Regulation of Antibiotic Resistance Genes on Agricultural Land Is Dependent on Both Choice of Organic Amendment and Prevalence of Predatory Bacteria
Abstract
:1. Introduction
2. Results
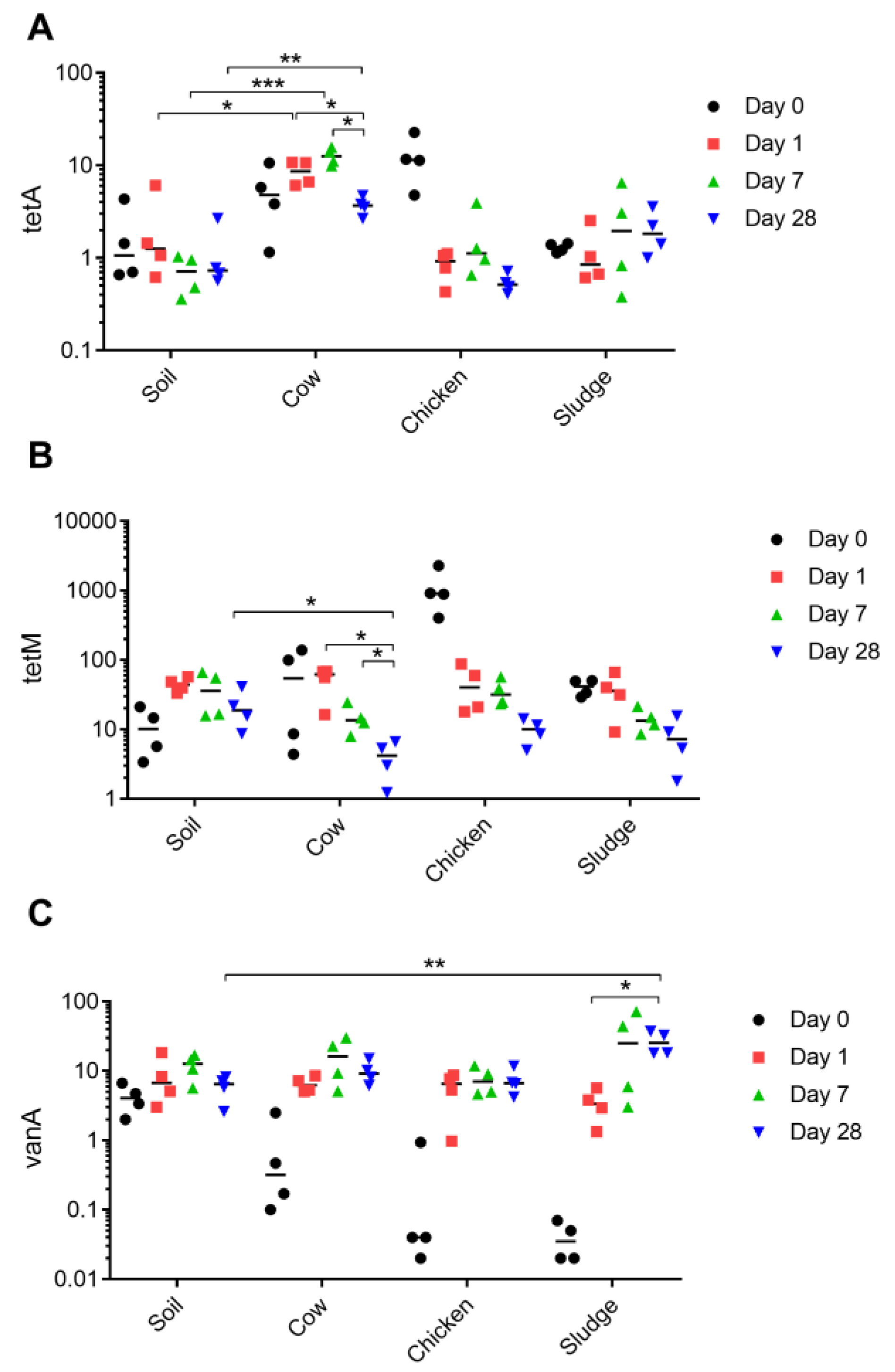
2.1. Choice of Manure Drives Prevalence of Unique Antibiotic Resistance Patterns in the Soil Microbiota
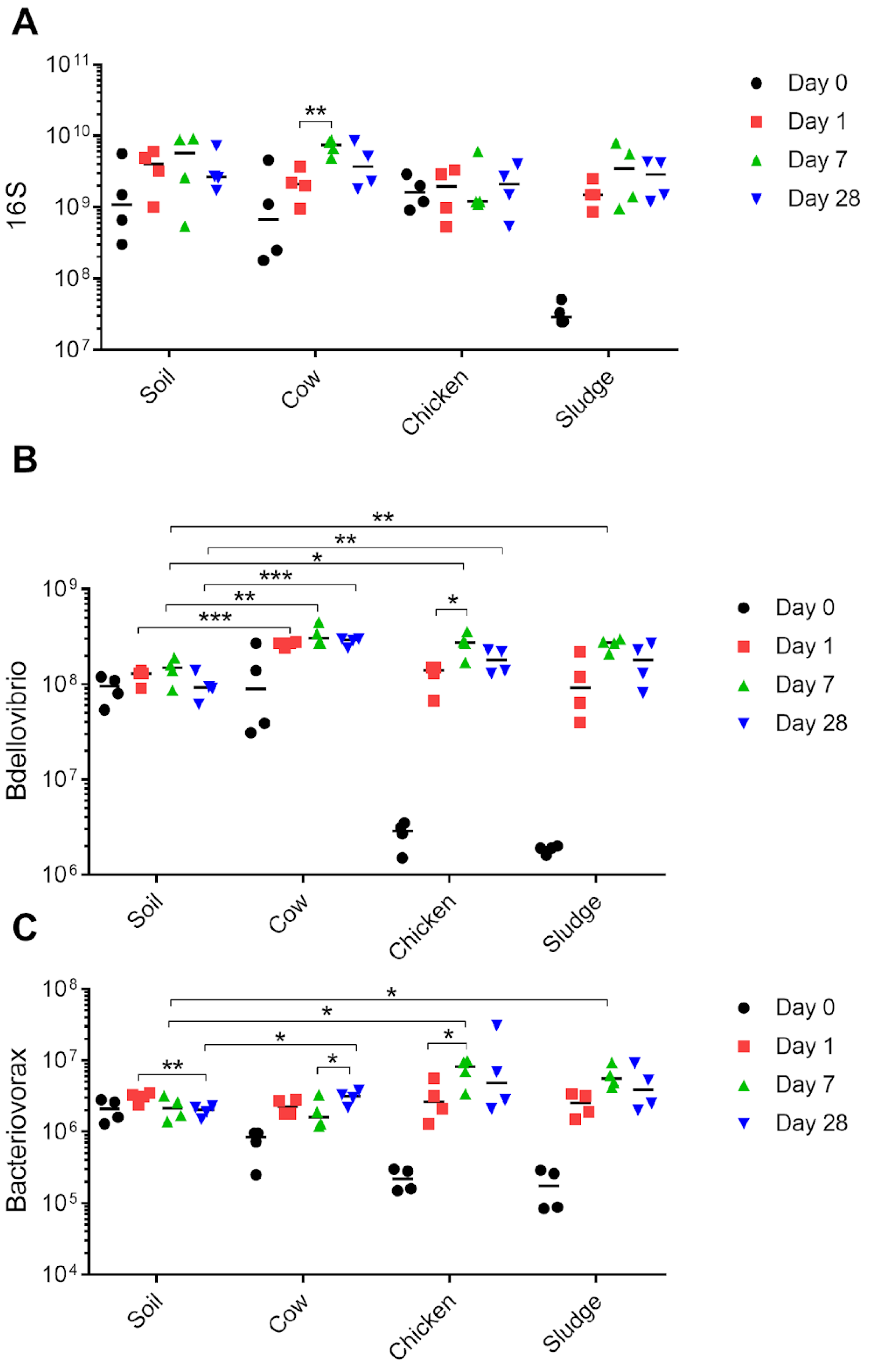
2.2. Predatory Bacteria Benefit from the Addition of Organic Amendments
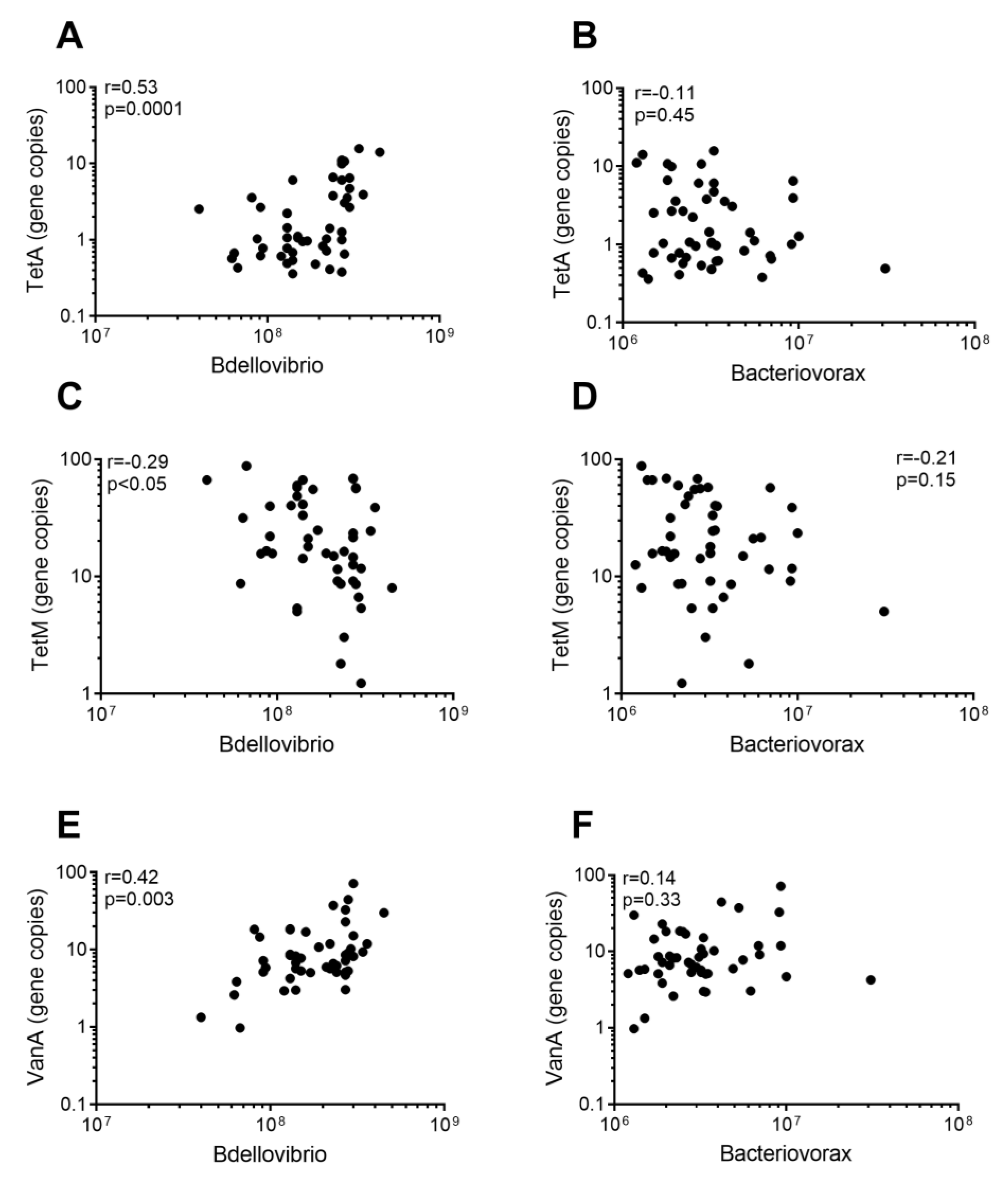
2.3. Presence of Predatory Bacteria Significantly Correlate with Prevalence of ARGs
2.4. Spurway Analysis
3. Discussion
4. Materials and Methods
4.1. Soil Incubation with Organic Amendments
4.2. DNA Extraction and Droplet Digital PCR Analysis of Microbiota (16S rRNA) and ARG
4.3. Nutrient Analysis of Soil
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zheng, D.; Yin, G.; Liu, M.; Hou, L.; Yang, Y.; Van Boeckel, T.P.; Zheng, Y.; Li, Y. Global Biogeography and Projection of Soil Antibiotic Resistance Genes. Sci. Adv. 2022, 8, eabq8015. [Google Scholar] [CrossRef]
