Assessment of Antibiotic Resistance in Pediatric Infections: A Romanian Case Study on Pathogen Prevalence and Effective Treatments
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
Tested Antibiotics
- Beta-lactams:
- ❖
- Penicillins:
- Ampicillin
- Penicillin
- Piperacillin
- Oxacillin
- Unasyn (ampicillin/sulbactam)
- Ticarcillin
- Augmentin (amoxicillin/clavulanate)
- Tazocin (piperacillin/tazobactam)
- ❖
- Cephalosporins:
- Cefepime (4th generation)
- Cefotaxime (3rd generation)
- Ceftriaxone (3rd generation)
- Cefoxitin (2nd generation)
- Ceftaroline (5th generation)
- Ceftazidime (3rd generation)
- Ceftazidime + avibactam (3rd generation with beta-lactamase inhibitor)
- Cefotaxime + avibactam
- Cefuroxime (2nd generation)
- Cefazolin (1st generation)
- Cefort (cefotetan, 2nd generation)
- ❖
- Carbapenems:
- Imipenem
- Meropenem
- Ertapenem
- ❖
- Monobactams:
- Aztreonam
- Macrolides:
- Clarithromycin
- Erythromycin
- Tetracyclines:
- Doxycycline
- Tetracycline
- Minocycline
- Tigecycline
- Aminoglycosides:
- Amikacin
- Gentamicin
- Tobramycin
- Fluoroquinolones:
- Ciprofloxacin (Ciprinol, Ciproxina)
- Levofloxacin
- Ofloxacin
- Moxifloxacin (Avelox)
- Sulfonamides:
- Biseptol (trimethoprim/sulfamethoxazole)
- Oxazolidinones:
- Linezolid
- Lincosamides:
- Clindamycin
- Polypeptides:
- Colistin
- Rifamycins:
- Rifampicin
- Phenicol:
- Chloramphenicol
- Phosphonate:
- Fosfomycin (Monural)
- Nitrofurans:
- Nitrofurantoin
- Lipopeptide:
- Daptomycin
- Others:
- Vancomycin (glycopeptide)
- Teicoplanin (glycopeptide)
- Sulcef (sultamicillin)
- Chlorhexidine
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial antibiotic resistance: The most critical pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist. 2014, 1–20. [Google Scholar]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [PubMed]
- Browne, A.J.; Chipeta, M.G.; Haines-Woodhouse, G.; Kumaran, E.P.A.; Hamadani, B.H.K.; Zaraa, S.; Henry, N.J.; Deshpande, A.; Reiner, R.C., Jr.; Day, N.P.J.; et al. Global antibiotic consumption and usage in humans, 2000–18: A spatial modelling study. Lancet Planet Health 2021, 5, e893–e904. [Google Scholar] [CrossRef] [PubMed]
- Malik, B.; Bhattacharyya, S. Antibiotic drug-resistance as a complex system driven by socio-economic growth and antibiotic misuse. Sci. Rep. 2019, 9, 9788. [Google Scholar]
- Ikuta, K.S.; Swetschinski, L.R.; Aguilar, G.R.; Sharara, F.; Mestrovic, T.; Gray, A.P.; Weaver, N.D.; Wool, E.E.; Han, C.; Hayoon, A.G.; et al. Global mortality associated with 33 bacterial pathogens in 2019: A systematic analysis for the global burden of disease study 2019. Lancet 2022, 400, 2221–2248. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Antibiotic resistance: Moving from individual health norms to social norms in one health and global health. Front. Microbiol. 2020, 11, 1914. [Google Scholar] [CrossRef]
- Sun, R.; Yao, T.; Zhou, X.; Harbarth, S.; Lin, L. Non-biomedical factors affecting antibiotic use in the community: A mixed-methods systematic review and meta-analysis. Clin. Microbiol. Infect. 2022, 28, 345–354. [Google Scholar] [CrossRef]
- Lucien, M.A.B.; Canarie, M.F.; Kilgore, P.E.; Jean-Denis, G.; Fénélon, N.; Pierre, M.; Cerpa, M.; Joseph, G.A.; Maki, G.; Zervos, M.J.; et al. Antibiotics and antimicrobial resistance in the COVID-19 era: Perspective from resource-limited settings. Int. J. Infect. Dis. 2021, 104, 250–254. [Google Scholar] [CrossRef]
- Truong, W.R.; Hidayat, L.; Bolaris, M.A.; Nguyen, L.; Yamaki, J. The antibiogram: Key considerations for its development and utilization. JAC Antimicrob. Resist. 2021, 3, dlab060. [Google Scholar] [CrossRef]
- Manga, M.; Ibrahim, M.; Isaac, E.; Hassan, M.D.; Muhammad, G.; Hassan, U.M.; Yunusa-Kaltungo, Z. Antibiogram of Pseudomonas species: An important tool to combat antibiotic resistance for patient safety in Gombe, Nigeria. Afr. J. Clin. Exp. Microbiol. 2021, 22, 279–283. [Google Scholar] [CrossRef]
- Klinker, K.P.; Hidayat, L.K.; DeRyke, C.A.; DePestel, D.D.; Motyl, M.; Bauer, K.A. Antimicrobial stewardship and antibiograms: Importance of moving beyond traditional antibiograms. Therap. Adv. Infect. Dis. 2021, 8, 20499361211011373. [Google Scholar] [CrossRef] [PubMed]
- The Burden of Antimicrobial Resistance (AMR) in Romania; University of Washington, Institute for Health Metrics and Evaluation (IHME): Washington, DC, USA, 2022.
