Machine Learning in FTIR Spectrum for the Identification of Antibiotic Resistance: A Demonstration with Different Species of Microorganisms
Abstract
1. Introduction
2. Results
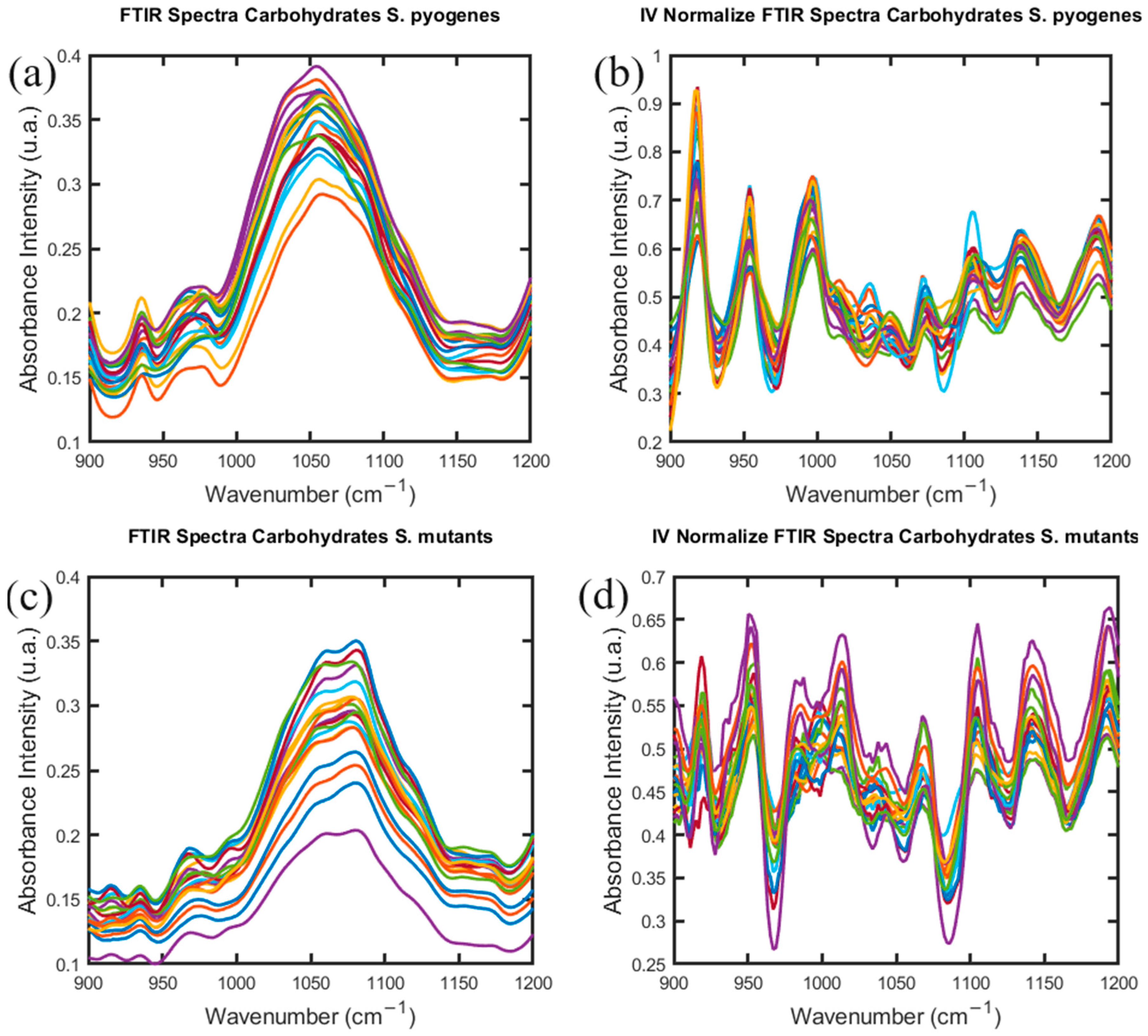
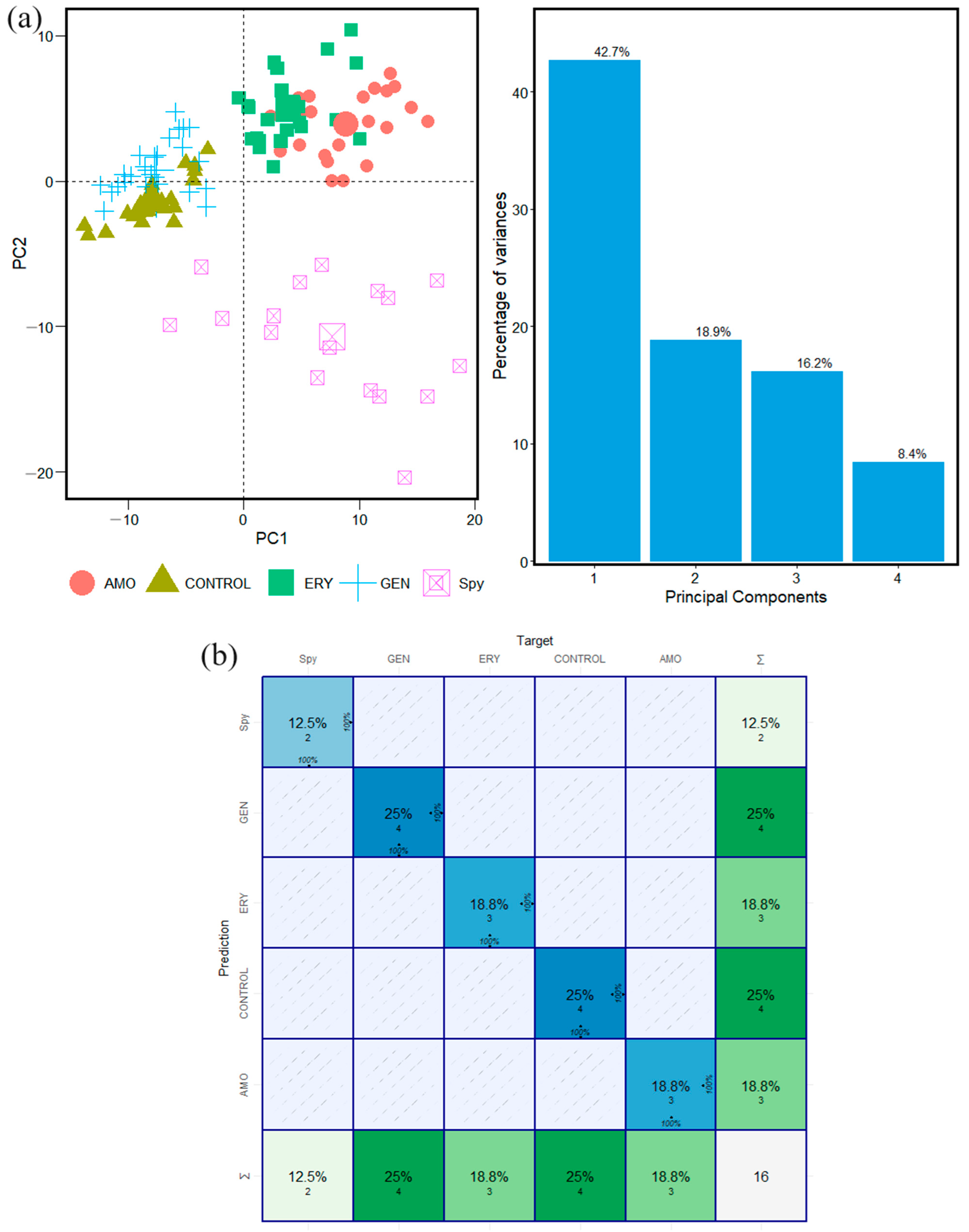
2.1. Gram-Positives Species: S. pyogenes and S. mutans
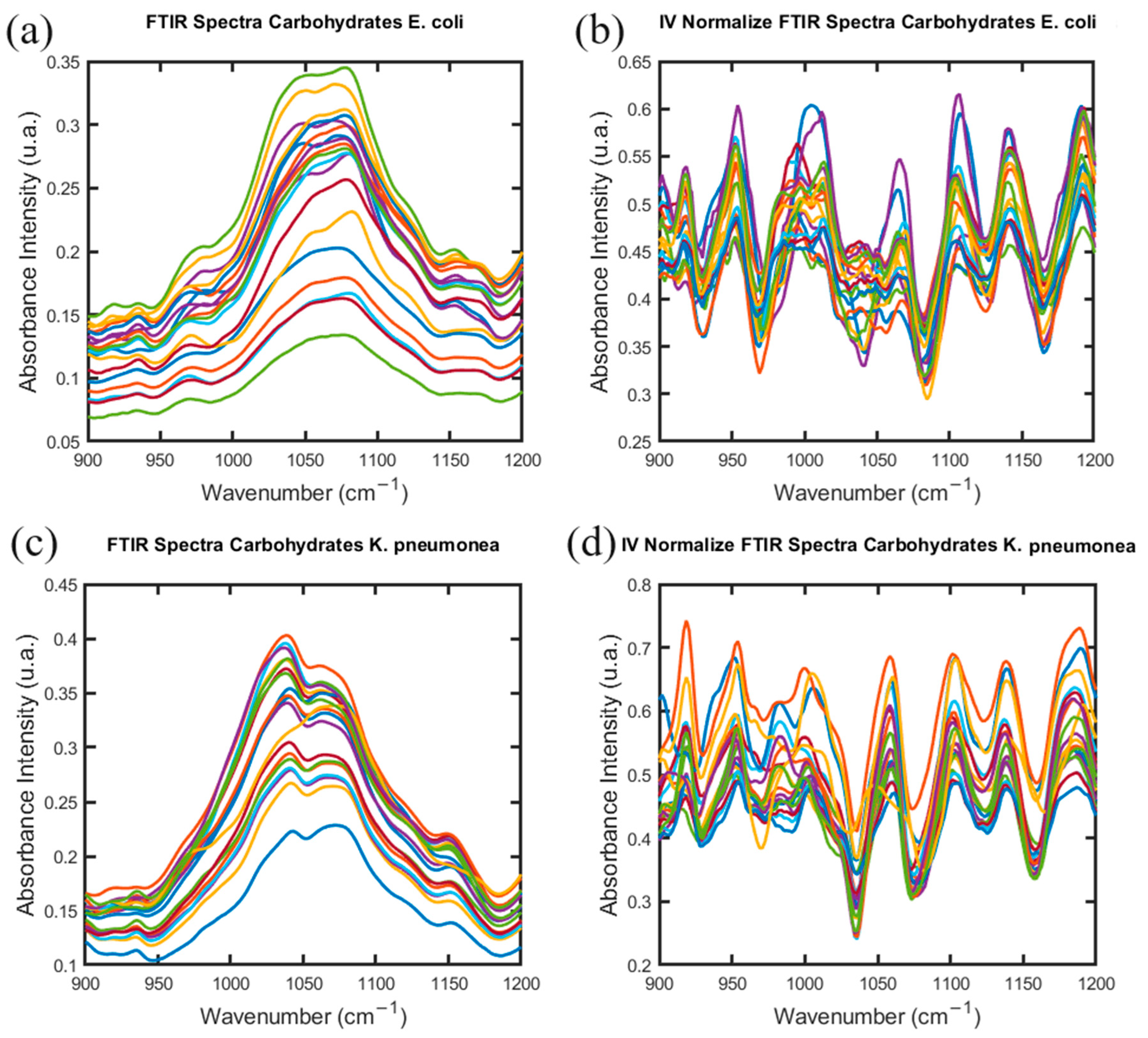
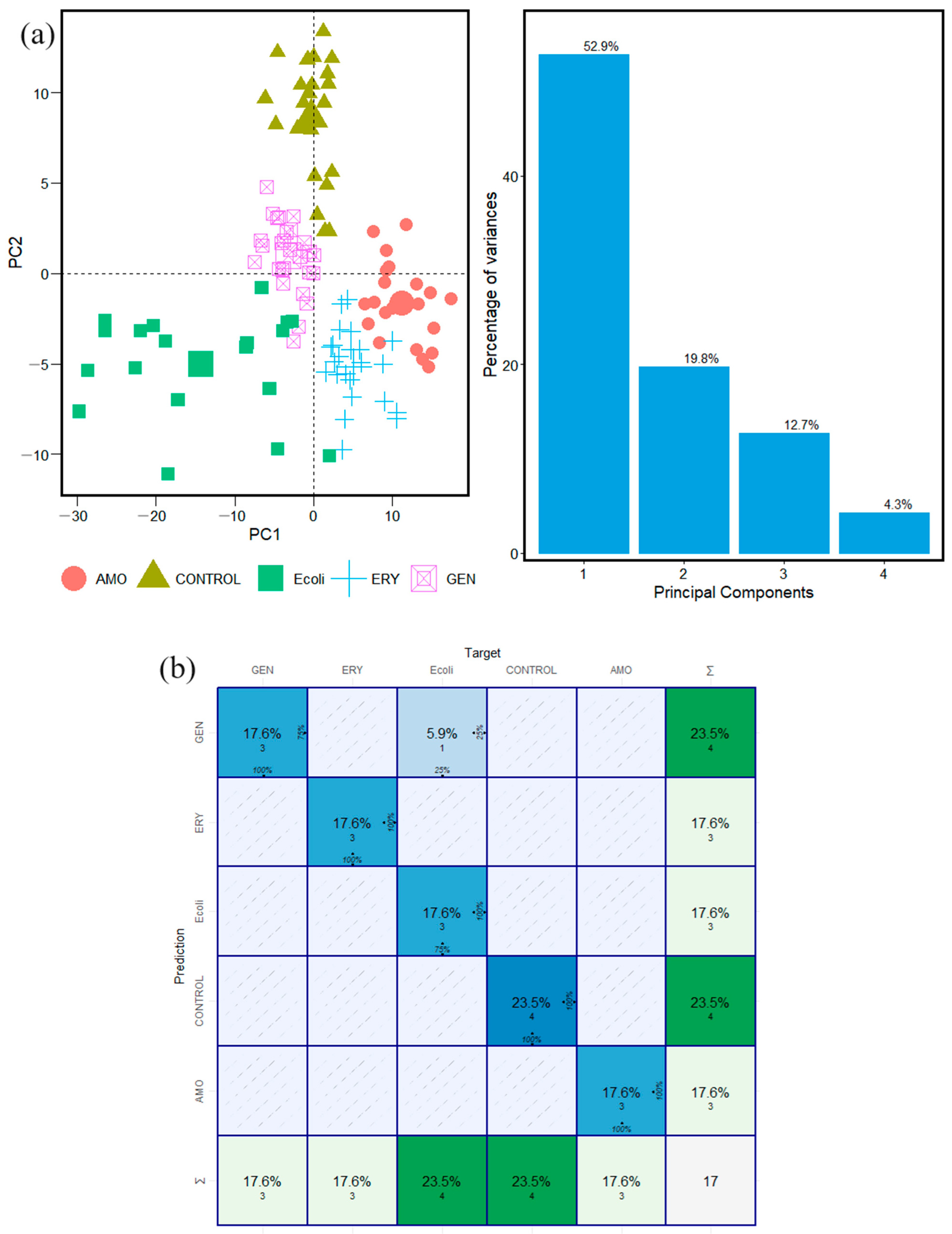
2.2. Gram-Negative Species: E. coli and K. pneumoniae
3. Discussion
4. Materials and Methods
4.1. Samples Preparation and FTIR Spectra Acquisition
4.2. Microorganisms
4.3. Fourier Transformation Infrared Spectroscopy
4.4. FTIR Spectra Database Analysis Process Overview
4.5. Methodology Developed to Find/Determine the Effect of the Antibiotics
4.6. Machine Learning Algorithms
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Antibiotic Resistance: Multi-Country Public Awareness Survey; World Health Organization: Geneva, Switzerland, 2015; pp. 1–51. [Google Scholar]
- Talebi Bezmin Abadi, A.; Rizvanov, A.A.; Haertlé, T.; Blatt, N.L. World Health Organization Report: Current Crisis of Antibiotic Resistance. Bionanoscience 2019, 9, 778–788. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial Resistance: A Global Multifaceted Phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Littmann, J.; Viens, A. The Ethical Significance of Antimicrobial Resistance. Public Health Ethics 2015, 8, phv025. [Google Scholar] [CrossRef]
- Martinez, J.L. General Principles of Antibiotic Resistance in Bacteria. Drug Discov. Today Technol. 2014, 11, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B.; Marshall, B. Antibacterial Resistance Worldwide: Causes, Challenges and Responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef]
- Li, H.; Sun, X.; Cui, W.; Xu, M.; Dong, J.; Ekundayo, B.E.; Ni, D.; Rao, Z.; Guo, L.; Stahlberg, H.; et al. Computational Drug Development for Membrane Protein Targets. Nat. Biotechnol. 2024, 42, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Wan, F.; Wong, F.; Collins, J.J.; de la Fuente-Nunez, C. Machine Learning for Antimicrobial Peptide Identification and Design. Nat. Rev. Bioeng. 2024, 2, 392–407. [Google Scholar] [CrossRef]
- Jiménez-Luna, J.; Grisoni, F.; Schneider, G. Drug Discovery with Explainable Artificial Intelligence. Nat. Mach. Intell. 2020, 2, 573–584. [Google Scholar] [CrossRef]
- Allenspach, S.; Hiss, J.A.; Schneider, G. Neural Multi-Task Learning in Drug Design. Nat. Mach. Intell. 2024, 6, 124–137. [Google Scholar] [CrossRef]