- Knapp, C.W.; Dolfing, J.; Ehlert, P.A.I.; Graham, D.W. Evidence of Increasing Antibiotic Resistance Gene Abundances in Archived Soils since 1940. Environ. Sci. Technol. 2010, 44, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Dcosta, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic Resistance Is Ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef]
- Wichmann, F.; Udikovic-Kolic, N.; Andrew, S.; Handelsman, J. Diverse Antibiotic Resistance Genes in Dairy Cow Manure. mBio 2014, 5, e01017. [Google Scholar] [CrossRef]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J.J. Antibiotic Resistance Genes from Livestock Waste: Occurrence, Dissemination, and Treatment. npj Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- Lima, T.; Domingues, S.; Silva, G.J. Da Manure as a Potential Hotspot for Antibiotic Resistance Dissemination by Horizontal Gene Transfer Events. Vet. Sci. 2020, 7, 110. [Google Scholar] [CrossRef]
- Zhang, Y.; Hao, X.; Thomas, B.W.; McAllister, T.A.; Workentine, M.; Jin, L.; Shi, X.; Alexander, T.W. Soil Antibiotic Resistance Genes Accumulate at Different Rates over Four Decades of Manure Application. J. Hazard. Mater. 2023, 443, 130136. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Gu, J.; Sun, W.; Wang, X.J.; Su, J.Q.; Stedfeld, R. Diversity, Abundance, and Persistence of Antibiotic Resistance Genes in Various Types of Animal Manure Following Industrial Composting. J. Hazard. Mater. 2018, 344, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; An, X.; Li, H.; Su, J.; Ma, Y.; Zhu, Y.G. Long-Term Field Application of Sewage Sludge Increases the Abundance of Antibiotic Resistance Genes in Soil. Environ. Int. 2016, 92–93, 1–10. [Google Scholar] [CrossRef]
- Urra, J.; Alkorta, I.; Mijangos, I.; Epelde, L.; Garbisu, C. Application of Sewage Sludge to Agricultural Soil Increases the Abundance of Antibiotic Resistance Genes without Altering the Composition of Prokaryotic Communities. Sci. Total Environ. 2019, 647, 1410–1420. [Google Scholar] [CrossRef]
- Jauregi, L.; Epelde, L.; Alkorta, I.; Garbisu, C. Agricultural Soils Amended with Thermally-Dried Anaerobically-Digested Sewage Sludge Showed Increased Risk of Antibiotic Resistance Dissemination. Front. Microbiol. 2021, 12, 666854. [Google Scholar] [CrossRef] [PubMed]
- Jauregi, L.; Epelde, L.; Alkorta, I.; Garbisu, C. Antibiotic Resistance in Agricultural Soil and Crops Associated to the Application of Cow Manure-Derived Amendments From Conventional and Organic Livestock Farms. Front. Vet. Sci. 2021, 8, 633858. [Google Scholar] [CrossRef] [PubMed]
- Publications Office of the European Union. Environmental, Economic and Social Impact of the Use of Spreading Sewage Sludge on Land: Final Report. Part III, Project Interim Reports; Publications Office of the European Union: Luxembourg, 2013. [Google Scholar]
- EUR-Lex-01991L0271-20140101-EN-EUR-Lex. Available online: https://eur-lex.europa.eu/eli/dir/1991/271/2014-01-01 (accessed on 4 July 2024).