- Bourgi, N.; Olaby, A.A.; Najdi, A.; Hatem, G. Predictors of antibiogram performance and antibiotic resistance patterns in the northern Syrian region: A cross-sectional investigation. Explor. Res. Clin. Soc. Pharm. 2024, 13, 1–6. [Google Scholar] [CrossRef] [PubMed]
- David, M.Z.; Daum, R.S. Update on epidemiology and treatment of MRSA infections in children. Curr. Pediatr. Rep. 2013, 1, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.; Lee, H.; Patel, S. Efficacy of carbapenems against Gram-negative bacteria in pediatric patients. Clin. Microbiol. Infect. 2023, 29, 45–53. [Google Scholar]
- Shi, Y.; Wu, H.L.; Wu, Y.H.; Li, S.; Zhang, L.Y.; Xu, S.S.; Huang, H.Y.; Zhang, C.H.; Yu, X.B.; Cai, K.; et al. Safety and clinical efficacy of linezolid in children: A systematic review and meta-analysis. World J. Pediatr. 2023, 19, 129–138. [Google Scholar] [CrossRef]
- Reynolds, D.; Burnham, J.P.; Vazquez Guillamet, C.; McCabe, M.; Yuenger, V.; Betthauser, K.; Micek, S.T.; Kollef, M.H. The threat of multidrug-resistant/extensively drug-resistant Gram-negative respiratory infections: Another pandemic. Eur. Respir. Rev. 2022, 31, 220068. [Google Scholar] [CrossRef]
- Lin, X.; Kück, U. Cephalosporins as key lead generation beta-lactam antibiotics. Appl. Microbiol. Biotechnol. 2022, 106, 8007–8020. [Google Scholar] [CrossRef]
- Gurung, R.R.; Maharjan, P.; Chhetri, G.G. Antibiotic resistance pattern of Staphylococcus aureus with reference to MRSA isolates from pediatric patients. Future Sci. OA 2020, 6, FSO464. [Google Scholar] [CrossRef]
- Williams, S.; Zhang, Q.; Yang, X. Role of carbapenems and linezolid in treating severe pediatric infections. Clin. Infect. Dis. 2023, 76, 789–798. [Google Scholar]
- Kim, J.; Oh, H.; Jung, Y. Resistance levels of lower-generation cephalosporins in pediatric infections. J. Clin. Microbiol. 2022, 60, e003. [Google Scholar]
- Nguyen, T.; Tran, L.; Vo, K. Efficacy of higher-generation cephalosporins against resistant strains. Infect. Drug Resist. 2023, 16, 109–119. [Google Scholar]
- Brown, P.; Green, S.; Evans, R. High resistance rates to oxacillin and ampicillin in pediatric patients. J. Glob. Antimicrob. Resist. 2023, 34, 100–107. [Google Scholar]