- Schneider, P.; Walters, W.P.; Plowright, A.T.; Sieroka, N.; Listgarten, J.; Goodnow, R.A.; Fisher, J.; Jansen, J.M.; Duca, J.S.; Rush, T.S.; et al. Rethinking Drug Design in the Artificial Intelligence Era. Nat. Rev. Drug Discov. 2020, 19, 353–364. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. In Virulence Mechanisms of Bacterial Pathogens; ASM Press: Washington, DC, USA, 2016; pp. 481–511. [Google Scholar]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular Mechanisms of Antibiotic Resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Opal, S.M.; Pop-Vicas, A. Molecular Mechanisms of Antibiotic Resistance in Bacteria. Mand. Douglas Bennett’s Princ. Pract. Infect. Dis. 2014, 1, 235–251. [Google Scholar] [CrossRef]
- Sodhi, K.K.; Kumar, M.; Balan, B.; Dhaulaniya, A.S.; Shree, P.; Sharma, N.; Singh, D.K. Perspectives on the Antibiotic Contamination, Resistance, Metabolomics, and Systemic Remediation. SN Appl. Sci. 2021, 3, 269. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Davis, M.F. Application of One Health Principles to the Control of Antimicrobial Resistance. In Oxford Research Encyclopedia of Global Public Health; Oxford University Press: Oxford, UK, 2024. [Google Scholar]
- Rzycki, M.; Gładysiewicz-Kudrawiec, M.; Kraszewski, S. Molecular Guidelines for Promising Antimicrobial Agents. Sci. Rep. 2024, 14, 4641. [Google Scholar] [CrossRef]
- Barrera-Patiño, C.P.; Soares, J.M.; Branco, K.C.; Inada, N.M.; Bagnato, V.S. Spectroscopic Identification of Bacteria Resistance to Antibiotics by Means of Absorption of Specific Biochemical Groups and Special Machine Learning Algorithm. Antibiotics 2023, 12, 1502. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Murugaiyan, J.; Kumar, P.A.; Rao, G.S.; Iskandar, K.; Hawser, S.; Hays, J.P.; Mohsen, Y.; Adukkadukkam, S.; Awuah, W.A.; Jose, R.A.M.; et al. Progress in Alternative Strategies to Combat Antimicrobial Resistance: Focus on Antibiotics. Antibiotics 2022, 11, 200. [Google Scholar] [CrossRef]
- Chellat, M.F.; Raguž, L.; Riedl, R. Targeting Antibiotic Resistance. Angew. Chemie Int. Ed. 2016, 55, 6600–6626. [Google Scholar] [CrossRef]
- Soares, J.M.; Guimarães, F.E.G.; Yakovlev, V.V.; Bagnato, V.S.; Blanco, K.C. Physicochemical Mechanisms of Bacterial Response in the Photodynamic Potentiation of Antibiotic Effects. Sci. Rep. 2022, 12, 21146. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.A.; Cheburkanov, V.; Chen, S.; Soares, J.M.; Kassab, G.; Blanco, K.C.; Bagnato, V.S.; de Figueiredo, P.; Yakovlev, V.V. Breaking down Antibiotic Resistance in Methicillin-Resistant Staphylococcus Aureus: Combining Antimicrobial Photodynamic and Antibiotic Treatments. Proc. Natl. Acad. Sci. USA 2022, 119, e2208378119. [Google Scholar] [CrossRef]
- Soares, J.M.; Yakovlev, V.V.; Blanco, K.C.; Bagnato, V.S. Recovering the Susceptibility of Antibiotic-Resistant Bacteria Using Photooxidative Damage. Proc. Natl. Acad. Sci. USA 2023, 120, e2311667120. [Google Scholar] [CrossRef] [PubMed]
