Antimicrobial and Phylogenomic Characterization of Bacillus cereus Group Strains Isolated from Different Food Sources in Italy
Abstract
1. Introduction
2. Results
2.1. Isolation of B. cereus Group Strains
2.2. MALDI-TOF Mass Spectrometry (MS) Analysis
2.3. WGS Analysis
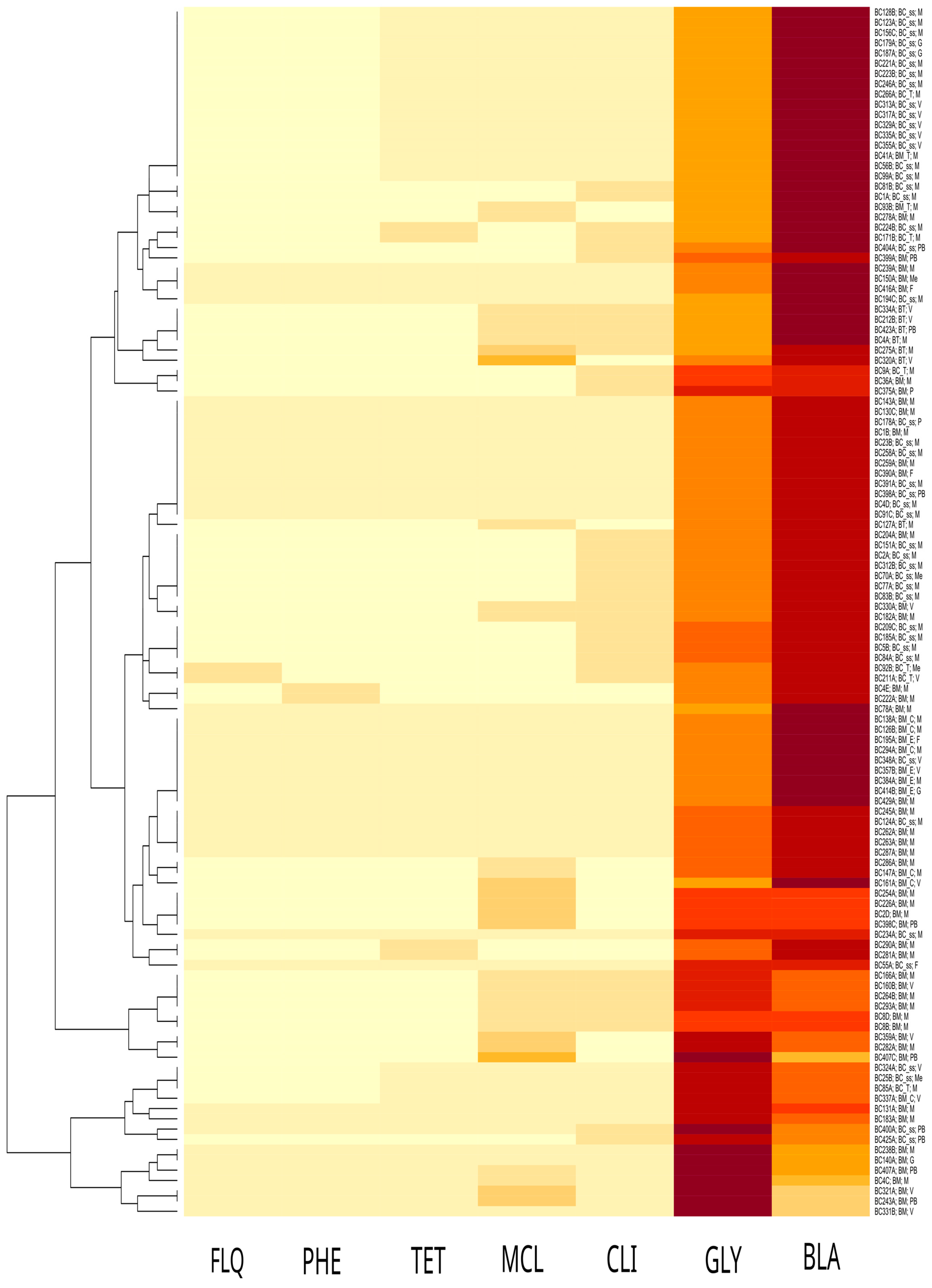
2.4. Antibiotic Susceptibility Analysis
3. Discussion
4. Materials and Methods
4.1. B. cereus Group Isolated Strains
4.2. MALDI-TOF Mass Spectrometry Analysis
4.3. WGS Sequencing and Bioinformatic Analyses
4.4. Antimicrobial Susceptibility Tests
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Drobniewski, F.A. Bacillus cereus and Related Species. Clin. Microbiol. Rev. 1993, 6, 324–338. [Google Scholar] [CrossRef]
- Ehling-Schulz, M.; Koehler, T.M.; Lereclus, D. The Bacillus cereus Group: Bacillus Species with Pathogenic Potential. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- El-Arabi, T.F.; Griffiths, M.W. Foodborne Infections and Intoxications: Chapter 29. Bacillus Cereus, 4th ed.; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Tirloni, E.; Stella, S.; Celandroni, F.; Mazzantini, D.; Bernardi, C.; Ghelardi, E. Bacillus cereus in Dairy Products and Production Plants. Foods 2022, 11, 2572. [Google Scholar] [CrossRef]
- Abee, T.; Groot, M.N.; Tempelaars, M.; Zwietering, M.; Moezelaar, R.; van der Voort, M. Germination and Outgrowth of Spores of Bacillus cereus Group Members: Diversity and Role of Germinant Receptors. Food Microbiol. 2011, 28, 199–208. [Google Scholar] [CrossRef]
- Logan, N.A. Bacillus and Relatives in Foodborne Illness. J. Appl. Microbiol. 2012, 112, 417–429. [Google Scholar] [CrossRef]
- Majed, R.; Faille, C.; Kallassy, M.; Gohar, M. Bacillus cereus Biofilms—Same, Only Different. Front. Microbiol. 2016, 7, 1054. [Google Scholar] [CrossRef]
- Ceuppens, S.; Boon, N.; Uyttendaele, M. Diversity of Bacillus cereus Group Strains Is Reflected in Their Broad Range of Pathogenicity and Diverse Ecological Lifestyles. FEMS Microbiol. Ecol. 2013, 84, 433–450. [Google Scholar] [CrossRef]
- Carroll, L.M.; Wiedmann, M.; Kovac, J. Proposal of a Taxonomic Nomenclature for the Bacillus cereus Group Which Reconciles Genomic Definitions of Bacterial Species with Clinical and Industrial Phenotypes. MBio 2020, 11. [Google Scholar] [CrossRef]
- Senesi, S.; Ghelardi, E. Production, Secretion and Biological Activity of Bacillus cereus Enterotoxins. Toxins 2010, 2, 1690–1703. [Google Scholar] [CrossRef]
- Rasko, D.A.; Altherr, M.R.; Han, C.S.; Ravel, J. Genomics of the Bacillus cereus Group of Organisms. FEMS Microbiol. Rev. 2005, 29, 303–329. [Google Scholar] [CrossRef]
- Bottone, E.J. Bacillus cereus, a Volatile Human Pathogen. Clin. Microbiol. Rev. 2010, 23, 382–398. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, R.; Jessberger, N.; Ehling-Schulz, M.; Märtlbauer, E.; Granum, P.E. The Food Poisoning Toxins of Bacillus cereus. Toxins 2021, 13, 98. [Google Scholar] [CrossRef]
