Evolution of the Antibiotic Resistance Levels, Multi-Resistance Patterns, and Presence of Antibiotic Resistance Genes in E. coli Isolates from the Feces of Breeding Hens during the Rearing Period
Abstract
1. Introduction
2. Results
2.1. Antibiotic Resistance Prevalence
2.1.1. Antibiotic Resistance Prevalence in E. coli Isolated from One-Day-Old Breeders
2.1.2. Antibiotic Resistance Prevalence in E. coli Isolated from Fecal Samples of Pullets and Adult Hens
2.1.3. Antibiotic Resistance Prevalence in E. coli Isolated from Boot Swab Samples
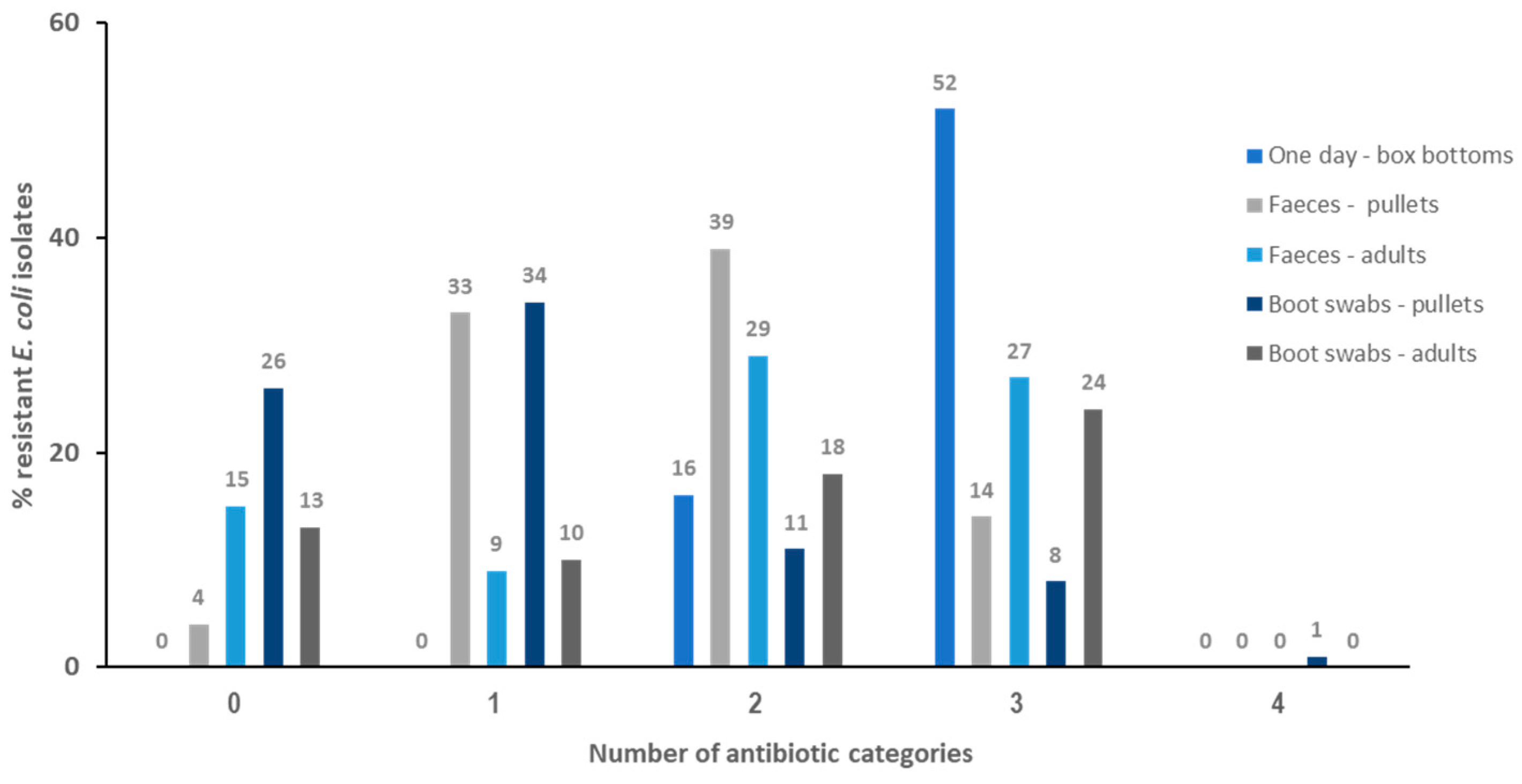