- Jubeh, B.; Breijyeh, Z.; Karaman, R. Resistance of Gram-Positive Bacteria to Current Antibacterial Agents and Overcoming Approaches. Molecules 2020, 25, 2888. [Google Scholar] [CrossRef] [PubMed]
- Exner, M.; Bhattacharya, S.; Christiansen, B.; Gebel, J.; Goroncy-Bermes, P.; Hartemann, P.; Heeg, P.; Ilschner, C.; Kramer, A.; Larson, E.; et al. Antibiotic Resistance: What Is so Special about Multidrug-Resistant Gram-Negative Bacteria? Antibiotikaresistenz: Was Ist so Besonders an Den Gram-Negativen. GMS Hyg. Infect. Control 2017, 12, 1–24. [Google Scholar]
- Schmitt, J.; Flemming, H.-C. FTIR-Spectroscopy in Microbial and Material Analysis. Int. Biodeterior. Biodegrad. 1998, 41, 1–11. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Atamanyuk, I.; Kondratenko, Y.; Havrysh, V.; Volosyuk, Y. Computational Method of the Cardiovascular Diseases Classification Based on a Generalized Nonlinear Canonical Decomposition of Random Sequences. Sci. Rep. 2023, 13, 59. [Google Scholar] [CrossRef]
- MATLAB App Building. MATLAB, (2021b); The MathWorks: Natick, MA, USA, 2021.
- Nguyen, G.; Dlugolinsky, S.; Bobák, M.; Tran, V.; López García, Á.; Heredia, I.; Malík, P.; Hluchý, L. Machine Learning and Deep Learning Frameworks and Libraries for Large-Scale Data Mining: A Survey. Artif. Intell. Rev. 2019, 52, 77–124. [Google Scholar] [CrossRef]
- Rizzo, M.L. Statistical Computing with R; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2021. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2013. [Google Scholar]
- Mair, P.; Hofmann, E.; Gruber, K.; Hatzinger, R.; Zeileis, A.; Hornik, K. Motivation, Values, and Work Design as Drivers of Participation in the R Open Source Project for Statistical Computing. Proc. Natl. Acad. Sci. USA 2015, 112, 14788–14792. [Google Scholar] [CrossRef]
- Fox, J. Aspects of the Social Organization and Trajectory of the r Project. R J. 2009, 1, 5. [Google Scholar] [CrossRef]
- Gareth, J.; Daniela, W.; Trevor, H.; Robert, T. An Introduction to Statistical Learning: With Applications in R; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Chambers, J.M. Software for Data Analysis: Programming with R; Springer: Berlin/Heidelberg, Germany, 2008; Volume 2. [Google Scholar]
- Ripley, B.D. The R Project in Statistical Computing. MSOR Connect. Newsl. LTSN Maths Stats Netw. 2001, 1, 23–25. [Google Scholar] [CrossRef]
- Germond, A.; Ichimura, T.; Horinouchi, T.; Fujita, H.; Furusawa, C.; Watanabe, T.M. Raman Spectral Signature Reflects Transcriptomic Features of Antibiotic Resistance in Escherichia Coli. Commun. Biol. 2018, 1, 85. [Google Scholar] [CrossRef] [PubMed]
