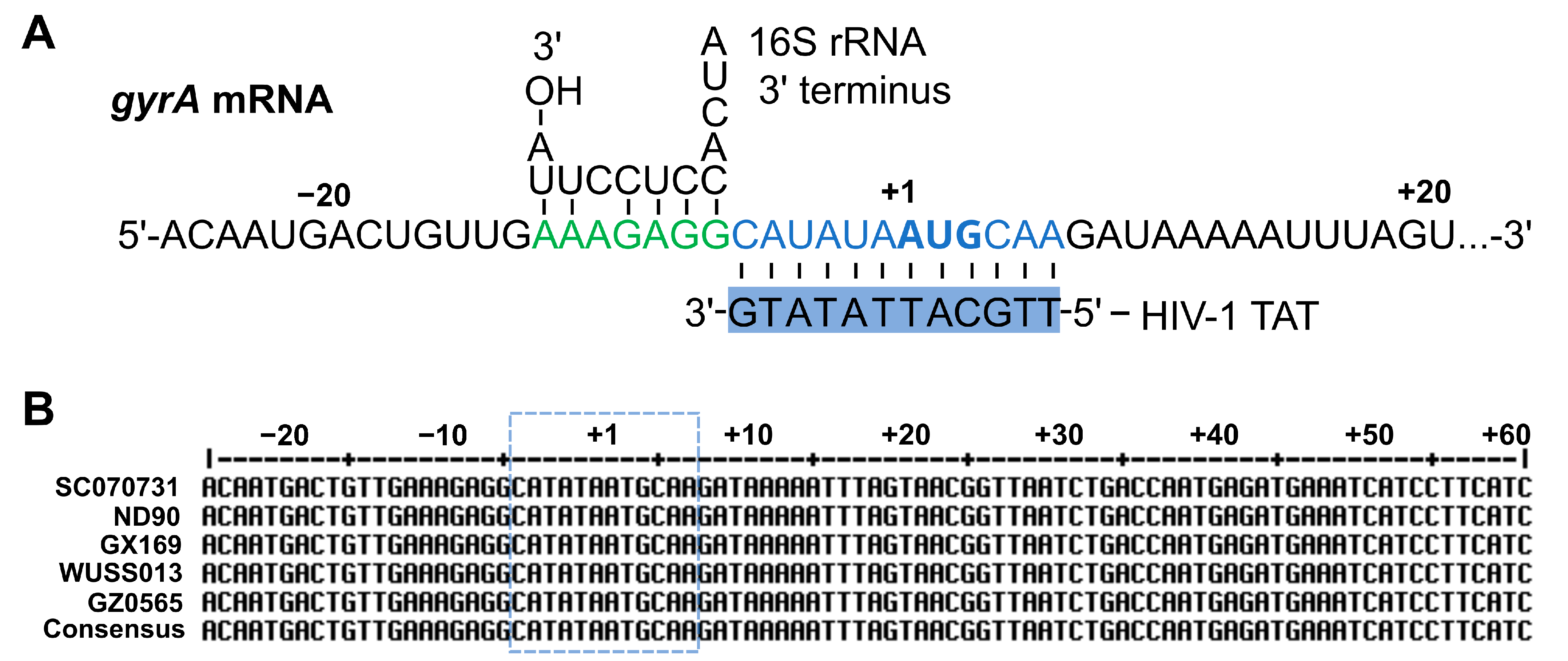
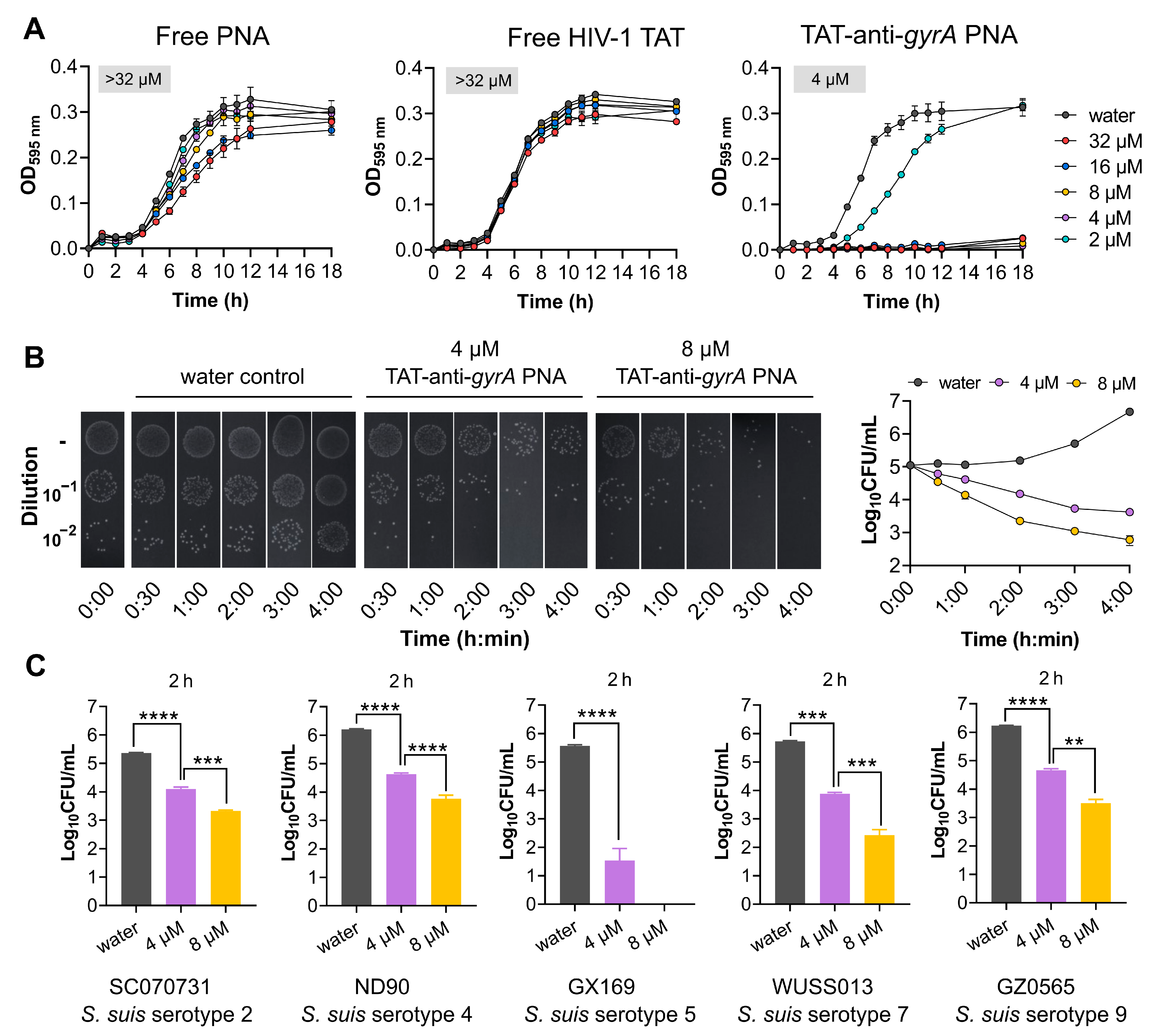
| Name | CPP Sequence | PNA Sequence | MIC (µM) | Reference |
|---|---|---|---|---|
| K8 | KKKKKKKK-NH2 | - | - | [18] |
| (KFF)3K | KFFKFFKFFK-NH2 | - | - | [35] |
| HIV-1 TAT | GRKKRRQRRRYK-NH2 | - | >128 | [36] |
| (RXR)4XB | RXRRXRRXRRXRXB-NH2 | - | 4 | [37] |
| Free PNA | - | ttgcattatatg | >32 | |
| TAT-anti-gyrA PNA | GRKKRRQRRRYK | ttgcattatatg | 4 |
| Treatment | 8 µM | 16 µM | 32 µM | 64 µM | ||||
|---|---|---|---|---|---|---|---|---|
| (Lg CFU) ± SD | Lg CFU Reduction | (Lg CFU) ± SD | Lg CFU Reduction | (Lg CFU) ± SD | Lg CFU Reduction | (Lg CFU) ± SD | Lg CFU Reduction | |
| HIV-1 TAT | 6.37 ± 0.06 | −0.09 ns | 6.32 ± 0.09 | −0.04 ns | 6.05 ± 0.27 | 0.23 ns | 6.00 ± 0.25 | 0.28 ns |
| Water control | 6.28 ± 0.08 | 0 | ||||||
| (RXR)4XB | 5.01 ± 0.14 | 1.34 * | 4.86 ± 0.02 | 1.49 * | 4.72 ± 0.03 | 1.63 * | 4.58 ± 0.01 | 1.77 * |
| Water control | 6.35 ± 0.13 | 0 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, J.; Liang, Z.; Yao, H.; Wu, Z. Identifying Cell-Penetrating Peptides for Effectively Delivering Antimicrobial Molecules into Streptococcus suis. Antibiotics 2024, 13, 725. https://doi.org/10.3390/antibiotics13080725
Zhu J, Liang Z, Yao H, Wu Z. Identifying Cell-Penetrating Peptides for Effectively Delivering Antimicrobial Molecules into Streptococcus suis. Antibiotics. 2024; 13(8):725. https://doi.org/10.3390/antibiotics13080725
Chicago/Turabian StyleZhu, Jinlu, Zijing Liang, Huochun Yao, and Zongfu Wu. 2024. "Identifying Cell-Penetrating Peptides for Effectively Delivering Antimicrobial Molecules into Streptococcus suis" Antibiotics 13, no. 8: 725. https://doi.org/10.3390/antibiotics13080725
APA StyleZhu, J., Liang, Z., Yao, H., & Wu, Z. (2024). Identifying Cell-Penetrating Peptides for Effectively Delivering Antimicrobial Molecules into Streptococcus suis. Antibiotics, 13(8), 725. https://doi.org/10.3390/antibiotics13080725







