Opportunities for Laboratory Testing to Inform Antimicrobial Use for Bovine Respiratory Disease: Application of Information Quality Value Stream Maps in Commercial Feedlots
Abstract
:1. Introduction
2. Results
2.1. Overview and Environmental Context: IQ-VSM for BRD Treatment Plans in Commercial Feedlots
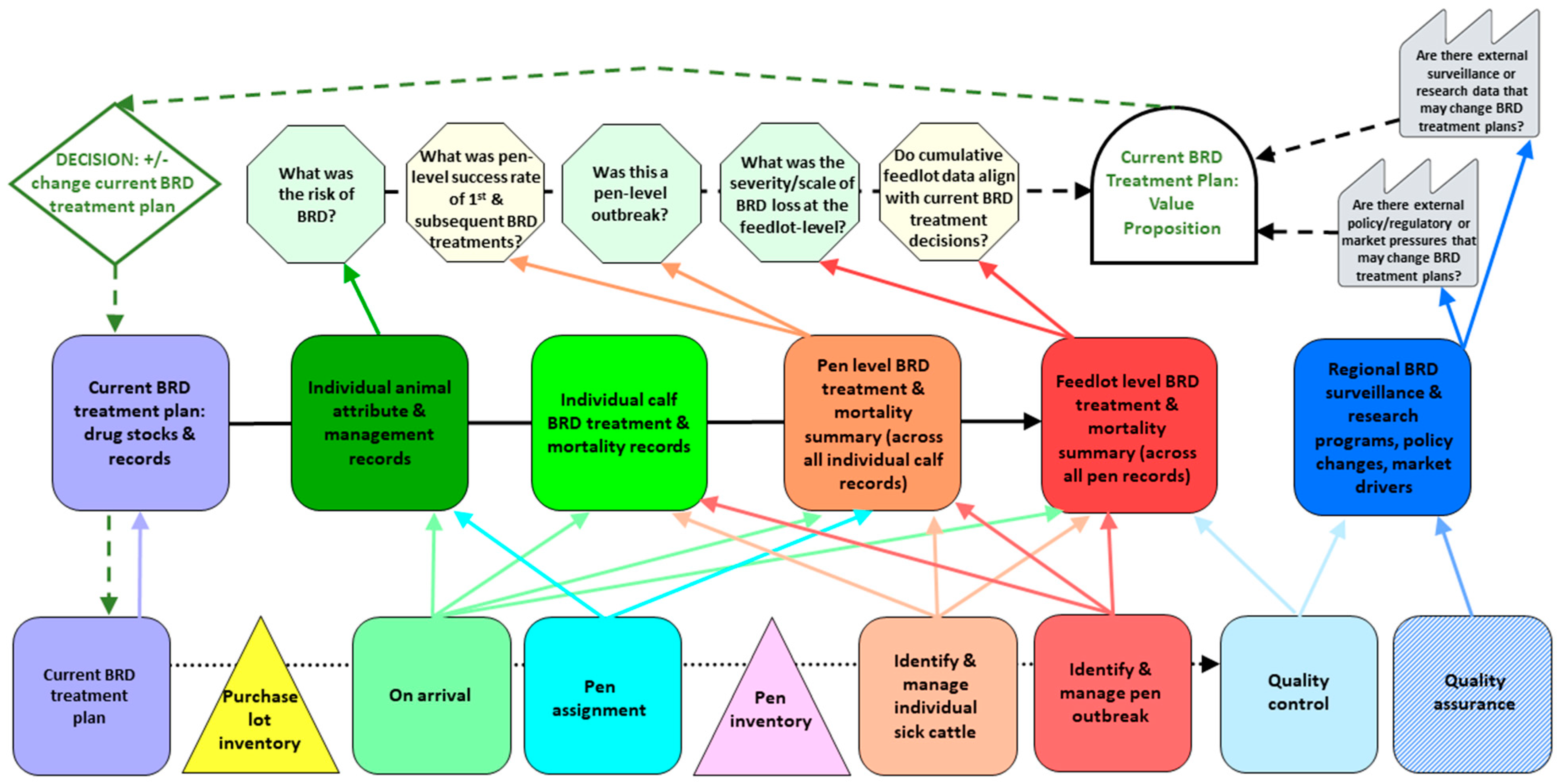
2.2. Current State IQ-VSM for BRD Treatment Plans in Commercial Feedlots
2.2.1. Process Lane (Bottom)
| Production Processes | Information Resources | Types of Static Data |
|---|---|---|
| Current BRD treatment plan | In-house proprietary protocols and algorithms:
| Data Type 1
|
| Material inventory 1: incoming purchase lot animal inventory |
| Data Type 2—purchase lot records to capture:
|
| On-arrival process | In-house proprietary protocols and algorithms:
| Data Type 3—calf records to capture:
|
| Pen assignment process | In-house proprietary protocols and algorithms:
| Data Type 4—pen records and tools to summarize: (Record of pen identification and membership)
|
| Material inventory 2: pen animal inventory | Pen records within feedlot management software | Data Type 5—tools to summarize:
|
| Process to identify and manage individual sick animals | In-house proprietary protocols and algorithms:
| Data Type 6—record system and tools to summarize:
|
| Process to identify and manage a pen outbreak | In-house proprietary protocols and algorithms:
| Data Type 7—process to collect and summarize:
|
| Quality control process | In-house proprietary protocols and algorithms:
| Data Type 8—process to collect and summarize:
|
| Quality assurance process | Shared resources (in-house +/− external):
| Data Type 9:
|
2.2.2. Information Lane (Middle)
| Information Processes | Static Information 1 | Dynamic Information |
|---|---|---|
| Current BRD treatment plan: drug stocks and records | Data Type 1 | Drug inventory/supply:
|
| Individual animal attribute and management records | Data Types 3 + 4 | Purchase history:
|
| Individual calf BRD treatment and mortality records | Data Types 3 + 6 + 7 |
|
| Pen-level BRD treatment and mortality summary (across all individual calf records) | Data Types 3, 4, 6, 7 | Pen-level summary of individual calf records for:
|
| Feedlot BRD treatment and mortality summary (analysis stratified for different classes of fed cattle) | Data Types 3, 6, 7, 8 | Feedlot-level summary across all pen records for:
|
| Regional BRD surveillance and research programs, policy changes and market drivers | Data Types 8 + 9 |
|
2.2.3. Information Processing Lane (Top)
| Information Assessment | Type of Data | Explanation: Information Processing Components to Inform Decisions | Degree of Uncertainty (Low/Medium/High) |
|---|---|---|---|
| What was the risk of BRD? | Internal data |
| Low |
| What was the pen-level success rate for 1st and and subsequent BRD treatment(s)? | Internal data |
| Moderate 1 |
| Was this a pen-level outbreak? | Internal data |
| Low |
| What was the severity/scale of the BRD loss at the feedlot level? | Internal data assessment |
| Low |
| Do the cumulative feedlot data align with the current BRD treatment decisions? | Internal data assessment |
| Moderate 2 |
| Are there surveillance or research data that would change BRD treatment plans? | External data assessment |
| High |
| Are there policy/regulatory changes or customer/market pressures that would change BRD treatment plans? | External data assessment |
| High |
| Current treatment plan: value proposition | Information processing that follows assessments |
| N/A |
| Control decision: +/− change current feedlot BRD treatment plan and feedback loop | Decision |
| N/A |
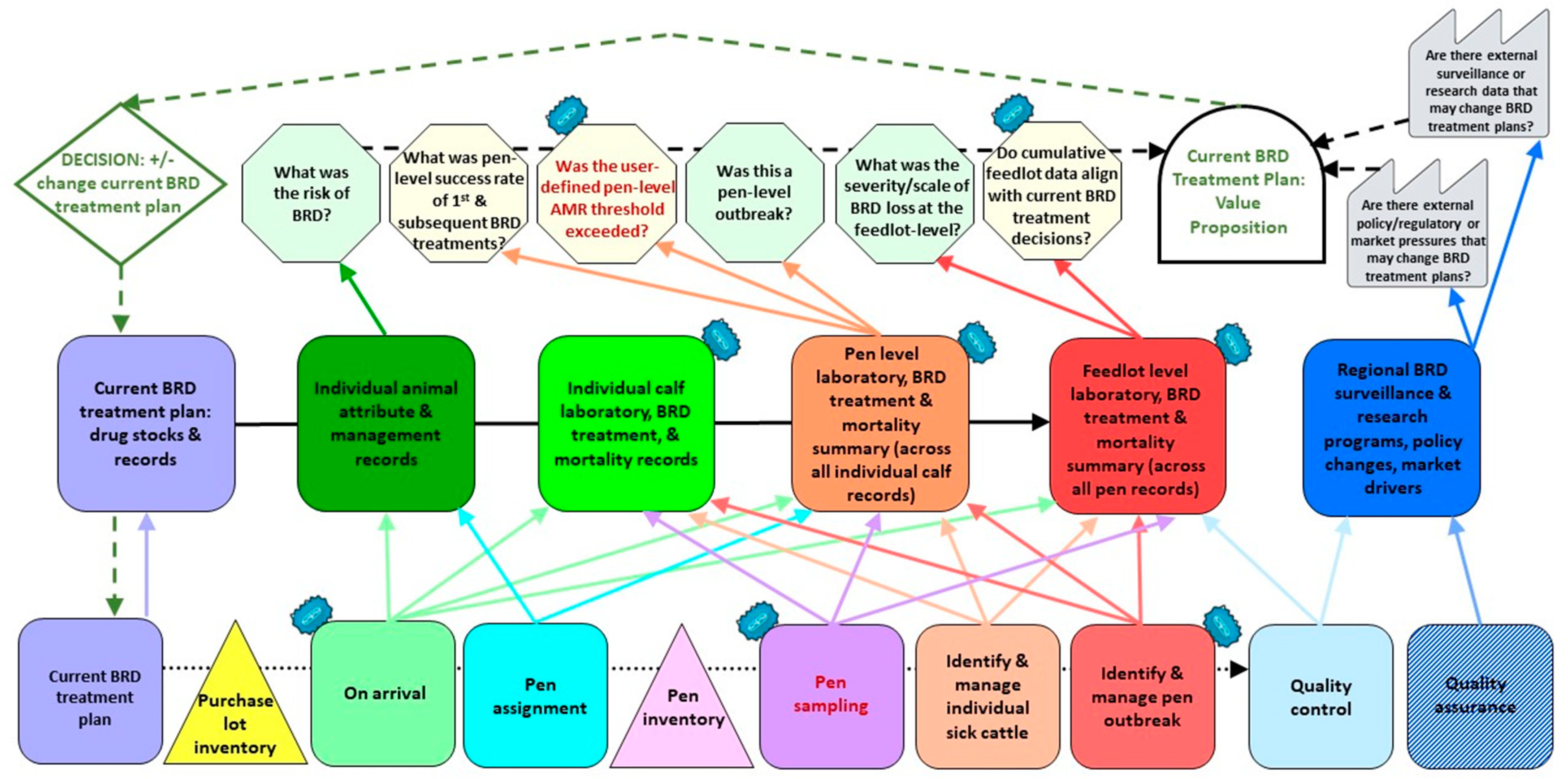
2.3. Future State IQ-VSM
| IQ-VSM Lane | Kaizen |
|---|---|
| Process Lane | Kaizen 1: On-arrival process (representative sample of calves from a pen at arrival) Kaizen 2: Pen sampling (representative sample of calves from a pen at 10–14 days on feed) * Kaizen 3: Identify and manage pen outbreak |
| Information Lane | Addition of laboratory data from sampling to: Kaizen 1: Individual calf laboratory, BRD treatment, and mortality records Kaizen 2: Pen-level summary of individual laboratory, BRD treatment, and mortality records Kaizen 3: Feedlot-level summary of pen-level laborato-ry, BRD treatment, and mortality records |