- Wenning, M.; Scherer, S. Identification of Microorganisms by FTIR Spectroscopy: Perspectives and Limitations of the Method. Appl. Microbiol. Biotechnol. 2013, 97, 7111–7120. [Google Scholar] [CrossRef] [PubMed]
- Lamprell, H.; Mazerolles, G.; Kodjo, A.; Chamba, J.F.; Noël, Y.; Beuvier, E. Discrimination of Staphylococcus Aureus Strains from Different Species of Staphylococcus Using Fourier Transform Infrared (FTIR) Spectroscopy. Int. J. Food Microbiol. 2006, 108, 125–129. [Google Scholar] [CrossRef]
- Vogt, S.; Löffler, K.; Dinkelacker, A.G.; Bader, B.; Autenrieth, I.B.; Peter, S.; Liese, J. Fourier-Transform Infrared (FTIR) Spectroscopy for Typing of Clinical Enterobacter Cloacae Complex Isolates. Front. Microbiol. 2019, 10, 2582. [Google Scholar] [CrossRef]
- Naumann, D. Infrared Spectroscopy in Microbiology. Encycl. Anal. Chem. 2000, 102, 131. [Google Scholar]
- Zhang, F.; Cheng, W. The Mechanism of Bacterial Resistance and Potential Bacteriostatic Strategies. Antibiotics 2022, 11, 1215. [Google Scholar] [CrossRef]
- Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef]
- Uysal Ciloglu, F.; Saridag, A.M.; Kilic, I.H.; Tokmakci, M.; Kahraman, M.; Aydin, O. Identification of Methicillin-Resistant: Staphylococcus Aureus Bacteria Using Surface-Enhanced Raman Spectroscopy and Machine Learning Techniques. Analyst 2020, 145, 7559–7570. [Google Scholar] [CrossRef]
- Asnicar, F.; Thomas, A.M.; Passerini, A.; Waldron, L.; Segata, N. Machine Learning for Microbiologists. Nat. Rev. Microbiol. 2024, 22, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Müllner, D. Modern Hierarchical, Agglomerative Clustering Algorithms. arXiv 2011, arXiv:1109.2378. [Google Scholar]
- Fillbrunn, A.; Berthold, M.R. Diversity-Driven Widening of Hierarchical Agglomerative Clustering; Springer: Berlin/Heidelberg, Germany, 2015; pp. 84–94. [Google Scholar]
- Rousseeuw, P.J. Silhouettes: A Graphical Aid to the Interpretation and Validation of Cluster Analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Steinley, D.; Brusco, M.J. Choosing the Number of Clusters in Κ-Means Clustering. Psychol. Methods 2011, 16, 285–297. [Google Scholar] [CrossRef] [PubMed]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning; Springer: Berlin/Heidelberg, Germany, 2013; Volume 112. [Google Scholar]
- Dunn, J.C. A Fuzzy Relative of the ISODATA Process and Its Use in Detecting Compact Well-Separated Clusters. J. Cybern. 1973, 3, 32–57. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, D.; Frangi, A.F.; Yang, J. Two-Dimensional PCA: A New Approach to Appearance-Based Face Representation and Recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2004, 26, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Buntine, W.L.; Jakulin, A. Applying Discrete PCA in Data Analysis. arXiv 2012, arXiv:1207.4125. [Google Scholar]