- Rahnama, H.; Azari, R.; Yousefi, M.H.; Berizi, E.; Mazloomi, S.M.; Hosseinzadeh, S.; Derakhshan, Z.; Ferrante, M.; Conti, G.O. A Systematic Review and Meta-Analysis of the Prevalence of Bacillus cereus in Foods. Food Control 2023, 143, 109250. [Google Scholar] [CrossRef]
- Jovanovic, J.; Ornelis, V.F.M.; Madder, A.; Rajkovic, A. Bacillus cereus Food Intoxication and Toxicoinfection. Compr. Rev. Food Sci. Food Saf. 2021, 20, 3719–3761. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Biological Hazards (BIOHAZ). Risks for Public Health Related to the Presence of Bacillus cereus and Other Bacillus Spp. Including Bacillus thuringiensis in Foodstuffs. EFSA J. 2016, 14, e04524. [Google Scholar] [CrossRef]
- Shawish, R.; Tarabees, R. Prevalence and Antimicrobial Resistance of Bacillus cereus Isolated from Beef Products in Egypt. Open Vet. J. 2017, 7, 337–341. [Google Scholar] [CrossRef]
- Granum, P.E.; Lund, T. Bacillus cereus and Its Food Poisoning Toxins. FEMS Microbiol. Lett. 1997, 157, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The Antibiotic Resistance Crisis: Part 1: Causes and Threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Cox, G.; Wright, G.D. Intrinsic Antibiotic Resistance: Mechanisms, Origins, Challenges and Solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef]
- Giedraitienė, A.; Vitkauskienė, A.; Naginienė, R.; Pavilonis, A. Antibiotic Resistance Mechanisms of Clinically Important Bacteria. Medicina 2011, 47, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Sornchuer, P.; Saninjuk, K.; Amonyingcharoen, S.; Ruangtong, J.; Thongsepee, N.; Martviset, P.; Chantree, P.; Sangpairoj, K. Whole Genome Sequencing Reveals Antimicrobial Resistance and Virulence Genes of Both Pathogenic and Non-Pathogenic B. cereus Group Isolates from Foodstuffs in Thailand. Antibiotics 2024, 13, 245. [Google Scholar] [CrossRef] [PubMed]
- Bianco, A.; Capozzi, L.; Monno, M.R.; Del Sambro, L.; Manzulli, V.; Pesole, G.; Loconsole, D.; Parisi, A. Characterization of Bacillus cereus Group Isolates From Human Bacteremia by Whole-Genome Sequencing. Front. Microbiol. 2020, 11, 599524. [Google Scholar] [CrossRef] [PubMed]
- Bartoszewicz, M.; Czyżewska, U. Comparison of the Antibiotic Resistance between Genetically Diverse and Toxigenic Bacillus cereus Sensu Lato from Milk, Pepper and Natural Habitats. J. Appl. Microbiol. 2021, 130, 370–381. [Google Scholar] [CrossRef]
- Kotiranta, A.; Lounatmaa, K.; Haapasalo, M. Epidemiology and Pathogenesis of Bacillus cereus Infections. Microbes Infect. 2000, 2, 189–198. [Google Scholar] [CrossRef]
- Parulekar, R.S.; Sonawane, K.D. Insights into the Antibiotic Resistance and Inhibition Mechanism of Aminoglycoside Phosphotransferase from Bacillus cereus: In Silico and in Vitro Perspective. J. Cell. Biochem. 2018, 119, 9444–9461. [Google Scholar] [CrossRef]