2.2. Resistance Profiles and Multi-Resistance (MDR) Patterns
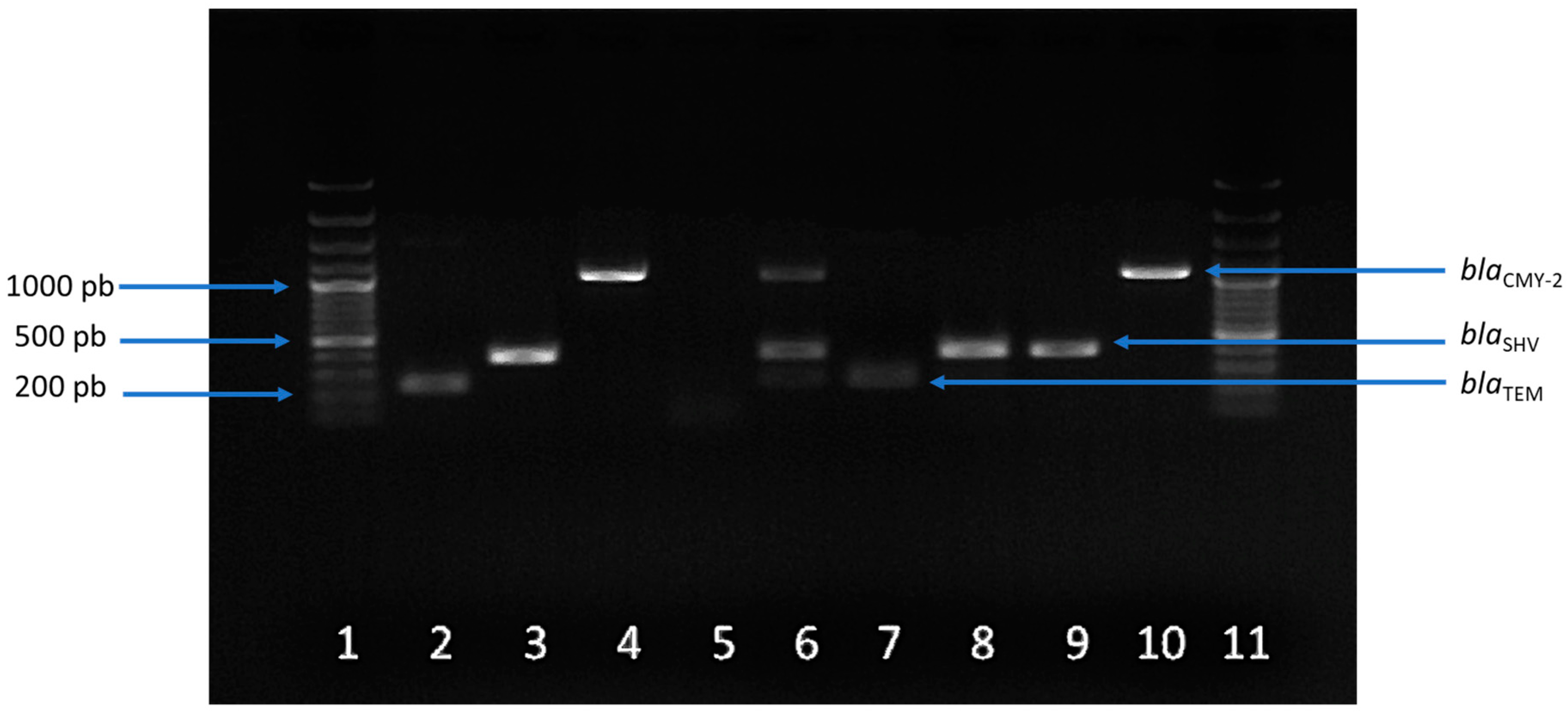
2.3. Prevalence and Profiles of Antibiotic Resistance Genes (ARGs)
3. Discussion
4. Materials and Methods
4.1. Sampling and Sample Preparation
4.2. Escherichia coli Isolation
4.3. Antimicrobial Susceptibility Testing of E. coli Isolates
4.4. DNA Extraction
4.5. Detection of Antibiotic Resistance Genes (ARGs)
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). Antimicrobial Resistance. 2024. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 20 July 2024).