| Information Processing Lane | Kaizen 1: Based on pen-level laboratory results, was the user-defined pen-level AMR threshold exceeded? Kaizen 2: Do cumulative feedlot-level data align with current BRD treatment decisions? |
- Granularity: The degree of resolution for which the considered information is available. The granularity influences the efficacy of control methods, as it defines the representation of the real-life phenomenon [11]:
- Process Lane: The number of calves in a purchase lot or pen that can be sampled at any of the three sampling time points as well as the number of purchase lots or pens sampled per feedlot in the current fall run of calves entering the feedlot;
- Information Lane: The 95% confidence intervals (CIs) for the resulting proportions of reported laboratory outcomes for individual pens (ARGs or phenotypic AMR for specific BRD pathogens) and how these data vary across pens within the feedlot;
- Information Processing Lane: The number of pens sampled and precision of the resulting 95% CIs that are sufficient to inform subsequent decisions on BRD treatment protocols at the pen and feedlot level relative to other information sources.
- Frequency: The time interval in which the information is acquired or has been updated [11]. Delayed information could reduce or eliminate the value of data for time-sensitive decisions:
- Process Lane: (i) timing of the sample collection (relative to when calves arrive), (ii) number of times a pen is sampled (individual pens may be sampled more than once), and (iii) not all pens will be sampled. The veterinarian will determine the pen sampling rate in order to select which pens will be sampled and what proportion of pens will be sampled throughout the current run;
- Information Lane: Timing of receipt and upload of laboratory results into the feedlot data management system;
- Information Processing Lane: Laboratory results are available in sufficient time for analysis to inform decisions on the appropriateness of current BRD treatment plans for (i) the sampled pen(s), (ii) other similar but non-sampled pens during the current run, (iii) and/or decisions regarding the appropriateness of feedlot-level treatment plans for the current and subsequent years.