- Lee, C.; Kim, S.; Park, J. Effectiveness of linezolid and ertapenem against resistant pediatric strains. Ann. Clin. Microbiol. Antimicrob. 2022, 21, 57. [Google Scholar]
- Garcia, R.; Martinez, J.; Gonzalez, L. Comparative efficacy of linezolid and ertapenem in pediatric infections. Pediatr. Clin. N. Am. 2023, 70, 103–115. [Google Scholar]
- Lyu, S.; Shi, W.; Dong, F.; Xu, B.P.; Liu, G.; Wang, Q.; Yao, K.H.; Yang, Y.H. Serotype distribution and antimicrobial resistance of pediatric Streptococcus pneumoniae isolated from inpatients and outpatients at Beijing Children’s Hospital. Braz. J. Infect. Dis. 2024, 28, 103734. [Google Scholar] [CrossRef]
- Su, Y.; Zhao, L.; Wang, J. Trends in antibiotic susceptibility and clonal distribution of Staphylococcus aureus. J. Pediatr. Infect. Dis. 2023, 18, 145–153. [Google Scholar]
- Wu, X.; Wang, C.; He, L.; Xu, H.; Jing, C.; Chen, Y.; Lin, A.; Deng, J.; Cao, Q.; Deng, H.; et al. Antimicrobial resistance profile of methicillin-resistant Staphylococcus aureus isolates in children reported from the ISPED surveillance of bacterial resistance, 2016-2021. Front. Cell Infect. Microbiol. 2023, 13, 1102779. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, H.; Huang, B.; Hu, X.; Chen, Y.; Zheng, L.; Yang, L.; Deng, J.; Wang, Q. Characterization of resistance genes and plasmids from sick children caused by Salmonella enterica resistance to azithromycin in Shenzhen, China. Front. Cell Infect. Microbiol. 2023, 13, 1116172. [Google Scholar] [CrossRef]
- Markovska, R.; Stankova, P.; Stoeva, T.; Ivanova, D.; Pencheva, D.; Kaneva, R.; Boyanova, L. Fecal carriage and epidemiology of extended-spectrum beta-lactamase/carbapenemases producing Enterobacterales isolates in Bulgarian hospitals. Antibiotics 2021, 10, 747. [Google Scholar] [CrossRef]
- Xiao, R.; Li, Y.; Liu, X.; Ding, Y.; Lai, J.; Li, Y.; Kang, W.; Zou, P.; Wang, J.; Du, Y.; et al. Antibiotic susceptibility of Escherichia coli isolated from neonates admitted to neonatal intensive care units across China from 2015 to 2020. Front. Cell Infect. Microbiol. 2023, 13, 1183736. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Hsu, W.Y.; Chang, T.H. Macrolide-resistant Mycoplasma pneumoniae infections in pediatric community-acquired pneumonia. Emerg. Infect. Dis. 2020, 26, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Weidmann, M.D.; Green, D.A.; Berry, G.J.; Wu, F. Assessing respiratory viral exclusion and affinity interactions through co-infection incidence in a pediatric population during the 2022 resurgence of influenza and RSV. Front. Cell Infect. Microbiol. 2023, 13, 1208235. [Google Scholar] [CrossRef] [PubMed]