- Pinto Jimenez, C.E.; Keestra, S.; Tandon, P.; Cumming, O.; Pickering, A.J.; Moodley, A.; Chandler, C.I.R. Biosecurity and water, sanitation, and hygiene (WASH) interventions in animal agricultural settings for reducing infection burden, antibiotic use, and antibiotic resistance: A One Health systematic review. Lancet Planet Health 2023, 7, e418–e434. [Google Scholar] [CrossRef] [PubMed]
- Skandalis, N.; Maeusli, M.; Papafotis, D.; Miller, S.; Lee, B.; Theologidis, I.; Luna, B. Environmental spread of antibiotic resistance. Antibiotics 2021, 10, 640. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA & ECDC). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2021-22. EFSA J. 2024, 22, e8583. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA); Amore, G.; Beloeil, P.-A.; García-Fierro, R.; Guerra, B.; Papanikolaou, A.; Rizzi, V.; Stoicescu, A.-V. Manual for Reporting 2022 Antimicrobial Resistance Data within the Framework of Directive 2003/99/EC and Decision 2020/1729/EU; EFSA Supporting Publication 2023: EN-7826; EFSA: Parma, Italy, 2023. [Google Scholar] [CrossRef]
- Morrow, C.J. Antimicrobial resistance (AMR): An important one health issue for layer and meat poultry industries worldwide. Poult. Sci. 2024, 103, 103690. [Google Scholar] [CrossRef] [PubMed]
- Sagar, P.; Aseem, A.; Banjara, S.K.; Veleri, S. The role of food chain in antimicrobial resistance spread and One Health approach to reduce risks. Int. J. Food Microbiol. 2023, 16, 110148. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). WHO List of Medically Important Antimicrobials: A Risk Management Tool for Mitigating Antimicrobial Resistance due to Non-Human Use. Geneva. 2024. Available online: https://cdn.who.int/media/docs/default-source/gcp/who-mia-list-2024-lv.pdf?sfvrsn=3320dd3d_2 (accessed on 30 May 2024).
- Aristizábal-Hoyos, A.M.; Rodriguez, E.A.; Arias, L.; Jiménez, J.N. High clonal diversity of multidrug-resistant and extended spectrum beta-lactamase-producing Escherichia coli in a wastewater treatment plant. J. Environ. Manag. 2019, 245, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Mourand, G.; Le Devendec, L.; Delannoy, S.; Fach, P.; Keita, A.; Amelot, M. Variations of the Escherichia coli population in the digestive tract of broilers. Avian Pathol. 2020, 49, 678–688. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA); Aerts, M.; Battisti, A.; Hendriksen, R.; Kempf, I.; Teale, C.; Tenhagen, B.A.; Veldman, K.; Wasyl, D.; Guerra, B.; et al. Technical specifications on harmonised monitoring of antimicrobial resistance in zoonotic and indicator bacteria from food-producing animals and food. EFSA J. 2019, 17, 05709. [Google Scholar] [CrossRef]
- Anjum, M.F.; Schmitt, H.; Börjesson, S.; Berendonk, T.U.; on behalf of the WAWES Network. The potential of using E. coli as an indicator for the surveillance of antimicrobial resistance (AMR) in the environment. Curr. Opinion Microbiol. 2021, 64, 152–158. [Google Scholar] [CrossRef]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef]
- Bergšpica, I.; Kaprou, G.; Alexa, E.A.; Prieto, M.; Alvarez-Ordóñez, A. Extended spectrum β-Lactamase (ESBL) producing Escherichia coli in pigs and pork meat in the European Union. Antibiotics 2020, 9, 678. [Google Scholar] [CrossRef]
- Fisher, J.F.; Meroueh, S.O.; Mobashery, S. Bacterial resistance to beta-lactam antibiotics: Compelling opportunism, compelling opportunity. Chem. Rev. 2005, 105, 395–424. [Google Scholar] [CrossRef]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A. AmpC beta-lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation (WHO). Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; World Health Organisation: Geneva, Switzerland, 2017; Available online: https://www.who.int/publications/i/item/WHO-EMP-IAU-2017.12 (accessed on 20 July 2024).
- EFSA Panel on Biological Hazards, EFSA Panel on Contaminants in the Food Chain & EFSA Panel on Animal Health and Welfare. Scientific Opinion on the public health hazards to be covered by inspection of meat (poultry). EFSA J. 2012, 10, 2741. [Google Scholar] [CrossRef]
- Jacoby, G.A. Plasmid-mediated quinolone resistance BT. In Antimicrobial Drug Resistance. Mechanisms of Drug Resistance, 2nd ed.; Mayers, D.L., Sobel, J.D., Ouellette, M., Kaye, K.S., Marchaim, D., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; Volume 1, pp. 265–268. [Google Scholar]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- De Koster, S.; Ringenier, M.; Xavier, B.B.; Lammens, C.; De Coninck, D.; De Bruyne, K.; Mensaert, K.; Kluytmans-van den Bergh, M.; Kluytmans, J.; Dewulf, J.; et al. Genetic characterization of ESBL-producing and ciprofloxacin-resistant Escherichia coli from Belgian broilers and pigs. Front. Microbiol. 2023, 14, 1150470. [Google Scholar] [CrossRef] [PubMed]