- EUR-Lex-52022PC0541-EN-EUR-Lex. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A52022PC0541 (accessed on 4 July 2024).
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Jackson, C.R.; Frye, J.G. Freshwater Environment as a Reservoir of Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae. J. Appl. Microbiol. 2023, 134, lxad034. [Google Scholar] [CrossRef] [PubMed]
- Wellington, E.M.H.; Boxall, A.B.A.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W.; et al. The Role of the Natural Environment in the Emergence of Antibiotic Resistance in Gram-Negative Bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Hu, H.W.; Chen, Q.L.; Singh, B.K.; Yan, H.; Chen, D.; He, J.Z. Transfer of Antibiotic Resistance from Manure-Amended Soils to Vegetable Microbiomes. Environ. Int. 2019, 130, 104912. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sui, Q.; Tong, J.; Zhong, H.; Wang, Y.; Chen, M.; Wei, Y. Soil Types Influence the Fate of Antibiotic-Resistant Bacteria and Antibiotic Resistance Genes Following the Land Application of Sludge Composts. Environ. Int. 2018, 118, 34–43. [Google Scholar] [CrossRef]
- Li, Y.W.; Wu, X.L.; Mo, C.H.; Tai, Y.P.; Huang, X.P.; Xiang, L. Investigation of Sulfonamide, Tetracycline, and Quinolone Antibiotics in Vegetable Farmland Soil in the Pearl River Delta Area, Southern China. J. Agric. Food Chem. 2011, 59, 7268–7276. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Valera, E.; Kyselková, M.; Ahmed, E.; Sladecek, F.X.J.; Goberna, M.; Elhottová, D. Native Soil Microorganisms Hinder the Soil Enrichment with Antibiotic Resistance Genes Following Manure Applications. Sci. Rep. 2019, 9, 6760. [Google Scholar] [CrossRef]
- Hungate, B.A.; Marks, J.C.; Power, M.E.; Schwartz, E.; Groenigen, K.J.v.; Blazewicz, S.J.; Chuckran, P.; Dijkstra, P.; Finley, B.K.; Firestone, M.K.; et al. The Functional Significance of Bacterial Predators. mBio 2021, 12, e00466-21. [Google Scholar] [CrossRef]
- Bratanis, E.; Andersson, T.; Lood, R.; Bukowska-Faniband, E. Biotechnological Potential of Bdellovibrio and Like Organisms and Their Secreted Enzymes. Front. Microbiol. 2020, 11, 662. [Google Scholar] [CrossRef] [PubMed]
- Bratanis, E.; Lood, R. A Novel Broad-Spectrum Elastase-Like Serine Protease From the Predatory Bacterium Bdellovibrio Bacteriovorus Facilitates Elucidation of Site-Specific IgA Glycosylation Pattern. Front. Microbiol. 2019, 10, 971. [Google Scholar] [CrossRef]
- Bratanis, E.; Molina, H.; Naegeli, A.; Collin, M.; Lood, R. BspK, a Serine Protease from the Predatory Bacterium Bdellovibrio Bacteriovorus with Utility for Analysis of Therapeutic Antibodies. Appl. Environ. Microbiol. 2017, 83, e03037-16. [Google Scholar] [CrossRef] [PubMed]
- Bukowska-Faniband, E.; Andersson, T.; Lood, R. Studies on Bd0934 and Bd3507, Two Secreted Nucleases from Bdellovibrio Bacteriovorus, Reveal Sequential Release of Nucleases during the Predatory Cycle. J. Bacteriol. 2020, 202, e00150-20. [Google Scholar] [CrossRef]