- Nahata, M.C. Vancomycin dosage regimens for pediatric patients. J. Pediatr. Pharmacol. Ther. 2009, 14, 64–65. [Google Scholar] [CrossRef] [PubMed]
- Romandini, A.; Pani, A.; Schenardi, P.A.; Pattarino, G.A.C.; De Giacomo, C.; Scaglione, F. Antibiotic Resistance in Pediatric Infections: Global Emerging Threats, Predicting the Near Future. Antibiotics 2021, 10, 393. [Google Scholar] [CrossRef]
- Pani, A.; Comandini, E.; De Socio, G. Challenges in treating pediatric infections with high resistance rates. J. Glob. Antimicrob. Resist. 2023, 35, 50–57. [Google Scholar]
- Schenardi, P.; Pattarino, G.; Falcone, M. Judicious use of antibiotics to combat resistance in pediatrics. Antibiotics 2022, 11, 1783. [Google Scholar]
- De Giacomo, M.; Scaglione, F.; Esposito, S. Strategic antibiotic use and ongoing research for new treatments. Pharmacol. Res. 2023, 187, 106568. [Google Scholar]
- Tian, J.; Li, Y.; Zhang, L. Efficacy of specific antibiotics against resistant pediatric bacteria. Antimicrob. Resist. Infect. Control 2023, 12, 31. [Google Scholar]
- Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of antibiotics and antibiotic resistance, and their impacts on drug development: A narrative review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef]
- Reta, A.; Wubie, M.; Mekuria, G. Nasal colonization and antimicrobial susceptibility pattern of Staphylococcus aureus among pre-school children in Ethiopia. BMC Res. Notes 2017, 10, 746. [Google Scholar] [CrossRef] [PubMed]
- Ansari, S.; Gautam, R.; Shrestha, S.; Ansari, S.R.; Subedi, S.N.; Chhetri, M.R. Risk factors assessment for nasal colonization of Staphylococcus aureus and its methicillin-resistant strains among pre-clinical medical students of Nepal. BMC Res. Notes 2016, 9, 214. [Google Scholar] [CrossRef] [PubMed]
- Fadare, F.T.; Adefisoye, M.A.; Okoh, A.I. Occurrence, identification, and antibiogram signatures of selected Enterobacteriaceae from Tsomo and Tyhume rivers in the Eastern Cape Province, Republic of South Africa. PLoS ONE 2020, 15, e0238084. [Google Scholar] [CrossRef] [PubMed]