- Accuracy: The degree to which the obtained information represents the real-life phenomenon. [11]. Discrepancies are related to the measurement unit described by the IQ dimension granularity and include all possible influencing factors responsible for deviations:
- Process Lane: (i) the degree to which the sample received by the laboratory can be analyzed to reflect the presence of BRD pathogens and the AMR status (e.g., the quality of the collected sample and shipping efficiency/impacts of shipping delays), and (ii) limitations of the laboratory procedures for bacterial isolation and AST or metagenomic sequencing and bioinformatics;
- Information Lane: (i) completeness of the list of BRD pathogens and types of ARGs or phenotypic AMR identified and reported by the laboratory, and (ii) what is known regarding the sample-level diagnostic sensitivity and specificity of these tests;
- Information Processing Lane: (i) How does the resulting information reflect the true antimicrobial susceptibility of BRD pathogens in fall-placed high-risk calves at the pen and feedlot level for the current production run? (ii) How can this information be used to inform decisions on the appropriateness of current BRD treatment protocols?
3. Discussion
4. Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Food and Agriculture Organization; United Nations Environment Programme; World Health Organization; World Organization for Animal Health. One Health Joint Plan of Action (2022–2026): Working Together for the Health of Humans, Animals, Plants and the Environment; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2022; Volume xii, pp. 1–72. [Google Scholar] [CrossRef]
- World Health Organization. Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015; pp. 1–28. [Google Scholar]
- McEwen, S.A.; Angulo, F.J.; Collignon, P.J.; Conly, J.M. Unintended consequences associated with national-level restrictions on antimicrobial use in food-producing animals. Lancet Planet. Health 2018, 2, e279–e282. [Google Scholar] [CrossRef] [PubMed]
- Adewusi, O.O.; Waldner, C.L.; Hanington, P.C.; Hill, J.E.; Freeman, C.N.; Otto, S.J.G. Laboratory tools for the direct detection of bacterial respiratory infections and antimicrobial resistance: A scoping review. J. Vet. Diagn. Investig. 2024, 36, 400–417. [Google Scholar] [CrossRef]
- Rother, M.; Shook, J. Learning to See: Value Stream Mapping to Create Value and Eliminate Muda; Lean Enterprise Institute: Boston, MA, USA, 1999. [Google Scholar]
- Jones, D.; Womack, J. Seeing the Whole: Mapping the Extended Value Stream; Lean Enterprise Institute, Brookline: Boston, MA, USA, 2002. [Google Scholar]
- Schwab, K. The Fourth Industrial Revolution: What It Means and How to Respond. Available online: https://www.weforum.org/agenda/2016/01/the-fourth-industrial-revolution-what-it-means-and-how-to-respond/ (accessed on 16 April 2024).
- Busert, T.; Fay, A. Information Quality Dimensions for Identifying and Handling Inaccuracy and Uncertainty in Production Planning and Control. In Proceedings of the 2018 IEEE 23rd International Conference on Emerging Technologies and Factory Automation (ETFA), Turin, Italy, 4–7 September 2018; pp. 581–588. [Google Scholar]
- Busert, T.; Fay, A. Extended Value Stream Mapping Method: Harmonizing Information Flows for the Control of Production Processes. IFAC-PapersOnLine 2019, 52, 54–59. [Google Scholar] [CrossRef]
- Busert, T.; Fay, A. Extended Value Stream Mapping Method for Information Based Improvement of Production Logistics Processes. IEEE Eng. Manag. Rev. 2019, 47, 119–127. [Google Scholar] [CrossRef]
- Busert, T.; Fay, A. Information quality focused value stream mapping for the coordination and control of production processes. Int. J. Prod. Res. 2021, 59, 4559–4578. [Google Scholar] [CrossRef]
- Buczinski, S.; Pardon, B. Bovine Respiratory Disease Diagnosis: What Progress Has Been Made in Clinical Diagnosis? Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 399–423. [Google Scholar] [CrossRef] [PubMed]
- Ives, S.E.; Richeson, J.T. Use of Antimicrobial Metaphylaxis for the Control of Bovine Respiratory Disease in High-Risk Cattle. Vet. Clin. N. Am. Food Anim. Pract. 2015, 31, 34–50. [Google Scholar] [CrossRef]