- Guinebretière, M.-H.; Thompson, F.L.; Sorokin, A.; Normand, P.; Dawyndt, P.; Ehling-Schulz, M.; Svensson, B.; Sanchis, V.; Nguyen-The, C.; Heyndrickx, M.; et al. Ecological Diversification in the Bacillus cereus Group. Environ. Microbiol. 2008, 10, 851–865. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of In-Frequently Isolated or Fastidious Bacteria. CLSI Guideline M45, 3rd ed.; CLSI: Wayne, PA, USA, 2016. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100, 27th ed.; CLSI: Wayne, PA, USA, 2017. [Google Scholar]
- Ghelardi, E.; Celandroni, F.; Salvetti, S.; Fiscarelli, E.; Senesi, S. Bacillus thuringiensis Pulmonary Infection: Critical Role for Bacterial Membrane-Damaging Toxins and Host Neutrophils. Microbes Infect. 2007, 9, 591–598. [Google Scholar] [CrossRef]
- Roca, A.; Cabeo, M.; Enguidanos, C.; Martínez-Checa, F.; Sampedro, I.; Llamas, I. Potential of the Quorum-quenching and Plant-growth Promoting Halotolerant Bacillus toyonensis AA1EC1 as Biocontrol Agent. Microb. Biotechnol. 2024, 17, e14420. [Google Scholar] [CrossRef]
- Andriūnaitė, E.; Tamošiūnė, I.; Aleksandravičiūtė, M.; Gelvonauskienė, D.; Vinskienė, J.; Rugienius, R.; Baniulis, D. Stimulation of Nicotiana Tabacum L. In Vitro Shoot Growth by Endophytic Bacillus cereus Group Bacteria. Microorganisms 2021, 9, 1893. [Google Scholar] [CrossRef]
- Hollensteiner, J.; Wemheuer, F.; Harting, R.; Kolarzyk, A.M.; Diaz Valerio, S.M.; Poehlein, A.; Brzuszkiewicz, E.B.; Nesemann, K.; Braus-Stromeyer, S.A.; Braus, G.H.; et al. Bacillus thuringiensis and Bacillus weihenstephanensis Inhibit the Growth of Phytopathogenic Verticillium Species. Front. Microbiol. 2017, 7, 2171. [Google Scholar] [CrossRef] [PubMed]
- Celandroni, F.; Salvetti, S.; Senesi, S.; Ghelardi, E. Bacillus thuringiensis Membrane-Damaging Toxins Acting on Mammalian Cells. FEMS Microbiol. Lett. 2014, 361, 95–103. [Google Scholar] [CrossRef]
- Guérin, A.; Rønning, H.T.; Dargaignaratz, C.; Clavel, T.; Broussolle, V.; Mahillon, J.; Granum, P.E.; Nguyen-The, C. Cereulide Production by Bacillus weihenstephanensis Strains during Growth at Different pH Values and Temperatures. Food Microbiol. 2017, 65, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.A.; Beno, S.M.; Kent, D.J.; Carroll, L.M.; Martin, N.H.; Boor, K.J.; Kovac, J. Bacillus Wiedmannii Sp. Nov., a Psychrotolerant and Cytotoxic Bacillus cereus Group Species Isolated from Dairy Foods and Dairy Environments. Int. J. Syst. Evol. Microbiol. 2016, 66, 4744–4753. [Google Scholar] [CrossRef] [PubMed]
- Hinnekens, P.; Fayad, N.; Gillis, A.; Mahillon, J. Conjugation across Bacillus cereus and Kin: A Review. Front. Microbiol. 2022, 13, 1034440. [Google Scholar] [CrossRef]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J.; Dou, T. Antibiotic Resistance Genes in Bacteria: Occurrence, Spread, and Control. J. Basic Microbiol. 2021, 61, 1049–1070. [Google Scholar] [CrossRef]