- Haeili, M.; Salehzeinali, H.; Mirzaei, S.; Pishnian, Z.; Ahmadi, A. Molecular characterization of quinolone resistance and antimicrobial resistance profiles of Klebsiella pneumoniae and Escherichia coli isolated from human and broiler chickens. Int. J. Environ. Health. Res. 2022, 32, 1382–1392. [Google Scholar] [CrossRef]
- Aworh, M.K.; Kwaga, J.K.P.; Hendriksen, R.S.; Okolocha, E.C.; Harrell, E.; Thakur, S. Quinolone-resistant Escherichia coli at the interface between humans, poultry and their shared environment—A potential public health risk. One Health Outlook 2023, 5, 2. [Google Scholar] [CrossRef]
- Truswell, A.; Lee, Z.Z.; Stegger, M.; Blinco, J.; Abraham, R.; Jordan, D.; Milotic, M.; Hewson, K.; Pang, S.; Abraham, S. Augmented surveillance of antimicrobial resistance with high-throughput robotics detects transnational flow of fluoroquinolone-resistant Escherichia coli strain into poultry. J. Antimicrob. Chemother. 2023, 78, 2878–2885. [Google Scholar] [CrossRef]
- Abreu, R.; Semedo-Lemsaddek, T.; Cunha, E.; Tavares, L.; Oliveira, M. Antimicrobial drug resistance in poultry production: Current status and innovative strategies for bacterial control. Microorganisms 2023, 11, 953. [Google Scholar] [CrossRef] [PubMed]
- OECD-FAO. OECD-FAO Agricultural Outlook 2021–2030; Paris: Political Science; OECD Publishing: Paris, France, 2021. [Google Scholar] [CrossRef]
- Korver, R.D. Review: Current challenges in poultry nutrition, health, and welfare. Animal 2023, 2, 100755. [Google Scholar] [CrossRef] [PubMed]
- Gržinić, G.; Piotrowicz-Cieślak, A.; Klimkowicz-Pawlas, A.; Górny, R.L.; Ławniczek-Wałczyk, A.; Piechowicz, L.; Olkowska, E.; Potrykus, M.; Tankiewicz, M.; Krupka, M.; et al. Intensive poultry farming: A review of the impact on the environment and human health. Sci. Total Environ. 2023, 858, 160014. [Google Scholar] [CrossRef] [PubMed]
- Mak, P.H.W.; Rehman, M.A.; Kiarie, E.G.; Topp, E.; Diarra, M.S. Production systems and important antimicrobial resistant-pathogenic bacteria in poultry: A review. J. Anim. Sci. Biotechnol. 2022, 13, 148. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.A.; García-Soto, S.; Hernández, M.; Bárcena, C.; Rodríguez-Lázaro, D.; Ugarte-Ruíz, M.; Domínguez, L. Day-old chicks are a source of antimicrobial resistant bacteria for laying hen farms. Vet. Microbiol. 2019, 230, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Belenguer, A.; Doménech, E.; Villagrá, A.; Fenollar, A.; Ferrús, M.A. Antimicrobial resistance of Escherichia coli isolated in newly-hatched chickens and effect of amoxicillin treatment during their growth. Avian Pathol. 2016, 45, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Wang, J.; Klumpp, J.; Nuesch-Inderbinen, M.; Fanning, S.; Stephan, R. Vertical transmission of highly similar blaCTX-M−1-harboring IncI1 plasmids in Escherichia coli with different MLST types in the poultry production pyramid. Front. Microbiol. 2014, 5, 519. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, O.; Börjesson, S.; Landén, A.; Bengtsson, B. Vertical transmission of Escherichia coli carrying plasmid-mediated AmpC (pAmpC) through the broiler production pyramid. J. Antimicrob. Chemother. 2014, 69, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Sodagari, H.R.; Varga, C.; Habib, I.; Sahibzada, S. Comparison of Antimicrobial Resistance among Commensal Escherichia coli Isolated from Retail Table Eggs Produced by Laying Hens from the Cage and Non-Cage Housing Systems in Western Australia. Antibiotics 2023, 12, 588. [Google Scholar] [CrossRef]
- Baron, S.; Jouy, E.; Larvor, E.; Eono, F.; Bougeard, S.; Kempf, I. Impact of third-generation cephalosporin administration in hatcheries on fecal Escherichia coli antimicrobial resistance in broilers and layers. Antimicrob. Agents Chemother. 2014, 58, 5428–5434. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA). Scientific Opinion on the public health risks of bacterial strains producing extended-spectrum β-lactamases and/or AmpC β-lactamases in food and food-producing animals. EFSA J. 2011, 9, 2322. [Google Scholar] [CrossRef]
- EU Directorate-General for Health and Food Safety. Council Recommendation on Stepping Up EU Actions to Combat Antimicrobial Resistance in a One Health Approach 2023/C 220/01. 2023. Available online: https://health.ec.europa.eu/publications/council-recommendation-stepping-eu-actions-combat-antimicrobial-resistance-one-health-approach_en (accessed on 13 June 2024).