- Brault, S.A.; Hannon, S.J.; Gow, S.P.; Warr, B.N.; Withell, J.; Song, J.; Williams, C.M.; Otto, S.J.G.; Booker, C.W.; Morley, P.S. Antimicrobial Use on 36 Beef Feedlots in Western Canada: 2008–2012. Front. Vet. Sci. 2019, 6, 329. [Google Scholar] [CrossRef]
- Fulton, R.W.; Confer, A.W. Laboratory test descriptions for Bovine Respiratory Disease diagnosis and their strengths and weaknesses: Gold standards for diagnosis, do they exist? Can. Vet. J. 2012, 53, 754–761. [Google Scholar]
- Panciera, R.J.; Confer, A.W. Pathogenesis and pathology of bovine pneumonia. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 191–214. [Google Scholar] [CrossRef]
- Kamel, M.S.; Davidson, J.L.; Verma, M.S. Strategies for Bovine Respiratory Disease (BRD) Diagnosis and Prognosis: A Comprehensive Overview. Animals 2024, 14, 627. [Google Scholar] [CrossRef]
- De Steur, H.; Wesana, J.; Dora, M.K.; Pearce, D.; Gellynck, X. Applying Value Stream Mapping to reduce food losses and wastes in supply chains: A systematic review. Waste Manag. 2016, 58, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.H. Strategic considerations in the development of lean agri-food supply chains: A case study of the UK pork sector. Supply Chain. Manag. Int. J. 2006, 11, 271–280. [Google Scholar] [CrossRef]
- Wesana, J.; Gellynck, X.; Dora, M.K.; Pearce, D.; De Steur, H. Measuring food and nutritional losses through value stream mapping along the dairy value chain in Uganda. Resour. Conserv. Recycl. 2019, 150, 104416. [Google Scholar] [CrossRef]
- Alberta Cattle Feeders Association. Have You Ever Wondered What Goes on in a Feedlot? Feedlot 101. Available online: https://cattlefeeders.ca/feedlot-101/#:~:text=Cattle%20in%20Canada%20spend%20most,cattle%20to%20forage%20for%20food (accessed on 26 June 2024).
- Canfax. Cattle on Feed Reports: Region Capacity. Available online: https://canfax.ca/resources/reports/cattle-on-feed-reports/region-capacity.html (accessed on 3 July 2024).
- Meudt, T.; Metternich, J.; Abele, E. Value stream mapping 4.0: Holistic examination of value stream and information logistics in production. CIRP Ann. 2017, 66, 413–416. [Google Scholar] [CrossRef]
- Abi Younes, J.N.; Campbell, J.R.; Otto, S.J.G.; Gow, S.P.; Woolums, A.R.; Jelinski, M.; Lacoste, S.; Waldner, C.L. Variation in Pen-Level Prevalence of BRD Bacterial Pathogens and Antimicrobial Resistance Following Feedlot Arrival in Beef Calves. Antibiotics 2024, 13, 322. [Google Scholar] [CrossRef] [PubMed]
- Herman, E.K.; Lacoste, S.R.; Freeman, C.N.; Otto, S.J.G.; McCarthy, E.L.; Links, M.G.; Stothard, P.; Waldner, C.L. Bacterial enrichment prior to third-generation metagenomic sequencing improves detection of BRD pathogens and genetic determinants of antimicrobial resistance in feedlot cattle. Front. Microbiol. 2024, 15, 1386319. [Google Scholar] [CrossRef]
- Andrés-Lasheras, S.; Ha, R.; Zaheer, R.; Lee, C.; Booker, C.W.; Dorin, C.; Van Donkersgoed, J.; Deardon, R.; Gow, S.; Hannon, S.J. Prevalence and risk factors associated with antimicrobial resistance in bacteria related to bovine respiratory disease—A broad cross-sectional study of beef cattle at entry into Canadian feedlots. Front. Vetinary Sci. 2021, 8, 119. [Google Scholar] [CrossRef]
- Abi Younes, J.N.; Campbell, J.R.; Gow, S.P.; Woolums, A.R.; Waldner, C.L. Association between respiratory disease pathogens in calves near feedlot arrival with treatment for bovine respiratory disease and subsequent antimicrobial resistance status. Front. Vetinary Sci. 2024, 11, 1416436. [Google Scholar] [CrossRef]
- Snyder, E.R.; Credille, B.; Berghaus, R.; Giguere, S. Prevalence of multi drug antimicrobial resistance in isolated from high-risk stocker cattle at arrival and two weeks after processing. J. Anim. Sci. 2017, 95, 1124–1131. [Google Scholar] [CrossRef]
- Snyder, E.R.; Alvarez-Narvaez, S.; Credille, B.C. Genetic characterization of susceptible and multi-drug resistant Mannheimia haemolytica isolated from high-risk stocker calves prior to and after antimicrobial metaphylaxis. Vet. Microbiol. 2019, 235, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Booker, C.W.; Abutarbush, S.M.; Morley, P.S.; Jim, G.K.; Pittman, T.J.; Schunicht, O.C.; Perrett, T.; Wildman, B.K.; Fenton, R.K.; Guichon, P.T.; et al. Microbiological and histopathological findings in cases of fatal bovine respiratory disease of feedlot cattle in Western Canada. Can. Vet. J. 2008, 49, 473–481. [Google Scholar] [PubMed]