- Zhai, Z.; Cui, C.; Li, X.; Yan, J.; Sun, E.; Wang, C.; Guo, H.; Hao, Y. Prevalence, Antimicrobial Susceptibility, and Antibiotic Resistance Gene Transfer of Bacillus Strains Isolated from Pasteurized Milk. J. Dairy Sci. 2023, 106, 75–83. [Google Scholar] [CrossRef]
- Mills, E.; Sullivan, E.; Kovac, J. Comparative Analysis of Bacillus cereus Group Isolates’ Resistance Using Disk Diffusion and Broth Microdilution and the Correlation between Antimicrobial Resistance Phenotypes and Genotypes. Appl. Environ. Microbiol. 2022, 88, e0230221. [Google Scholar] [CrossRef]
- Ikeda, M.; Yagihara, Y.; Tatsuno, K.; Okazaki, M.; Okugawa, S.; Moriya, K. Clinical Characteristics and Antimicrobial Susceptibility of Bacillus cereus Blood Stream Infections. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 43. [Google Scholar] [CrossRef]
- Turnbull, P.C.B.; Sirianni, N.M.; LeBron, C.I.; Samaan, M.N.; Sutton, F.N.; Reyes, A.E.; Peruski, L.F. MICs of Selected Antibiotics for Bacillus anthracis, Bacillus cereus, Bacillus thuringiensis, and Bacillus mycoides from a Range of Clinical and Environmental Sources as Determined by the Etest. J. Clin. Microbiol. 2004, 42, 3626–3634. [Google Scholar] [CrossRef]
- Fiedler, G.; Schneider, C.; Igbinosa, E.O.; Kabisch, J.; Brinks, E.; Becker, B.; Stoll, D.A.; Cho, G.-S.; Huch, M.; Franz, C.M.A.P. Antibiotics Resistance and Toxin Profiles of Bacillus cereus-Group Isolates from Fresh Vegetables from German Retail Markets. BMC Microbiol. 2019, 19, 250. [Google Scholar] [CrossRef] [PubMed]
- Yim, J.-H.; Kim, K.-Y.; Chon, J.-W.; Kim, D.-H.; Kim, H.-S.; Choi, D.-S.; Choi, I.-S.; Seo, K.-H. Incidence, Antibiotic Susceptibility, and Toxin Profiles of Bacillus cereus Sensu Lato Isolated from Korean Fermented Soybean Products. J. Food Sci. 2015, 80, M1266–M1270. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, G.; Russo, G.; Bonfiglio, G. Recent Developments in Carbapenems. Expert Opin. Investig. Drugs 2002, 11, 529–544. [Google Scholar] [CrossRef] [PubMed]
- Materon, I.C.; Queenan, A.M.; Koehler, T.M.; Bush, K.; Palzkill, T. Biochemical Characterization of Beta-Lactamases bla1 and bla2 from Bacillus anthracis. Antimicrob. Agents Chemother. 2003, 47, 2040–2042. [Google Scholar] [CrossRef]
- Wagner, T.M.; Howden, B.P.; Sundsfjord, A.; Hegstad, K. Transiently Silent Acquired Antimicrobial Resistance: An Emerging Challenge in Susceptibility Testing. J. Antimicrob. Chemother. 2023, 78, 586–598. [Google Scholar] [CrossRef]
- Deekshit, V.K.; Srikumar, S. ‘To Be, or Not to Be’-The Dilemma of “silent” Antimicrobial Resistance Genes in Bacteria. J. Appl. Microbiol. 2022, 133, 2902–2914. [Google Scholar] [CrossRef]