- Page, J.A.; Lubbers, B.; Maher, J.; Ritsch, L.; Gragg, S.E. Investigation into the Efficacy of Bdellovibrio Bacteriovorus as a Novel Preharvest Intervention To Control Escherichia Coli O157, H7 and Salmonella in Cattle Using an In Vitro Model. J. Food Prot. 2015, 78, 1745–1749. [Google Scholar] [CrossRef]
- Havenga, B.; Reyneke, B.; Waso-Reyneke, M.; Ndlovu, T.; Khan, S.; Khan, W. Biological Control of Acinetobacter Baumannii: In Vitro and In Vivo Activity, Limitations, and Combination Therapies. Microorganisms 2022, 10, 1052. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, W.; Wang, W.; Zhang, H.; Jin, Y. In Situ Forming Hydrogel Loaded with Predatory Bacteria Treats Drug-Resistant Corneal Infection. J. Control. Release 2023, 364, 393–405. [Google Scholar] [CrossRef]
- Romanowski, E.G.; Brothers, K.M.; Calvario, R.C.; Stella, N.A.; Kim, T.; Elsayed, M.; Kadouri, D.E.; Shanks, R.M.Q. Intra-Ocular Predation of Fluoroquinolone-Resistant Pseudomonas Aeruginosa and Serratia Marcescens by Predatory Bacteria. Microbiology 2024, 170, 001433. [Google Scholar] [CrossRef] [PubMed]
- Upatissa, S.; Mun, W.; Mitchell, R.J. Pairing Colicins B and E5 with Bdellovibrio Bacteriovorus To Eradicate Carbapenem- and Colistin-Resistant Strains of Escherichia coli. Microbiol. Spectr. 2023, 11, e00173-23. [Google Scholar] [CrossRef]
- Jurkevitch, E.; Minz, D.; Ramati, B.; Barel, G. Prey Range Characterization, Ribotyping, and Diversity of Soil and Rhizosphere Bdellovibrio Spp. Isolated on Phytopathogenic Bacteria. Appl. Environ. Microbiol. 2000, 66, 2365. [Google Scholar] [CrossRef]
- Andersson, T.; Adell, A.D.; Moreno-Switt, A.I.; Spégel, P.; Turner, C.; Overballe-Petersen, S.; Fuursted, K.; Lood, R. Biogeographical Variation in Antimicrobial Resistance in Rivers Is Influenced by Agriculture and Is Spread through Bacteriophages. Environ. Microbiol. 2022, 24, 4869–4884. [Google Scholar] [CrossRef]
- Baraka, V.; Andersson, T.; Makenga, G.; Francis, F.; Minja, D.T.R.; Overballe-Petersen, S.; Tang, M.H.E.; Fuursted, K.; Lood, R. Unveiling Rare Pathogens and Antibiotic Resistance in Tanzanian Cholera Outbreak Waters. Microorganisms 2023, 11, 2490. [Google Scholar] [CrossRef] [PubMed]
- Andersson, T.; Makenga, G.; Francis, F.; Minja, D.T.R.; Overballe-Petersen, S.; Tang, M.H.E.; Fuursted, K.; Baraka, V.; Lood, R. Enrichment of Antibiotic Resistance Genes within Bacteriophage Populations in Saliva Samples from Individuals Undergoing Oral Antibiotic Treatments. Front. Microbiol. 2022, 13, 1049110. [Google Scholar] [CrossRef]
- Tiwari, A.; Krolicka, A.; Tran, T.T.; Räisänen, K.; Ásmundsdóttir, Á.M.; Wikmark, O.G.; Lood, R.; Pitkänen, T. Antibiotic Resistance Monitoring in Wastewater in the Nordic Countries: A Systematic Review. Environ. Res. 2024, 246, 118052. [Google Scholar] [CrossRef]
- Han, B.; Ma, L.; Yu, Q.; Yang, J.; Su, W.; Hilal, M.G.; Li, X.; Zhang, S.; Li, H. The Source, Fate and Prospect of Antibiotic Resistance Genes in Soil: A Review. Front. Microbiol. 2022, 13, 976657. [Google Scholar] [CrossRef]