- Rotaru, L.T.; Istratoaie, O.; Udrescu, L.; Varut, R.M. TLC, GC-MS, HPLC Analyses and Testing the Antibacterial Effect of Tragopogon pratensis and Vaccinium myrtillus. Rev. Chim. 2018, 69, 1939–1943. [Google Scholar] [CrossRef]
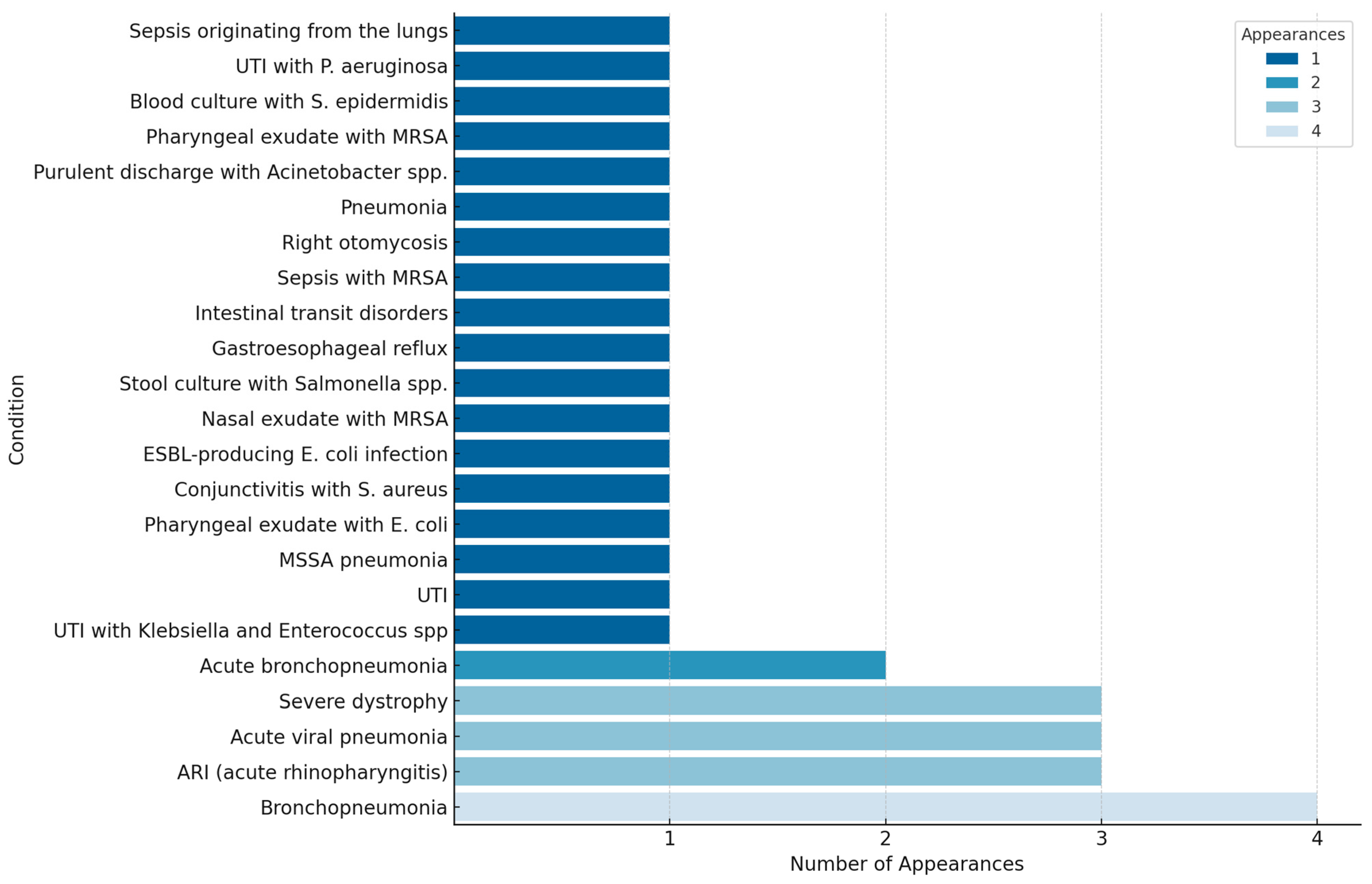
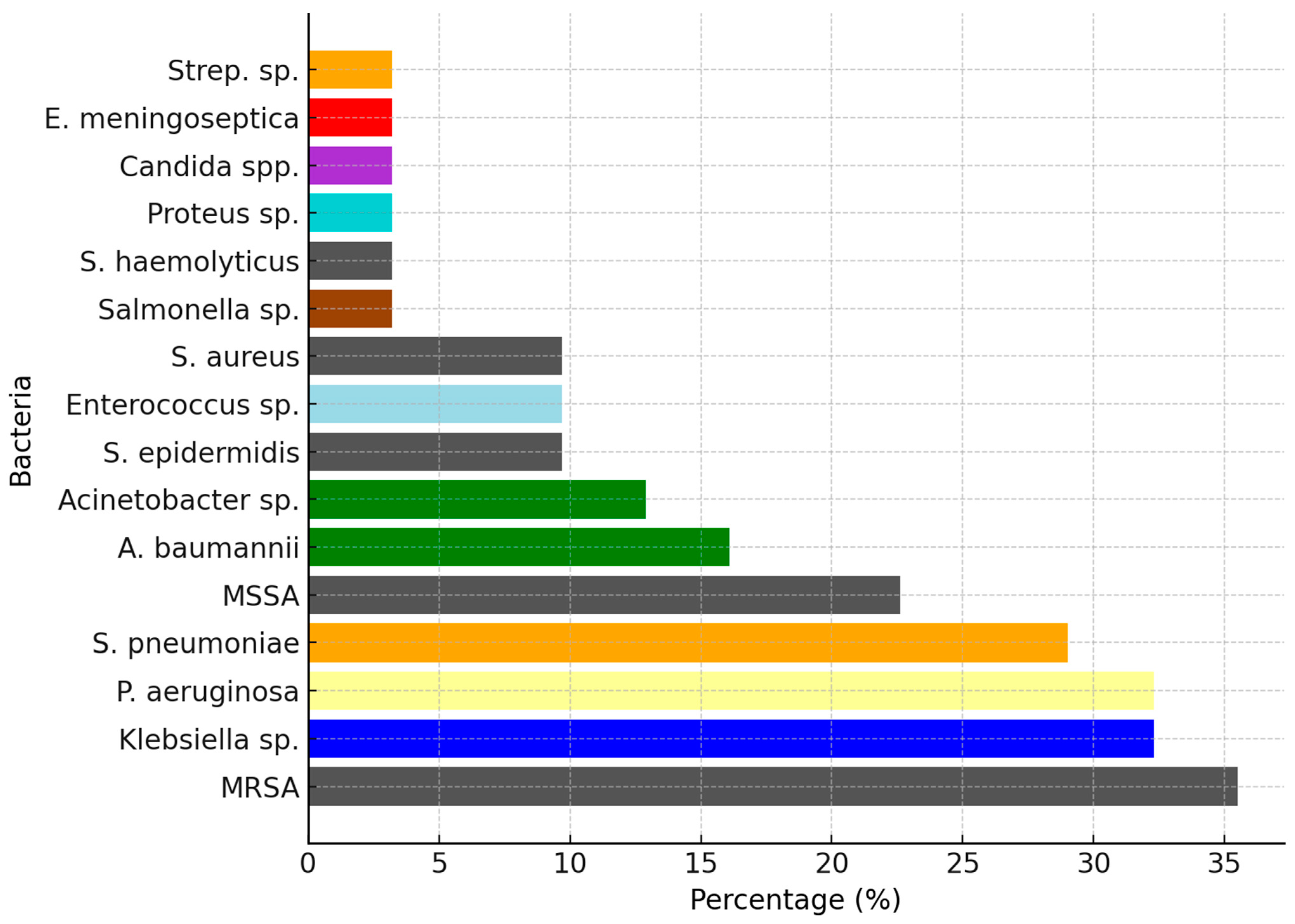
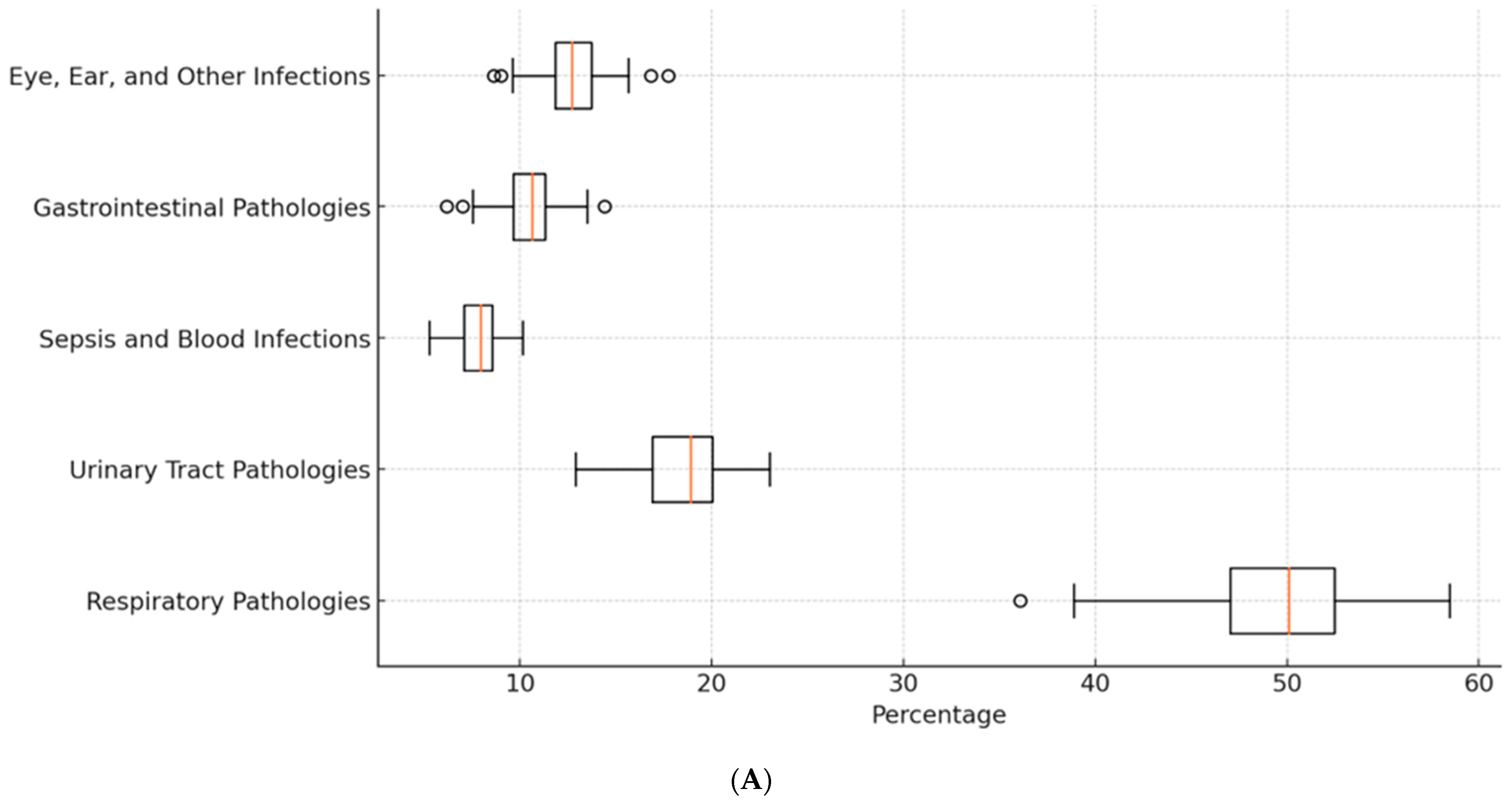
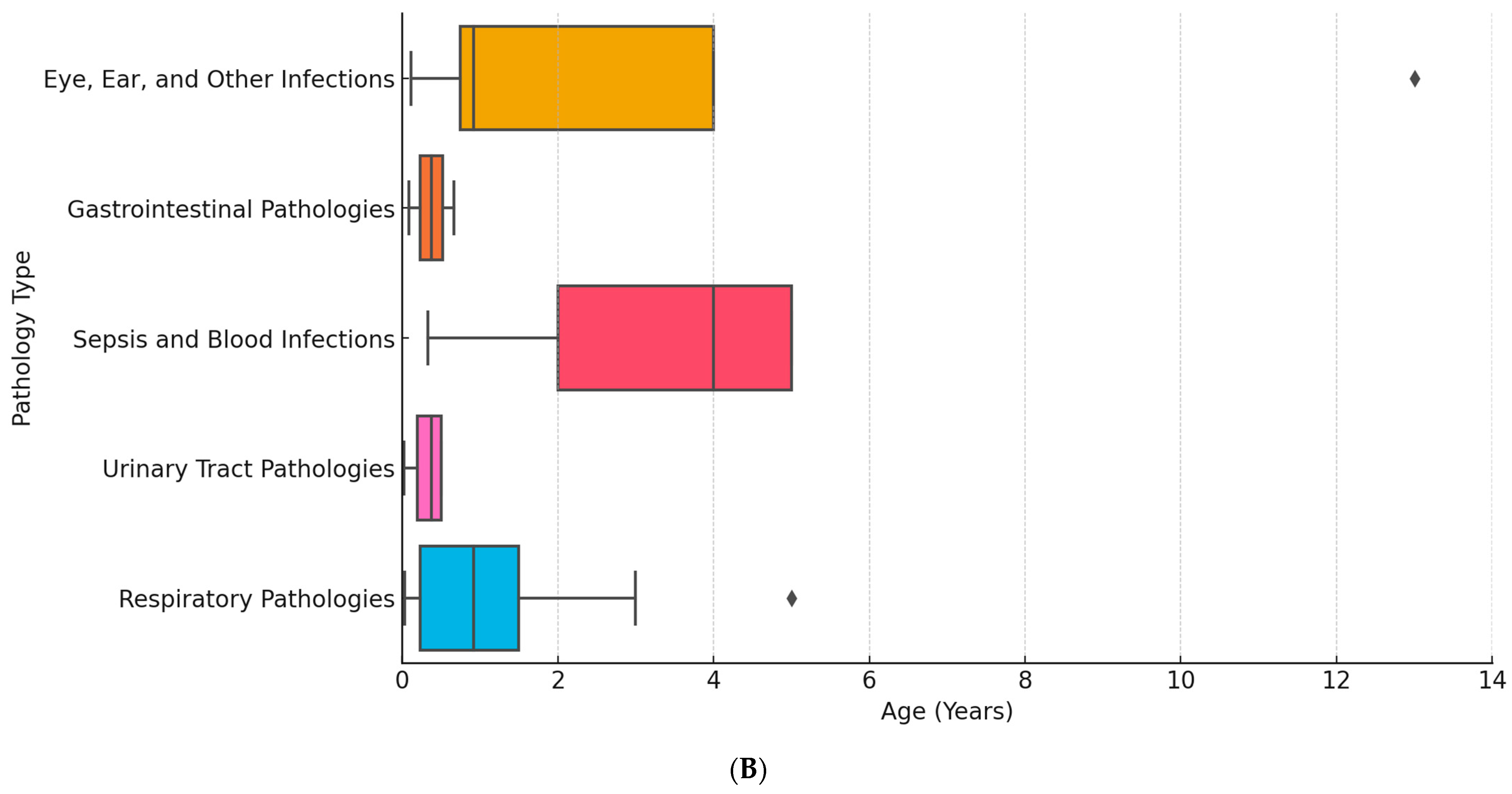

| Age | Diagnosis | Pathogens |
|---|---|---|
| 2 M | Bronchopneumonia, ARI | E. coli, S. pneumoniae, P. aeruginosa |
| 12 D | Bronchopneumonia, ARI, acute rhinopharyngitis | MSSA, S. pneumoniae, Streptococcus sp. |
| 1 Y | Acute viral pneumonia, ARI, severe dystrophy | A. baumanii, MRSA, P. aeruginosa, E. meningoseptica |
| 2 M | Acute bronchopneumonia, ARI | E. coli, S. pneumoniae |
| 3 M | UTI, acute diarrheal illness | Klebisiella, MRSA |
| 5 M | MSSA pneumonia, pharyngeal exudate with E. coli | MSSA, E. coli |
| 12 D | Pneumonia, conjunctivitis with S. aureus | Acinetobacter spp., S. aureus |
| 2 M | Acute bronchopneumonia | S. pneumoniae, MSSA |
| 6 M | ESBL-producing E. coli infection | E. coli |
| 8 D | UTI with Klebsiella and Enterococcus spp. | Klebsiella, Enterococcus spp. |
| 4 M | Sepsis originating from the lungs, bronchopneumonia, ARI | Enteroccocus spp., A. baumanni, S. aureus |
| 8 M | Gastroesophageal reflux, intestinal transit disorders | Klebsiella spp., E. coli, S. pneumoniae, Candida spp. |
| 1 Y | Bronchopneumonia, ARI, sepsis with MRSA | S. pneumoniae, P. aeruginosa, MRSA |
| 9 M | Gastroesophageal reflux, right otomycosis | S. pneumoniae, P. aeruginosa, A. baumanni |
| 11 M | Bullous impetigo, acute conjunctivitis, diarrhea | Enteroccocus spp., Klebsiella spp., S. aureus |
| 18 M | Acute pneumonia, acute rhinopharyngitis | MRSA, Staphilococcus epiderdimitis, Staphilococcus haemoliticus, P. aeruginosa |
| 10 M | Acute pneumonia, diarrhea, Ebstein’s anomaly | MRSA, P. aeruginosa |
| 4 Y | Sepsis with MRSA, anemia, secondary dyspepsia, UTI | MRSA, Acinetobacter spp., Klebsiella |
| 3 Y | Acute pneumonia, unspecified sepsis | Preoteus spp., Klebsiella |
| 2 Y | Sepsis with S. aureus, bronchopneumonia | S. pneumonie, MRSA, P. aeruginosa |