- Yang, J.; Yang, J. Why Can LDA Be Performed in PCA Transformed Space? Pattern Recognit. 2003, 36, 563–566. [Google Scholar] [CrossRef]
- Hoffmann, H. Kernel PCA for Novelty Detection. Pattern Recognit. 2007, 40, 863–874. [Google Scholar] [CrossRef]
- Daffertshofer, A.; Lamoth, C.J.C.; Meijer, O.G.; Beek, P.J. PCA in Studying Coordination and Variability: A Tutorial. Clin. Biomech. 2004, 19, 415–428. [Google Scholar] [CrossRef]
- Batista, G.E.A.P.A.; Prati, R.C.; Monard, M.C. A Study of the Behavior of Several Methods for Balancing Machine Learning Training Data. ACM SIGKDD Explor. Newsl. 2004, 6, 20–29. [Google Scholar] [CrossRef]
- Wierzchoń, S.T.; Kłopotek, M.A. Modern Algorithms of Cluster Analysis; Springer: Berlin/Heidelberg, Germany, 2018; Volume 34. [Google Scholar]
- Jaeger, A.; Banks, D. Cluster Analysis: A Modern Statistical Review. WIREs Comput. Stat. 2023, 15, e1597. [Google Scholar] [CrossRef]
- Schaeffer, S.E. Graph Clustering. Comput. Sci. Rev. 2007, 1, 27–64. [Google Scholar] [CrossRef]
- Lancichinetti, A.; Fortunato, S. Consensus Clustering in Complex Networks. Sci. Rep. 2012, 2, 336. [Google Scholar] [CrossRef] [PubMed]
- Dubes, R.; Jain, A.K. Clustering Methodologies in Exploratory Data Analysis; Elsevier: Amsterdam, The Netherlands, 1980; pp. 113–228. [Google Scholar]
- Aha, D.W.; Kibler, D. Noise-Tolerant Instance-Based Learning Algorithms; University of California: Irvine, CA, USA, 1989; Volume 1, pp. 794–799. [Google Scholar]
- Sandrin, T.R.; Goldstein, J.E.; Schumaker, S. MALDI TOF MS Profiling of Bacteria at the Strain Level: A Review. Mass Spectrom. Rev. 2013, 32, 188–217. [Google Scholar] [CrossRef]
- Palma, S.I.C.J.; Traguedo, A.P.; Porteira, A.R.; Frias, M.J.; Gamboa, H.; Roque, A.C.A. Machine Learning for the Meta-Analyses of Microbial Pathogens’ Volatile Signatures. Sci. Rep. 2018, 8, 3360. [Google Scholar] [CrossRef]
- Oyewole, G.J.; Thopil, G.A. Data Clustering: Application and Trends. Artif. Intell. Rev. 2022, 6, 6439–6475. [Google Scholar] [CrossRef]
- Sammut, C.; Webb, G.I. (Eds.) Encyclopedia of Machine Learning and Data Mining; Springer: Boston, MA, USA, 2017; ISBN 978-1-4899-7685-7. [Google Scholar]
- Monti, S.; Tamayo, P.; Mesirov, J.; Golub, T. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Mach. Learn. 2003, 52, 91–118. [Google Scholar] [CrossRef]
- King, R.S. Cluster Analysis and Data Mining: An Introduction; Mercury Learning and Information: Herndon, VA, USA, 2015. [Google Scholar]
- Dudoit, S.; Fridlyand, J. A Prediction-Based Resampling Method for Estimating the Number of Clusters in a Dataset. Genome Biol. 2002, 3, research0036.1. [Google Scholar] [CrossRef]