- Rutgersson, C.; Ebmeyer, S.; Lassen, S.B.; Karkman, A.; Fick, J.; Kristiansson, E.; Brandt, K.K.; Flach, C.F.; Larsson, D.G.J. Long-Term Application of Swedish Sewage Sludge on Farmland Does Not Cause Clear Changes in the Soil Bacterial Resistome. Environ. Int. 2020, 137, 105339. [Google Scholar] [CrossRef]
- Radu, E.; Woegerbauer, M.; Rab, G.; Oismüller, M.; Strauss, P.; Hufnagl, P.; Gottsberger, R.A.; Krampe, J.; Weyermair, K.; Kreuzinger, N. Resilience of Agricultural Soils to Antibiotic Resistance Genes Introduced by Agricultural Management Practices. Sci. Total Environ. 2021, 756, 143699. [Google Scholar] [CrossRef] [PubMed]
- Kyselková, M.; Jirout, J.; Vrchotová, N.; Schmitt, H.; Elhottová, D. Spread of Tetracycline Resistance Genes at a Conventional Dairy Farm. Front. Microbiol. 2015, 6, 536. [Google Scholar] [CrossRef]
- Miller, D.N.; Jurgens, M.E.; Durso, L.M.; Schmidt, A.M. Simulated Winter Incubation of Soil With Swine Manure Differentially Affects Multiple Antimicrobial Resistance Elements. Front. Microbiol. 2020, 11, 611912. [Google Scholar] [CrossRef] [PubMed]
- Browne, A.J.; Chipeta, M.G.; Haines-Woodhouse, G.; Kumaran, E.P.A.; Hamadani, B.H.K.; Zaraa, S.; Henry, N.J.; Deshpande, A.; Reiner, R.C.; Day, N.P.J.; et al. Global Antibiotic Consumption and Usage in Humans, 2000–2018, A Spatial Modelling Study. Lancet Planet Health 2021, 5, e893–e904. [Google Scholar] [CrossRef]
- Mulchandani, R.; Wang, Y.; Gilbert, M.; Van Boeckel, T.P. Global Trends in Antimicrobial Use in Food-Producing Animals: 2020 to 2030. PLOS Glob. Public Health 2023, 3, e0001305. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, N.; Li, C.; Shao, L. Diversity of Antibiotic Resistance Genes in Soils with Four Different Fertilization Treatments. Front. Microbiol. 2023, 14, 1291599. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.; Negus, D.; Raghunathan, D.; Radford, P.; Moore, C.; Clark, G.; Diggle, M.; Tyson, J.; Twycross, J.; Sockett, R.E. Measuring and Modelling the Response of Klebsiella Pneumoniae KPC Prey to Bdellovibrio Bacteriovorus Predation, in Human Serum and Defined Buffer. Sci. Rep. 2017, 7, 8329. [Google Scholar] [CrossRef] [PubMed]
- Cohen, Y.; Pasternak, Z.; Müller, S.; Hübschmann, T.; Schattenberg, F.; Sivakala, K.K.; Abed-Rabbo, A.; Chatzinotas, A.; Jurkevitch, E. Community and Single Cell Analyses Reveal Complex Predatory Interactions between Bacteria in High Diversity Systems. Nat. Commun. 2021, 12, 5481. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, Y.; Jurkevitch, E. Plastic Phenotypic Resistance to Predation by Bdellovibrio and like Organisms in Bacterial Prey. Environ. Microbiol. 2004, 6, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Varon, M. Interaction OfBdellovibrio with Its Prey in Mixed Microbial Populations. Microb. Ecol. 1981, 7, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Waso-Reyneke, M.; Khan, S.; Khan, W. Interaction of Bdellovibrio Bacteriovorus with Gram-Negative and Gram-Positive Bacteria in Dual Species and Polymicrobial Communities. Microorganisms 2022, 10, 793. [Google Scholar] [CrossRef]