| 5 Y | Acute lymphoblastic leukemia, conjunctivitis, sepsis | Acinetobacter baumanii, Acinetobacter spp. |
| 6 W | Pharyngitis with MSSA | MSSA, E. coli, P. aeruginosa |
| 5 Y | Pneumonia | MRSA, S. pneumonie |
| 13 Y | Purulent discharge with Acinetobacter spp. | Acinetobactr spp. |
| 4 Y | Pharyngeal exudate with MRSA | MRSA, Staphylococcus epiderdimitis |
| 5 Y | Blood culture with S. epidermidis | MSSA, Staphylococcus epiderdimitis |
| 6 M | UTI with P. aeruginosa | Klebsiella spp., P. aeruginosa |
| 1 M | Stool culture with Salmonella spp., pharyngeal exudate with MRSA, nasal exudate with MRSA | Salmonella spp., MRSA, MSSA |
| 18 M | Bronchopneumonia, ARI | E. coli, P. Aerugonisa, MSSA, Klebsiella spp. |
| 2 Y | Acute pneumonia, UTI | Klebsiella, MRSA |
| 6 M | Acute pneumonia | Acinetobacter baumanii, Klebsiella |
| Patient nr. | Age | Diagnosis | Pathogens | PN | C | CB | M | T | A | F | S | O | P | N | G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. | 2M | Bronchopneumonia, ARI | E. coli, S. pneumoniae, P. aeruginosa | H | H | H | H | R | H | H | R | H | H | / | H |
| 2. | 12 D | Bronchopneumonia, ARI, Acute Rhinopharyngitis | MSSA, S. pneumoniae, Streptococcus sp. | H | H | H | H | R | I | I | R | H | / | / | H |
| 3. | 1 Y | Acute Viral Pneumonia, ARI, Severe Dystrophy | Acinetobacter baumanii, MRSA, P. aeruginosa, Elizabethkingia meningoseptica * | I | I | H | H | I | H | H | H | H | H | / | / |
| 4. | 2 M | Acute bronchopneumonia, ARI | E. coli, S. pneumoniae | I | H | H | H | H | H | H | R | H | H | H | H |
| 5. | 3 M | UTI, acute diarrheal illness | Klebisiella, MRSA | R | R | I | R | H | I | H | R | H | / | R | / |
| 6. | 5 M | MSSA pneumonia, Pharyngeal exudate with E. coli | MSSA, E. coli | H | H | H | R | H | H | / | I | H | / | H | / |
| 7. | 12 D | Pneumonia, Conjunctivitis with S. aureus | Acinetobacter spp., S. aureus | I | R | R | H | H | H | H | H | / | H | / | / |
| 8. | 2M | Acute bronchopneumonia | S. pneumoniae, MSSA | H | H | H | I | H | H | R | R | H | / | / | H |
| 9. | 6 M | ESBL-producing E. coli infection | E. coli | R | R | H | / | H | R | / | H | / | H | / | / |
| 10. | 8 D | UTI with Klebsiella and Enterococcus spp. | Klebsiella spp., Enterococcus spp. | R | H | H | / | H | H | I | H | H | H | R | H |