- Mo, S.S.; Kristoffersen, A.B.; Sunde, M.; Nødtvedt, A.; Norström, M. Risk factors for occurrence of cephalosporin-resistant Escherichia coli in Norwegian broiler flocks. Prev. Vet. Med. 2016, 130, 112–118. [Google Scholar] [CrossRef]
- Roth, N.; Käsbohrer, A.; Mayrhofer, S.; Zitz, U.; Hofacre, C.; Domig, K.J. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: A global overwiew. Polt. Sci. 2019, 98, 1791–1804. [Google Scholar] [CrossRef] [PubMed]
- Roosmarijn, E.C.L.; Heederik, D.J.J.; Scherpenisse, P.; Van Gompel, L.; van Heijnsbergen, E.; Greve, G.D.; Jongerius-Gortemaker, B.G.M.; Tersteeg-Zijderveld, M.H.G.; Fischer, J.; Juraschek, K.; et al. Determinants for antimicrobial resistance genes in farm dust on 333 poultry and pig farms in nine European countries. Environ. Res. 2022, 208, 112715. [Google Scholar] [CrossRef]
- Ceccarelli, D.; Kant, A.; van Essen-Zandbergen, A.; Dierikx, C.; Hordijk, J.; Wit, B.; Mevius, D.J.; Veldman, K.T. Diversity of plasmids and genes encoding resistance to extended spectrum cephalosporins in commensal Escherichia coli from Dutch livestock in 2007–2017. Front. Microbiol. 2019, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Harisberger, M.; Gobeli, S.; Hoop, R.; Dewulf, J.; Perreten, V.; Regula, G. Antimicrobial resistance in Swiss laying hens, prevalence and risk factors. Zoonoses Public Health 2011, 58, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Kaspersen, H.; Urdahl, A.M.; Simm, R.; Slettemeås, J.S.; Lagesen, K.; Norström, M. Occurrence of quinolone resistant E. coli 508 originating from different animal species in Norway. Vet. Microbiol. 2018, 217, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Laube, H.; Friese, A.; von Salviati, C.; Guerra, B.; Käsbohrer, A.; Kreienbrock, L.; Roesler, U. Longitudinal monitoring of Extended-Spectrum-Beta-Lactamase/AmpC-producing Escherichia coli at German broiler chicken fattening farms. Appl. Environ. Microbiol. 2013, 79, 4815–4820. [Google Scholar] [CrossRef] [PubMed]
- Projahn, M.; Daehre, K.; Roesler, U.; Friese, A. Extended-spectrum-beta-lactamase and plasmid-encoded cephamycinase-producing enterobacteria in the broiler hatchery as a potential mode of pseudo-vertical transmission. Appl. Environ. Microbiol. 2017, 83, e02364-16. [Google Scholar] [CrossRef]
- Dierikx, C.M.; van der Goot, J.A.; Smith, H.E.; Kant, A.; Mevius, D.J. Presence of ESBL/AmpC -Producing Escherichia coli in the broiler production pyramid: A descriptive study. PLoS ONE 2013, 8, e79005. [Google Scholar] [CrossRef]
- Lu, J.; Idris, U.; Harmon, B.; Hofacre, C.; Maurer, J.J.; Lee, M.D. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 2003, 69, 6816–6824. [Google Scholar] [CrossRef] [PubMed]
- Huo, M.; Xu, X.; Mi, K.; Ma, W.; Zhou, Q.; Lin, X.; Cheng, G.; Huang, L. Co-selection mechanism for bacterial resistance to major chemical pollutants in the environment. Sci. Total Environ. 2024, 912, 169223. [Google Scholar] [CrossRef] [PubMed]
- Dame-Korevaar, A.; Fischer, E.; Stegeman, A.; Mevius, D.; van Essen-Zandbergen, A.; Velkers, F.; van der Goot, J. Dynamics of CMY-2 producing E. coli in a broiler parent flock. Vet. Microbiol. 2017, 203, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance. Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Cantón, R.; Ruiz-Garbajosa, P. Co-resistance: An opportunity for the bacteria and resistance genes. Curr. Opin. Pharmacol. 2011, 11, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Apostolakos, I.; Mughini-Gras, L.; Fasolato, L.; Piccirillo, A. Assessing the occurrence and transfer dynamics of ESBL/pAmpC-producing Escherichia coli across the broiler production pyramid. PLoS ONE 2019, 14, e0217174. [Google Scholar] [CrossRef] [PubMed]