- Nguyen, T.B.A.; Bonkowski, M.; Dumack, K.; Chen, Q.L.; He, J.Z.; Hu, H.W. Protistan Predation Selects for Antibiotic Resistance in Soil Bacterial Communities. ISME J. 2023, 17, 2182–2189. [Google Scholar] [CrossRef]


| Analysis | Unit | Soil (d0) | Soil (d28) | Chicken | Cow | Sludge |
|---|---|---|---|---|---|---|
| pH | 5.8 | 6.0 | 5.85 | 6.35 | 5.45 | |
| Electrical conductivity | mS/cm | 3.4 | 2.75 | 3.8 | 2.45 | 2.9 |
| Nitrogen (N) | mg/L | 160 | 145 | 180 | 140 | 210 |
| Nitrate (N) | mg/L | 160 | 145 | 180 | 140 | 210 |
| Ammonium (N) | mg/L | 3 | 1 | 1 | 1 | 1 |
| Phosphorous (P) | mg/L | 13 | 15 | 23 | 110 | 28.5 |
| Potassium (K) | mg/L | 52 | 54 | 130 | 420 | 88 |
| Magnesium (Mg) | mg/L | 78 | 77 | 91.5 | 135 | 50 |
| Sulfur (S) | mg/L | 250 | 145 | 230 | 73.5 | 135 |
| Calcium (Ca) | mg/L | 790 | 675 | 715 | 510 | 705 |
| Manganese (Mn) | mg/L | 1.9 | 0.66 | 1.5 | 0.95 | 1.65 |
| Boron (B) | mg/L | 0.54 | 0.6 | 0.6 | 1.2 | 1.05 |
| Iron (Fe) | mg/L | 1.3 | 1.8 | 1.35 | 1.55 | 1.58 |
| Sodium (Na) | mg/L | 180 | 170 | 205 | 105 | 72.5 |
| Aluminum (Al) | mg/L | 3.2 | 4.25 | 3.15 | 3.1 | 4.5 |
| Target | Forward Primer | Reverse Primer | Probe | Ref |
|---|---|---|---|---|
| 16S | AGAGTTTGATCCTGGCTCAGGA | CGTGTTACTCACCCGTCCG | CGCTGGCGGCGTGCCTAATACATGC | [34,35] |
| Bdellovibrio | GGAGGCAGCAGTAGGGAATA | GCTAGGATCCCTCGTCTTACC | CGCGTGAGTGATGAAGGCCTTCGGGTCG | This study |
| Bacteriovorax | CAGCCGCGGTAATACGAA | CGGATTTTACCCCTACATGC | GGGTGCAAGCGTTGTTCGGATTTATTGGGC | This study |
| tetA | TTGAACGGCCTCAATTTCCT | GATGAAGAAGACCGCCATCA | GCATGACCGTCGTCGCCGCCC | [34,35,36] |
| tetM | TGCAAGAAAAGTATCATGTGGAG | AAACCGAGCTCTCATACTGC | TGCCGCCAAATCCTTTCTGGGCTTCCA | [34,35,36] |
| vanA | GTTGTGCGGTATTGGGAAAC | GTTTCCTGTATCCGTCCTCG | GCCGCGTTAGCTGTTGGCGAGGT | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosberg, A.K.; Silva, M.J.; Skøtt Feidenhans’l, C.; Cytryn, E.; Jurkevitch, E.; Lood, R. Regulation of Antibiotic Resistance Genes on Agricultural Land Is Dependent on Both Choice of Organic Amendment and Prevalence of Predatory Bacteria. Antibiotics 2024, 13, 750. https://doi.org/10.3390/antibiotics13080750
Rosberg AK, Silva MJ, Skøtt Feidenhans’l C, Cytryn E, Jurkevitch E, Lood R. Regulation of Antibiotic Resistance Genes on Agricultural Land Is Dependent on Both Choice of Organic Amendment and Prevalence of Predatory Bacteria. Antibiotics. 2024; 13(8):750. https://doi.org/10.3390/antibiotics13080750
Chicago/Turabian StyleRosberg, Anna Karin, Maria João Silva, Cecilie Skøtt Feidenhans’l, Eddie Cytryn, Edouard Jurkevitch, and Rolf Lood. 2024. "Regulation of Antibiotic Resistance Genes on Agricultural Land Is Dependent on Both Choice of Organic Amendment and Prevalence of Predatory Bacteria" Antibiotics 13, no. 8: 750. https://doi.org/10.3390/antibiotics13080750