| 11. | 4 M | Sepsis originating from the lungs, bronchopneumonia, ARI | Enteroccocus spp., Acinetobacter baumanni, S. aureus | R | R | R | R | I | I | I | H | H | I | I | / |
| 12. | 8M | Gastroesophageal reflux, intestinal transit disorders. | Klebsiella spp., E. coli, S. pneumoniae, Candida spp. * | H | H | H | H | R | H | H | H | H | / | H | H |
| 13. | 1Y | Bronchopneumonia, ARI, sepsis with MRSA | S. pneumoniae, P. aeruginosa, MRSA | I | / | R | R | R | I | H | I | H | / | / | H |
| 14. | 9 M | Gastroesophageal reflux, right otomycosis | S. pneumoniae, P. aeruginosa, Acinetobacter baumanni | I | H | I | R | R | I | I | R | H | H | / | H |
| 15. | 11 M | Bullous impetigo, acute conjunctivitis, diarrhea | Enteroccocus spp., Klebsiella spp., S. aureus | H | H | H | H | I | H | H | H | H | H | / | / |
| 16. | 10 M | Acute pneumonia, diarrhea, Ebstein’s anomaly | MRSA, P. aeruginosa | I | H | R | R | R | I | H | R | H | H | / | / |
| 17. | 4Y | Sepsis with MRSA, anemia, secondary dyspepsia, UTI | MRSA, Acinetobacter spp., Klebsiella spp. | R | I | I | R | R | R | R | I | H | H | / | H |
| 18. | 3Y | Acute pneumonia, Unspecified sepsis | Preoteus spp., Klebsiella | I | I | H | / | I | R | / | H | / | I | / | / |
| 19. | 2Y | Sepsis with Staphylococcus aureus, bronchopneumonia | S. pneumonie, MRSA, P. aeruginosa | I | / | H | R | R | I | H | / | / | / | / | H |
| 20. | 5Y | Acute lymphoblastic leukemia, conjunctivitis, sepsis | Acinetobacter baumanii, Acinetobacter spp. | I | R | R | / | R | I | / | H | / | H | / | / |
| 21. | 6 W | Pharyngitis with MSSA | MSSA, E. coli, P. aeruginosa | I | I | H | R | H | H | H | H | H | H | / | / |
| 22. | 5 Y | Pnemunonia | MRSA, S. pneumonie | I | H | H | I | H | I | H | I | H | / | / | H |
| 23. | 13 Y | Purulent discharge with Acinetobacter spp. | Acinetobacter spp. | I | I | R | / | / | I | / | H | / | H | / | / |
| 24. | 4 Y | Pharyngeal exudate with MRSA | MRSA, Staphylococcus epiderdimitis | I | / | / | R | / | I | H | H | I | / | / | H |
| 25. | 5 Y | Blood culture with S. epidermidis | MSSA, Staphylococcus epiderdimitis | I | / | / | H | H | H | H | H | H | / | H | H |
| 26. | 6 M | UTI with P. aeruginosa | Klebsiella spp., P. aeruginosa | I | R | H | / | R | H | I | R | / | / | / | / |