- Manageiro, V.; Clemente, L.; Graça, R.; Correia, I.; Albuquerque, T.; Ferreira, E.; Caniça, M. New insights into resistance to colistin and third-generation cephalosporins of Escherichia coli in poultry, Portugal: Novel blaCTX-M-166 and blaESAC genes. Int. J. Food Microbiol. 2017, 263, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Blaak, H.; van Hoek, A.H.; Hamidjaja, R.A.; van der Plaats, R.Q.; Kerkhof-de Heer, L.; de Roda Husman, A.M.; Schets, F.M. Distribution, numbers, and diversity of ESBL-producing E. coli in the poultry farm environment. PLoS ONE 2015, 10, e0135402. [Google Scholar] [CrossRef] [PubMed]
- Jones-Dias, D.; Manageiro, V.; Francisco, A.P.; Martins, A.P.; Domingues, G.; Louro, D.; Ferreira, E.; Caniça, M. Assessing the molecular basis of transferable quinolone resistance in Escherichia coli and Salmonella spp. from food-producing animals and food products. Vet. Microbiol. 2013, 167, 523–531. [Google Scholar] [CrossRef]
- Börjesson, S.; Guillard, T.; Landén, A.; Bengtsson, B.; Nilsson, O. Introduction of quinolone resistant Escherichia coli to Swedish broiler population by imported breeding animals. Vet. Microbiol. 2016, 194, 74–78. [Google Scholar] [CrossRef]
- Niero, G.; Bortolaia, V.; Vanni, M.; Intorre, L.; Guardabassi, L.; Piccirillo, A. High diversity of genes and plasmids encoding resistance to third-generation cephalosporins and quinolones in clinical Escherichia coli from commercial poultry flocks in Italy. Vet. Microbiol. 2018, 216, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Laube, H.; Friese, A.; von Salviati, C.; Guerra, B.; Rösler, U. Transmission of ESBL/AmpC-producing Escherichia coli from broiler chicken farms to surrounding areas. Vet. Microbiol. 2014, 172, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Scicchitano, D.; Babbi, G.; Palladino, G.; Turroni, S.; Mekonnen, Y.T.; Laczny, C.; Wilmes, P.; Leekitcharoenphon, P.; Castagnetti, A.; D’Amico, F.; et al. Routes of dispersion of antibiotic resistance genes from the poultry farm system. Sci. Total. Environ. 2024, 912, 169086. [Google Scholar] [CrossRef] [PubMed]
- Merchant, L.E.; Rempel, H.; Forge, T.; Kannangara, T.; Bittman, S.; Delaquis, P.; Topp, E.; Ziebell, K.A.; Diarra, M.S. Characterization of antibiotic-resistant and potentially pathogenic Escherichia coli from soil fertilized with litter of broiler chickens fed antimicrobial-supplemented diets. Can. J. Microbiol. 2012, 58, 1084–1098. [Google Scholar] [CrossRef] [PubMed]
- Lautenbach, E.; Strom, B.L.; Bilker, W.B.; Patel, J.B.; Edelstein, P.H.; Fishman, N.O. Epidemiological investigation of fluoroquinolone resistance in infections due to extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Clin. Infect. Dis. 2001, 33, 1288–1294. [Google Scholar] [CrossRef]
- Paterson, D.L.; Mulazimoglu, L.; Casellas, J.M.; Ko, W.C.; Goossens, H.; Von Gottberg, A.; Mohapatra, S.; Trenholme, G.M.; Klugman, K.P.; McCormack, J.G.; et al. Epidemiology of ciprofloxacin resistance and its relationship to Extended-Spectrum Beta-Lactamase production in Klebsiella pneumoniae isolates causing bacteremia. Clin. Infect. Dis. 2000, 30, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Husna, A.; Rahman, M.M.; Badruzzaman, A.T.M.; Sikder, M.H.; Islam, M.R.; Rahman, M.T.; Alam, J.; Ashour, H.M. Extended-Spectrum β-Lactamases (ESBL): Challenges and Opportunities. Biomedicines 2023, 11, 2937. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk Susceptibility Tests, 14th ed.; CLSI Document M02; Committee for Clinical Laboratory Standards: Wayne, PA, USA, 2024. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Glasel, J.A. Validity of nucleic acid purities monitored by 260nm/280nm absorbance ratios. Biotechniques 1995, 18, 62–63. [Google Scholar] [PubMed]