- Anderson, P.W. Absence of Diffusion in Certain Random Lattices. Phys. Rev. 1958, 109, 1492–1505. [Google Scholar] [CrossRef]
- Chambon, S.; Galtier, M.N.; Arnal, P.J.; Wainrib, G.; Gramfort, A. A Deep Learning Architecture for Temporal Sleep Stage Classification Using Multivariate and Multimodal Time Series. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 758–769. [Google Scholar] [CrossRef]
- Ciloglu, F.U.; Caliskan, A.; Saridag, A.M.; Kilic, I.H.; Tokmakci, M.; Kahraman, M.; Aydin, O. Drug-Resistant Staphylococcus Aureus Bacteria Detection by Combining Surface-Enhanced Raman Spectroscopy (SERS) and Deep Learning Techniques. Sci. Rep. 2021, 11, 18444. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Dong, H.; Shen, W.; Yang, Y.; Li, Z.; Liu, Y.; Wang, C.; Gu, B.; Zhang, L. Rapid SERS Identification of Methicillin-Susceptible and Methicillin-Resistant: Staphylococcus Aureus via Aptamer Recognition and Deep Learning. RSC Adv. 2021, 11, 34425–34431. [Google Scholar] [CrossRef]
- Damrich, S.; Berens, P.; Kobak, D. Persistent Homology for High-Dimensional Data Based on Spectral Methods. arXiv 2023, arXiv:2311.03087. [Google Scholar]
- Kulkarni, S.R.; Harman, G. Statistical Learning Theory: A Tutorial. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 543–556. [Google Scholar] [CrossRef]
- Bruce, P.; Bruce, A.; Gedeck, P. Practical Statistics for Data Scientists: 50+ Essential Concepts Using R and Python; O’Reilly Media: Newton, MA, USA, 2020. [Google Scholar]
- Lancichinetti, A.; Radicchi, F.; Ramasco, J.J.; Fortunato, S. Finding Statistically Significant Communities in Networks. PLoS ONE 2011, 6, e18961. [Google Scholar] [CrossRef]
- Tibshirani, R.; Walther, G.; Hastie, T. Estimating the Number of Clusters in a Data Set Via the Gap Statistic. J. R. Stat. Soc. Ser. B Stat. Methodol. 2001, 63, 411–423. [Google Scholar] [CrossRef]
- Jain, A.K.; Murty, M.N.; Flynn, P.J. Data Clustering. ACM Comput. Surv. 1999, 31, 264–323. [Google Scholar] [CrossRef]
- Tokuda, E.K.; Comin, C.H.; Costa, L. da F. Revisiting Agglomerative Clustering. Phys. A Stat. Mech. Its Appl. 2022, 585, 126433. [Google Scholar] [CrossRef]
- Wackerly, D.; Mendenhall, W.; Scheaffer, R.L. Mathematical Statistics with Applications; Cengage Learning: Boston, MA, USA, 2014. [Google Scholar]
- Anderson, T.W. Introduction to Multivariate Statistical Analysis; Wiley: New York, NY, USA, 1958. [Google Scholar]
- Kong, K.-F.; Schneper, L.; Mathee, K. Beta-Lactam Antibiotics: From Antibiosis to Resistance and Bacteriology. APMIS 2010, 118, 1–36. [Google Scholar] [CrossRef]