- Burciaga-Robles, L.O.; McMullen, C.; Bras, A.; Guerra-Maupome, M.; Nava-Gaspar, R.A.; Rademacher, R.D.; Hannon, S.J.; Mercado-Talamantes, V.E.; Booker, C.W. An evaluation of arrival metaphylaxis with enrofloxacin compared to tulathromycin in feedlot cattle at high risk of developing bovine respiratory disease in Mexico. Bov. Pract. 2022, 56, 53–59. [Google Scholar] [CrossRef]
- Schunicht, O.C.; Booker, C.W.; Guichon, P.T.; Jim, G.K.; Wildman, B.K.; Pittman, T.J.; Perrett, T. An evaluation of the relative efficacy of tulathromycin for the treatment of undifferentiated fever in feedlot calves in Nebraska. Can. Vet. J. 2007, 48, 600–606. [Google Scholar]
- Wildman, B.K.; Perrett, T.; Abutarbush, S.M.; Guichon, P.T.; Pittman, T.J.; Booker, C.W.; Schunicht, O.C.; Fenton, R.K.; Jim, G.K. A comparison of 2 vaccination programs in feedlot calves at ultra-high risk of developing undifferentiated fever/bovine respiratory disease. Can. Vet. J. 2008, 49, 463–472. [Google Scholar]
- Morrone, S.; Dimauro, C.; Gambella, F.; Cappai, M.G. Industry 4.0 and Precision Livestock Farming (PLF): An up to Date Overview across Animal Productions. Sensors 2022, 22, 4319. [Google Scholar] [CrossRef] [PubMed]
- Conrad, C.C.; Funk, T.; Andres-Lasheras, S.; Yevtushenko, C.; Claassen, C.; Otto, S.J.G.; Waldner, C.; Zaheer, R.; McAllister, T.A. Improving the detection of integrative conjugative elements in bovine nasopharyngeal swabs using multiplex recombinase polymerase amplification. J. Microbiol. Methods 2024, 221, 106943. [Google Scholar] [CrossRef]
- Hannon, S.J.; Brault, S.A.; Otto, S.J.G.; Morley, P.S.; McAllister, T.A.; Booker, C.W.; Gow, S.P. Feedlot cattle antimicrobial use surveillance network: A Canadian journey. Front. Vet. Sci. 2020, 7, 59604. [Google Scholar] [CrossRef] [PubMed]
- CFAASP. Canadian Feedlot Antimicrobial Use/Antimicrobial Resistance Surveillance Program. Available online: https://cfaasp.ca/ (accessed on 26 June 2024).
- Waldner, C.; Windeyer, M.C.; Rousseau, M.; Campbell, J. The Canadian Cow-Calf Surveillance Network—Productivity and health summary 2018 to 2022. Front. Vetinary Sci. 2024, 11, 1392166. [Google Scholar] [CrossRef]
- Fossen, J.D.; Campbell, J.R.; Gow, S.P.; Erickson, N.; Waldner, C.L. Antimicrobial Use in Canadian Cow-Calf Herds. Vet. Sci. 2023, 10, 366. [Google Scholar] [CrossRef]
- Fossen, J.D.; Campbell, J.R.; Gow, S.P.; Erickson, N.; Waldner, C.L. Antimicrobial resistance in generic E. coli isolated from western Canadian cow-calf herds. Can. Vet. J. 2024, 65, 146–155. [Google Scholar] [PubMed]
- Fossen, J.D.; Campbell, J.R.; Gow, S.P.; Erickson, N.; Waldner, C.L. Antimicrobial resistance in Enterococcus isolated from western Canadian cow-calf herds. BMC Vet. Res. 2024, 20, 6. [Google Scholar] [CrossRef] [PubMed]
- Freeman, C.N.; Herman, E.K.; Abi Younes, J.; Ramsay, D.E.; Erikson, N.; Stothard, P.; Links, M.G.; Otto, S.J.G.; Waldner, C. Evaluating the potential of third generation metagenomic sequencing for the detection of BRD pathogens and genetic determinants of antimicrobial resistance in chronically ill feedlot cattle. BMC Vet. Res. 2022, 18, 211. [Google Scholar] [CrossRef] [PubMed]
- McGregor, J.C.; Fitzpatrick, M.A.; Suda, K.J. Expanding Antimicrobial Stewardship Through Quality Improvement. JAMA Netw. Open 2021, 4, e211072. [Google Scholar] [CrossRef]
- Endalamaw, A.; Khatri, R.B.; Mengistu, T.S.; Erku, D.; Wolka, E.; Zewdie, A.; Assefa, Y. A scoping review of continuous quality improvement in healthcare system: Conceptualization, models and tools, barriers and facilitators, and impact. BMC Health Serv. Res. 2024, 24, 487. [Google Scholar] [CrossRef]
- Hill, J.E.; Stephani, A.M.; Sapple, P.; Clegg, A.J. The effectiveness of continuous quality improvement for developing professional practice and improving health care outcomes: A systematic review. Implement. Sci. 2020, 15, 23. [Google Scholar] [CrossRef]
- European Union. Regulation (EU) 2019/6 of the European Parliament and of the Council of 11 December 2019 on Veterinary Medicinal Products and Repealing Directive 2001/83/EC. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A02019R0006-20220128 (accessed on 18 June 2024).