| 27. | 1 M | Stool culture with Salmonella spp., Pharyngeal exudate with MRSA, Nasal exudate with MRSA | Salmonella spp., MRSA, MSSA | I | H | H | H | H | H | H | I | H | H | / | H |
| 28. | 18M | Bronchopneumonia, ARI | E. coli, P. Aerugonisa, MSSA, Klebsiella spp. | I | H | I | H | H | H | H | H | H | H | / | / |
| 29. | 2Y | Acute pneumonia, UTI | Klebsiella spp., MRSA | R | H | H | R | I | H | / | H | H | / | H | / |
| 30. | 6 M | Acute pneumonia | Acinetobacter baumanii, Klebsiella spp. | I | H | I | / | H | I | I | I | / | H | / | / |
| 31. | 18 M | Acute pneumonia, Acute Rhinopharyngitis | MRSA, Staph. Epiderdimitis, Staph. Haemoliticus, P. aeruginosa | I | H | H | I | I | H | H | I | H | I | / | H |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singer, M.M.; Văruț, R.M.; Popescu, C.; Radivojevic, K.; Rotaru, L.T.; Octavian, D.R.; Mihai-Covei, B.; Popescu, M.; Irina, O.A.; Oancea, D.; et al. Assessment of Antibiotic Resistance in Pediatric Infections: A Romanian Case Study on Pathogen Prevalence and Effective Treatments. Antibiotics 2024, 13, 879. https://doi.org/10.3390/antibiotics13090879
Singer MM, Văruț RM, Popescu C, Radivojevic K, Rotaru LT, Octavian DR, Mihai-Covei B, Popescu M, Irina OA, Oancea D, et al. Assessment of Antibiotic Resistance in Pediatric Infections: A Romanian Case Study on Pathogen Prevalence and Effective Treatments. Antibiotics. 2024; 13(9):879. https://doi.org/10.3390/antibiotics13090879
Chicago/Turabian StyleSinger, Maria Madalina, Renata Maria Văruț, Cristina Popescu, Kristina Radivojevic, Luciana Teodora Rotaru, Damian Roni Octavian, Banicioiu Mihai-Covei, Mihaela Popescu, Oancea Andreea Irina, Dragos Oancea, and et al. 2024. "Assessment of Antibiotic Resistance in Pediatric Infections: A Romanian Case Study on Pathogen Prevalence and Effective Treatments" Antibiotics 13, no. 9: 879. https://doi.org/10.3390/antibiotics13090879
APA StyleSinger, M. M., Văruț, R. M., Popescu, C., Radivojevic, K., Rotaru, L. T., Octavian, D. R., Mihai-Covei, B., Popescu, M., Irina, O. A., Oancea, D., Popescu, A. I. S., & Singer, C. E. (2024). Assessment of Antibiotic Resistance in Pediatric Infections: A Romanian Case Study on Pathogen Prevalence and Effective Treatments. Antibiotics, 13(9), 879. https://doi.org/10.3390/antibiotics13090879