- Vale de Macedo, G.H.R.; Costa, G.D.E.; Oliveira, E.R.; Damasceno, G.V.; Mendonça, J.S.P.; Silva, L.d.S.; Chagas, V.L.; Bazán, J.M.N.; Aliança, A.S.d.S.; Miranda, R.d.C.M.d.; et al. Interplay between ESKAPE Pathogens and Immunity in Skin Infections: An Overview of the Major Determinants of Virulence and Antibiotic Resistance. Pathogens 2021, 10, 148. [Google Scholar] [CrossRef]
- Liu, C.Y.; Han, Y.Y.; Shih, P.H.; Lian, W.N.; Wang, H.H.; Lin, C.H.; Hsueh, P.R.; Wang, J.K.; Wang, Y.L. Rapid Bacterial Antibiotic Susceptibility Test Based on Simple Surface-Enhanced Raman Spectroscopic Biomarkers. Sci. Rep. 2016, 6, 23375. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.; Kaur, P. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How Antibiotics Kill Bacteria: From Targets to Networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef]
- Wang, K.; Li, S.; Petersen, M.; Wang, S.; Lu, X. Detection and Characterization of Antibiotic-Resistant Bacteria Using Surface-Enhanced Raman Spectroscopy. Nanomaterials 2018, 8, 762. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Fu, Y.; Zhao, H.; Liu, X.; Wu, X.; Lin, T.; Wang, H.; Song, L.; Fang, Y.; Lu, W.; et al. Dynamic Insights into Increasing Antibiotic Resistance in Staphylococcus Aureus by Label-Free SERS Using a Portable Raman Spectrometer. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 273, 121070. [Google Scholar] [CrossRef]
- Salman, A.; Sharaha, U.; Rodriguez-Diaz, E.; Shufan, E.; Riesenberg, K.; Bigio, I.J.; Huleihel, M. Detection of Antibiotic Resistant: Escherichia Coli Bacteria Using Infrared Microscopy and Advanced Multivariate Analysis. Analyst 2017, 142, 2136–2144. [Google Scholar] [CrossRef]
- Carbonell, J.G.; Michalski, R.S.; Mitchell, T.M. Machine Learning: A Historical and Methodological Analysis. AI Mag. 1983, 4, 69. [Google Scholar] [CrossRef]
- Mitra, P.; Murthy, C.A.; Pal, S.K. Unsupervised Feature Selection Using Feature Similarity. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 301–312. [Google Scholar] [CrossRef]
- Liu, L.; Özsu, M.T. (Eds.) Encyclopedia of Database Systems; Springer: Boston, MA, USA, 2009; ISBN 978-0-387-35544-3. [Google Scholar]
- Lam, W.; Keung, C.-K.; Liu, D. Discovering Useful Concept Prototypes for Classification Based on Filtering and Abstraction. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 1075–1090. [Google Scholar] [CrossRef]
- Aha, D.W.; Kibler, D.; Albert, M.K. Instance-Based Learning Algorithms. Mach. Learn. 1991, 6, 37–66. [Google Scholar] [CrossRef]
- Sammut, C.; Webb, G.I. (Eds.) Encyclopedia of Machine Learning; Springer: Boston, MA, USA, 2010; ISBN 978-0-387-30768-8. [Google Scholar]
- Yager, R.R. Golden Rule and Other Representative Values for Atanassov Type Intuitionistic Membership Grades. IEEE Trans. Fuzzy Syst. 2015, 23, 2260–2269. [Google Scholar] [CrossRef]