- McDonald’s. McDonald’s Antibiotic Policy for our Beef Supply Chain. Available online: https://corporate.mcdonalds.com/content/dam/sites/corp/nfl/pdf/McDonalds_Beef_and_Dairy%20_Antibiotic_Policy.pdf (accessed on 18 June 2024).
- Roy, J.P.; Archambault, M.; Desrochers, A.; Dubuc, J.; Dufour, S.; Francoz, D.; Paradis, M.E.; Rousseau, M. New Quebec regulation on the use of antimicrobials of very high importance in food animals: Implementation and impacts in dairy cattle practice. Can. Vet. J. 2020, 61, 193–196. [Google Scholar]
- Government of Canada. Categorization of Antimicrobial Drugs Based on Importance in Human Medicine. Available online: https://www.canada.ca/en/health-canada/services/drugs-health-products/veterinary-drugs/antimicrobial-resistance/categorization-antimicrobial-drugs-based-importance-human-medicine.html (accessed on 15 September 2024).
- World Health Organization. WHO List of Medically Important Antimicrobials: A Risk Management Tool for Mitigating Antimicrobial Resistance Due to Non-Human Use; World Health Organization: Geneva, Switzerland, 2024; pp. 1–31. [Google Scholar]
- O’Connor, A.M.; Hu, D.; Totton, S.C.; Scott, N.; Winder, C.B.; Wang, B.; Wang, C.; Glanville, J.; Wood, H.; White, B.; et al. A systematic review and network meta-analysis of injectable antibiotic options for the control of bovine respiratory disease in the first 45 days post arrival at the feedlot. Anim. Health Res. Rev. 2019, 20, 163–181. [Google Scholar] [CrossRef]
- Capik, S.F.; White, B.J.; Lubbers, B.V.; Apley, M.D.; DeDonder, K.D.; Larson, R.L.; Harhay, G.P.; Chitko-McKown, C.G.; Harhay, D.M.; Kalbfleisch, T.S.; et al. Comparison of the diagnostic performance of bacterial culture of nasopharyngeal swab and bronchoalveolar lavage fluid samples obtained from calves with bovine respiratory disease. Am. J. Vet. Res. 2017, 78, 350–358. [Google Scholar] [CrossRef]
- Doyle, D.; Credille, B.; Lehenbauer, T.W.; Berghaus, R.; Aly, S.S.; Champagne, J.; Blanchard, P.; Crossley, B.; Berghaus, L.; Cochran, S.; et al. Agreement among 4 sampling methods to identify respiratory pathogens in dairy calves with acute bovine respiratory disease. J. Vet. Intern. Med. 2017, 31, 954–959. [Google Scholar] [CrossRef]
- Credille, B.C.; Capik, S.F.; Credille, A.; Crossley, B.M.; Blanchard, P.; Woolums, A.R.; Lehenbauer, T.W. Agreement of antimicrobial susceptibility testing of Pasteurella multocida and Mannheimia haemolytica isolates from preweaned dairy calves with bovine respiratory disease. Am. J. Vet. Res. 2023, 84, ajvr.23.06.0140. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.W.; Viel, L.; Bateman, K.G.; Rosendal, S.; Shewen, P.E.; Physick-Sheard, P. The microbial flora of the respiratory tract in feedlot calves: Associations between nasopharyngeal and bronchoalveolar lavage cultures. Can. J. Vet. Res. 1991, 55, 341–346. [Google Scholar]
- Pardon, B.; Buczinski, S. Bovine Respiratory Disease Diagnosis: What Progress Has Been Made in Infectious Diagnosis? Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 425–444. [Google Scholar] [CrossRef] [PubMed]
- Sarchet, J.J.; Pollreisz, J.P.; Bechtol, D.T.; Blanding, M.R.; Saltman, R.L.; Taube, P.C. Limitations of bacterial culture, viral PCR, and tulathromycin susceptibility from upper respiratory tract samples in predicting clinical outcome of tulathromycin control or treatment of bovine respiratory disease in high-risk feeder heifers. PLoS ONE 2022, 17, e0247213. [Google Scholar] [CrossRef]
- McClary, D.G.; Loneragan, G.H.; Shryock, T.R.; Carter, B.L.; Guthrie, C.A.; Corbin, M.J.; Mechor, G.D. Relationship of in vitro minimum inhibitory concentrations of tilmicosin against Mannheimia haemolytica and Pasteurella multocida and in vivo tilmicosin treatment outcome among calves with signs of bovine respiratory disease. J. Am. Vet. Med. Assoc. 2011, 239, 129–135. [Google Scholar] [CrossRef]