- Stasiak, M.; Maćkiw, E.; Kowalska, J.; Kucharek, K.; Postupolski, J. Silent Genes: Antimicrobial Resistance and Antibiotic Production. Pol. J. Microbiol. 2021, 70, 421–429. [Google Scholar] [CrossRef]
- Bravo, A.; Moreno-Blanco, A.; Espinosa, M. One Earth: The Equilibrium between the Human and the Bacterial Worlds. Int. J. Mol. Sci. 2023, 24, 15047. [Google Scholar] [CrossRef]
- Enne, V.I.; Delsol, A.A.; Roe, J.M.; Bennett, P.M. Evidence of Antibiotic Resistance Gene Silencing in Escherichia Coli. Antimicrob. Agents Chemother. 2006, 50, 3003–3010. [Google Scholar] [CrossRef] [PubMed]
- Deekshit, V.K.; Kumar, B.K.; Rai, P.; Srikumar, S.; Karunasagar, I.; Karunasagar, I. Detection of Class 1 Integrons in Salmonella Weltevreden and Silent Antibiotic Resistance Genes in Some Seafood-Associated Nontyphoidal Isolates of Salmonella in South-West Coast of India. J. Appl. Microbiol. 2012, 112, 1113–1122. [Google Scholar] [CrossRef]
- Kime, L.; Randall, C.P.; Banda, F.I.; Coll, F.; Wright, J.; Richardson, J.; Empel, J.; Parkhill, J.; O’Neill, A.J. Transient Silencing of Antibiotic Resistance by Mutation Represents a Significant Potential Source of Unanticipated Therapeutic Failure. MBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Manzulli, V.; Rondinone, V.; Buchicchio, A.; Serrecchia, L.; Cipolletta, D.; Fasanella, A.; Parisi, A.; Difato, L.; Iatarola, M.; Aceti, A.; et al. Discrimination of Bacillus cereus Group Members by MALDI-TOF Mass Spectrometry. Microorganisms 2021, 9, 1202. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Carroll, L.M.; Kovac, J.; Miller, R.A.; Wiedmann, M. Rapid, High-Throughput Identification of Anthrax-Causing and Emetic Bacillus cereus Group Genome Assemblies via BTyper, a Computational Tool for Virulence-Based Classification of Bacillus cereus Group Isolates by Using Nucleotide Sequencing Data. Appl. Environ. Microbiol. 2017, 83, e01096-17. [Google Scholar] [CrossRef]
- Carroll, L.M.; Cheng, R.A.; Kovac, J. No Assembly Required: Using BTyper3 to Assess the Congruency of a Proposed Taxonomic Framework for the Bacillus cereus Group With Historical Typing Methods. Front. Microbiol. 2020, 11, 580691. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a New Bioinformatic Tool to Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.-H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Florensa, A.F.; Kaas, R.S.; Clausen, P.T.L.C.; Aytan-Aktug, D.; Aarestrup, F.M. ResFinder—An Open Online Resource for Identification of Antimicrobial Resistance Genes in next-Generation Sequencing Data and Prediction of Phenotypes from Genotypes. Microb. Genom. 2022, 8, 000748. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Tourasse, N.J.; Jolley, K.A.; Kolstø, A.-B.; Økstad, O.A. Core Genome Multilocus Sequence Typing Scheme for Bacillus cereus Group Bacteria. Res. Microbiol. 2023, 174, 104050. [Google Scholar] [CrossRef] [PubMed]