- Reynolds, D. Gaussian Mixture Models. In Encyclopedia of Biometrics; Springer: Boston, MA, USA, 2009; pp. 659–663. [Google Scholar]
- Kearns, M.J.; Vazirani, U. An Introduction to Computational Learning Theory; MIT Press: Cambridge, MA, USA, 1994. [Google Scholar]
- Kurt, I.; Ture, M.; Kurum, A.T. Comparing Performances of Logistic Regression, Classification and Regression Tree, and Neural Networks for Predicting Coronary Artery Disease. Expert Syst. Appl. 2008, 34, 366–374. [Google Scholar] [CrossRef]
- Wang, F.K.; Du, T.C.T. Using Principal Component Analysis in Process Performance for Multivariate Data. Omega 2000, 28, 185–194. [Google Scholar] [CrossRef]


| Sample Group | Bacteria | Molecular Window Interval Groups | Antibiotic Resistance Susceptibility Identified | Result Machine Learning Identification | Real Antibiotic Susceptibility |
|---|---|---|---|---|---|
| Gram-positive | Streptococcus pyogenes | Carbohydrates | AMO | AMO | Accurate |
| Fatty acids | AMO | ||||
| Proteins | AMO | ||||
| Streptococcus mutans | Carbohydrates | GEN | ERY | Accurate | |
| Fatty acids | ERY | ||||
| Proteins | ERY | ||||
| Gram-negative | Escherichia coli | Carbohydrates | GEN | GEN, ERY, and AMO | Accurate |
| Fatty acids | ERY | ||||
| Proteins | AMO | ||||
| Klebsiella pneumoniae | Carbohydrates | CONTROL | ERY | Accurate | |
| Fatty acids | ERY | ||||
| Proteins | ERY |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barrera Patiño, C.P.; Soares, J.M.; Blanco, K.C.; Bagnato, V.S. Machine Learning in FTIR Spectrum for the Identification of Antibiotic Resistance: A Demonstration with Different Species of Microorganisms. Antibiotics 2024, 13, 821. https://doi.org/10.3390/antibiotics13090821
Barrera Patiño CP, Soares JM, Blanco KC, Bagnato VS. Machine Learning in FTIR Spectrum for the Identification of Antibiotic Resistance: A Demonstration with Different Species of Microorganisms. Antibiotics. 2024; 13(9):821. https://doi.org/10.3390/antibiotics13090821
Chicago/Turabian StyleBarrera Patiño, Claudia Patricia, Jennifer Machado Soares, Kate Cristina Blanco, and Vanderlei Salvador Bagnato. 2024. "Machine Learning in FTIR Spectrum for the Identification of Antibiotic Resistance: A Demonstration with Different Species of Microorganisms" Antibiotics 13, no. 9: 821. https://doi.org/10.3390/antibiotics13090821
APA StyleBarrera Patiño, C. P., Soares, J. M., Blanco, K. C., & Bagnato, V. S. (2024). Machine Learning in FTIR Spectrum for the Identification of Antibiotic Resistance: A Demonstration with Different Species of Microorganisms. Antibiotics, 13(9), 821. https://doi.org/10.3390/antibiotics13090821