- Colom, K.; Perez, J.; Alonso, R.; Fernandez-Aranguiz, A.; Larino, E.; Cisterna, R. Simple and reliable multiplex PCR assay for detection of blaTEM, blaSHV and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol. Lett. 2003, 223, 147–151. [Google Scholar] [CrossRef]
- Kozak, G.K.; Boerlin, P.; Janecko, N.; Reid-Smith, R.J.; Jardine, C. Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 2009, 75, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V.; Weill, F.X.; Poirel, L.; Fabre, L.; Soussy, C.J.; Nordmann, P. Prevalence of qnr genes in Salmonella in France. J. Antimicrob. Chemother. 2007, 59, 751–754. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.J.; Nordmann, P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef] [PubMed]

| Origin | No. of Tested Isolates | Number of Resistant Isolates (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CTX | CAZ | CIP | NA | C | CN | S | TE | ||
| Box bottoms One-day old | 68 | 68 (100) | 28 (41.2) | 7 (10.3) | 0 | 0 | 0 | 39 (57.4) | 45 (66.2) | 67(98.5) |
| Feces | ||||||||||
| Pullets | 90 | 72 (80) | 0 | 0 | 0 | 9 (10) | 0 | 2 (2.2) | 7 (7.8) | 63 (70) |
| Adults | 80 | 59 (73.8) | 3 (3.8) | 2 (2.5) | 2 (2.5) | 9 (11.3) | 3 (3.8) | 0 | 36 (40) | 45 (56.3) |
| % of resistant isolates | 76.9 | 1.9 | 1.2 | 1.2 | 10.7 | 1.9 | 1.1 | 23.9 | 63.1 | |
| Boot swabs | ||||||||||
| Pullets | 80 | 42 (52.5) | 4 (5) | 1 (1.3) | 2 (2.5) | 15 (18.8) | 6 (7.5) | 7 (8.8) | 4 (5) | 11 (13.8) |
| Adults | 65 | 45 (69.2) | 5 (7.7) | 1 (1.5) | 1 (1.5) | 21(32.3) | 6 (9.2) | 0 | 13 (20) | 34 (52.3) |
| % of resistant isolates | 60.9 | 6.4 | 1.4 | 2.0 | 25.6 | 8.4 | 4.4 | 12.5 | 33.1 | |
| TOTAL % of resistant isolates | 74.7 | 10.4 | 2.9 | 1.3 | 14.1 | 3.9 | 12.5 | 27.4 | 57.4 | |
| Profile | No. Isolates (%) | ||||
|---|---|---|---|---|---|
| Box Bottoms | Feces | Boot Swabs | |||
| One-Day-Old (n = 68) | Pullets (n = 90) | Adults (n = 80) | Pullets (n = 80) | Adults (n = 65) | |
| AMP | - | 20 (22.2) | 4 (5.0) | 22 (27.5) | 6 (9.2) |
| NA | - | - | 1 (1.3) | 9 (11.3) | - |
| S | - | - | - | 1 (1.3) | - |
| TE | - | 13 (14.4) | 3 (3.8) | 1 (1.3) | 2 (3.1) |
| AMP/C | - | - | 1 (1.3) | - | 1 (1.5) |
| AMP/CTX | - | - | - | - | 1 (1.5) |
| AMP/CN | - | 2 (2.2) | - | - | - |
| AMP/NA | - | 1 (1.1) | 4 (5.0) | 3 (3.8) | 9 (13.9) |
| AMP/S | - | - | 8 (10.0) | 1 (1.3) | - |
| AMP/TE | 13 (19.1) | 35 (38.9) | 13 (16.3) | 5 (6.3) | 6 (9.2) |
| CN/TE | - | - | - | 1 (1.3) | - |
| NA/TE | - | 1 (1.1) | 1 (1.3) | - | - |
| S/TE | - | - | 1 (1.3) | - | 1 (1.5) |
| AMP/NA/CIP | - | - | 1 (1.3) | 1 (1.3) | - |
| AMP/CTX/CAZ | - | - | 1 (1.3) | 1 (1.3) | 1 (1.5) |
| AMP/CTX/S | 1 (1.5) | - | - | - | - |
| AMP/CTX/TE | 2 (2.9) | - | - | - | 1 (1.5) |
| AMP/C/CN | - | - | - | 2 (2.5) | - |
| AMP/CN/TE | 2 (2.9) | - | - | - | - |
| AMP/S/TE | 4 (5.9) | 7 (7.8) | 23 (28.8) | 1 (1.3) | 8 (12.3) |
| AMP/NA/TE | - | 7 (7.8) | 1 (1.3) | 1 (1.3) | 10 (15.4) |
| C/S/TE | - | - | - | - | 4 (6.2) |
| AMP/CTX/C/CN | - | - | - | 3 (3.8) | - |
| AMP/CTX/C/TE | - | - | 1 (1.3) | - | 1 (1.5) |
| AMP/CTX/CN/TE | 4 (5.9) | - | - | - | - |
| AMP/CTX/S/TE | 6 (8.8) | - | - | - | - |
| AMP/CN/S/TE | 21 (30.9) | - | - | - | - |
| AMP/NA/CIP/TE | - | - | 1 (1.3) | 1 (1.3) | 1 (1.5) |