- Glasset, B.; Herbin, S.; Guillier, L.; Cadel-Six, S.; Vignaud, M.-L.; Grout, J.; Pairaud, S.; Michel, V.; Hennekinne, J.-A.; Ramarao, N.; et al. Bacillus cereus-Induced Food-Borne Outbreaks in France, 2007 to 2014: Epidemiology and Genetic Characterisation. Eurosurveillance 2016, 21, 30413. [Google Scholar] [CrossRef] [PubMed]

| Drug Class | Antibiotic | MIC Breakpoints (µg/mL) ** | MIC Value Results (%) | ||||
|---|---|---|---|---|---|---|---|
| S * (≤) | I * | R * (≥) | S * | I * | R * | ||
| Beta-lactam | Penicillin G | 0.12 | na *** | 0.25 | na | na | 100 |
| Beta-lactam | Meropenem | 4 | 8 | 16 | 99.1 | 0.9 | na |
| Aminoglycoside | Gentamicin | 4 | 8 | 16 | 100 | na | na |
| Tetracycline | Doxycycline | 4 | 8 | 16 | 100 | na | na |
| Tetracycline | Tetracycline | 4 | 8 | 16 | 97.5 | 2.5 | na |
| Fluoroquinolone | Ciprofloxacin | 1 | 2 | 4 | 100 | na | na |
| Rifamycin | Rifampicin | 1 | 2 | 4 | 100 | na | na |
| Amphenicol | Chloramphenicol | 8 | 16 | 32 | 100 | na | na |
| Macrolide | Erythromycin | 0.5 | 1–4 | 8 | 93.2 | 6.8 | na |
| Lincosamide | Clindamycin | 0.5 | 1–2 | 4 | 80.5 | 19.5 | na |
| Glycopeptide | Vancomycin | 4 | na | na | 100 | na | na |
| Oxazolidinone | Linezolid | 4 | na | 8 | 100 | na | na |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farina, D.; Bianco, A.; Manzulli, V.; Castellana, S.; Parisi, A.; Caruso, M.; Fraccalvieri, R.; Serrecchia, L.; Rondinone, V.; Pace, L.; et al. Antimicrobial and Phylogenomic Characterization of Bacillus cereus Group Strains Isolated from Different Food Sources in Italy. Antibiotics 2024, 13, 898. https://doi.org/10.3390/antibiotics13090898
Farina D, Bianco A, Manzulli V, Castellana S, Parisi A, Caruso M, Fraccalvieri R, Serrecchia L, Rondinone V, Pace L, et al. Antimicrobial and Phylogenomic Characterization of Bacillus cereus Group Strains Isolated from Different Food Sources in Italy. Antibiotics. 2024; 13(9):898. https://doi.org/10.3390/antibiotics13090898
Chicago/Turabian StyleFarina, Donatella, Angelica Bianco, Viviana Manzulli, Stefano Castellana, Antonio Parisi, Marta Caruso, Rosa Fraccalvieri, Luigina Serrecchia, Valeria Rondinone, Lorenzo Pace, and et al. 2024. "Antimicrobial and Phylogenomic Characterization of Bacillus cereus Group Strains Isolated from Different Food Sources in Italy" Antibiotics 13, no. 9: 898. https://doi.org/10.3390/antibiotics13090898
APA StyleFarina, D., Bianco, A., Manzulli, V., Castellana, S., Parisi, A., Caruso, M., Fraccalvieri, R., Serrecchia, L., Rondinone, V., Pace, L., Fasanella, A., Vetritto, V., Difato, L. M., Cipolletta, D., Iatarola, M., & Galante, D. (2024). Antimicrobial and Phylogenomic Characterization of Bacillus cereus Group Strains Isolated from Different Food Sources in Italy. Antibiotics, 13(9), 898. https://doi.org/10.3390/antibiotics13090898