| AMP/CTX/CAZ/C/TE | - | - | 1 (1.3) | - | - |
| AMP/CTX/CAZ/CN/TE | 2 (2.9) | - | - | - | - |
| AMP/CTX/CN/S/TE | 8 (11.8) | - | - | - | - |
| AMP/CTX/CAZ/S/TE | 3 (4.4) | - | - | - | - |
| AMP/CTX/CAZ/CN/S/TE | 2 (2.9) | - | - | - | - |
| AMP/C/CN/S/TE | - | - | - | 1 (1.3) | - |
| Number of Isolates (%) | |||||
|---|---|---|---|---|---|
| Origin | blaTEM | blaSHV | blaCMY-2 | qnrB | qnrS |
| Box bottoms (n = 68) | 68 (100) | ||||
| Feces (n = 41) | 37 (90.2) | 36 (87.8) | 1 (2.4) | 35 (85.4) | 10 (24.4) |
| Boot swabs (n = 23) | 11 (47.8) | 10 (43.5) | 5 (21.7) | 8 (34.8) | 9 (39.1) |
| Total (%) | 48 (36.4) | 46 (34.8) | 74 (56.1) | 43 (32.6) | 19 (14.4) |
| Origin | Profile | Number of Isolates (%) |
|---|---|---|
| Box bottoms (n = 68) | blaCMY-2 | 68 (100) |
| Feces (n = 41) | blaSHV-blaTEM-qnrB | 29 (70.7) |
| blaSHV-blaTEM-qnrB-qnrS | 2 (4.9) | |
| blaTEM-qnrB-qnrS | 2 (4.9) | |
| blaTEM-qnrS | 2 (4.9) | |
| blaSHV-blaCMY-2-qnrS | 1 (2.4) | |
| blaSHV-blaTEM-qnrS | 1 (2.4) | |
| blaSHV-qnrB | 1 (2.4) | |
| blaSHV-qnrB-qnrS | 1 (2.4) | |
| blaSHV-qnrS | 1 (2.4) | |
| blaTEM-qnrB | 1 (2.4) | |
| Boot swabs (n = 23) | none | 2 (8.7) |
| blaSHV-blaTEM-qnrB | 6 (26.1) | |
| blaCMY-2 | 4 (17.4) | |
| qnrS | 4 (17.4) | |
| blaSHV-qnrS | 2 (8.7) | |
| blaTEM | 2 (8.7) | |
| blaSHV-blaTEM-blaCMY-2-qnrB-qnrS | 1 (4.4) | |
| blaSHV-blaTEM-qnrB-qnrS | 1 (4.4) | |
| blaTEM-qnrS | 1 (4.4) |
| Primers | Sequence | Product Size (bp) | Reference |
|---|---|---|---|
| blaSHV-f | AGGATTGACTGCCTTTTTG | 393 | [68] |
| blaSHV-r | ATTTGCTGATTTCGCTCG | ||
| blaTEM-f | TTAACTGGCGAACTACTTAC | 247 | [69] |
| blaTEM-r | GTCTATTTCGTTCATCCATA | ||
| blaCMY-2-f | GACAGCCTCTTTCTCCACA | 1000 | |
| blaCMY-2-r | TGGACACGAAGGCTACGTA | ||
| qnrB-f | GGMATHGAAATTCGCCACTG | 264 | [70] |
| qnrB-r | TTTGCYGYYCGCCAGTCGAA | ||
| qnrS-f | GCAAGTTCATTGAACAGGGT | 428 | [71] |
| qnrS-r | TCTAAACCGTCGAGTTCGGCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fenollar-Penadés, A.; Catalá-Gregori, P.; Tallá-Ferrer, V.; Castillo, M.Á.; García-Ferrús, M.; Jiménez-Belenguer, A. Evolution of the Antibiotic Resistance Levels, Multi-Resistance Patterns, and Presence of Antibiotic Resistance Genes in E. coli Isolates from the Feces of Breeding Hens during the Rearing Period. Antibiotics 2024, 13, 753. https://doi.org/10.3390/antibiotics13080753
Fenollar-Penadés A, Catalá-Gregori P, Tallá-Ferrer V, Castillo MÁ, García-Ferrús M, Jiménez-Belenguer A. Evolution of the Antibiotic Resistance Levels, Multi-Resistance Patterns, and Presence of Antibiotic Resistance Genes in E. coli Isolates from the Feces of Breeding Hens during the Rearing Period. Antibiotics. 2024; 13(8):753. https://doi.org/10.3390/antibiotics13080753
Chicago/Turabian StyleFenollar-Penadés, Alejandro, Pablo Catalá-Gregori, Vicente Tallá-Ferrer, María Ángeles Castillo, Miguel García-Ferrús, and Ana Jiménez-Belenguer. 2024. "Evolution of the Antibiotic Resistance Levels, Multi-Resistance Patterns, and Presence of Antibiotic Resistance Genes in E. coli Isolates from the Feces of Breeding Hens during the Rearing Period" Antibiotics 13, no. 8: 753. https://doi.org/10.3390/antibiotics13080753
APA StyleFenollar-Penadés, A., Catalá-Gregori, P., Tallá-Ferrer, V., Castillo, M. Á., García-Ferrús, M., & Jiménez-Belenguer, A. (2024). Evolution of the Antibiotic Resistance Levels, Multi-Resistance Patterns, and Presence of Antibiotic Resistance Genes in E. coli Isolates from the Feces of Breeding Hens during the Rearing Period. Antibiotics, 13(8), 753. https://doi.org/10.3390/antibiotics13080753