- Godinho, K.S.; Rae, A.; Windsor, G.D.; Tilt, N.; Rowan, T.G.; Sunderland, S.J. Efficacy of tulathromycin in the treatment of bovine respiratory disease associated with induced Mycoplasma bovis infections in young dairy calves. Vet. Ther. Res. Appl. Vet. Med. 2005, 6, 96–112. [Google Scholar]
- Jobman, E.; Hagenmaier, J.; Meyer, N.; Harper, L.B.; Taylor, L.; Lukasiewicz, K.; Thomson, D.; Lowe, J.; Terrell, S. Cross-Section observational study to assess antimicrobial resistance prevalence among bovine respiratory disease bacterial isolates from commercial US feedlots. Antibiotics 2023, 12, 215. [Google Scholar] [CrossRef]
- Neal, K.; Amachawadi, R.G.; White, B.J.; Shippy, T.D.; Theurer, M.E.; Larson, R.L.; Lubbers, B.V.; Kleinhenz, M. Nasopharyngeal bacterial prevalence and microbial diversity at first treatment for bovine respiratory disease (BRD) and its associations with health and mortality outcomes in feedyard cattle. Microorganisms 2024, 12, 33. [Google Scholar] [CrossRef]
- Carter, H.F.; Wills, R.W.; Scott, M.A.; Thompson, A.C.; Singer, R.S.; Loy, J.D.; Karisch, B.B.; Epperson, W.B.; Woolums, A.R. Assessment of diversity of antimicrobial resistance phenotypes and genotypes of Mannheimia haemolytica isolates from bovine nasopharyngeal swabs. Front. Vetinary Sci. 2022, 9, 883389. [Google Scholar] [CrossRef]
- Woolums, A.R.; Karisch, B.B.; Frye, J.G.; Epperson, W.; Smith, D.R.; Blanton Jr, J.; Austin, F.; Kaplan, R.; Hiott, L.; Woodley, T.; et al. Multidrug resistant Mannheimia haemolytica isolated from high-risk beef stocker cattle after antimicrobial metaphylaxis and treatment for bovine respiratory disease. Vet. Microbiol. 2018, 221, 143–152. [Google Scholar] [CrossRef]
- Noyes, N.R.; Benedict, K.M.; Gow, S.P.; Booker, C.W.; Hannon, S.J.; McAllister, T.A.; Morley, P.S. Mannheimia haemolytica in feedlot cattle: Prevalence of recovery and associations with antimicrobial use, resistance, and health outcomes. J. Vet. Intern. Med. 2015, 29, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.A.; Ambroset, C.; Huleux, A.; Vialatte, A.; Colin, A.; Tricot, A.; Arcangioli, M.-A.; Tardy, F. Monitoring Mycoplasma bovis diversity and antimicrobial susceptibility in calf feedlots undergoing a respiratory disease outbreak. Pathogens 2020, 9, 593. [Google Scholar] [CrossRef] [PubMed]
- Cagnetti, C.; Gallo, T.; Silvestri, C.; Ruggieri, A. Lean production and Industry 4.0: Strategy/management or technique/implementation? A systematic literature review. Procedia Comput. Sci. 2021, 180, 404–413. [Google Scholar] [CrossRef]
- Gallo, T.; Cagnetti, C.; Silvestri, C.; Ruggieri, A. Industry 4.0 tools in lean production: A systematic literature review. Procedia Comput. Sci. 2021, 180, 394–403. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Otto, S.J.G.; Pollock, C.M.; Relf-Eckstein, J.-A.; McLeod, L.; Waldner, C.L. Opportunities for Laboratory Testing to Inform Antimicrobial Use for Bovine Respiratory Disease: Application of Information Quality Value Stream Maps in Commercial Feedlots. Antibiotics 2024, 13, 903. https://doi.org/10.3390/antibiotics13090903
Otto SJG, Pollock CM, Relf-Eckstein J-A, McLeod L, Waldner CL. Opportunities for Laboratory Testing to Inform Antimicrobial Use for Bovine Respiratory Disease: Application of Information Quality Value Stream Maps in Commercial Feedlots. Antibiotics. 2024; 13(9):903. https://doi.org/10.3390/antibiotics13090903
Chicago/Turabian StyleOtto, Simon J. G., Colleen M. Pollock, Jo-Anne Relf-Eckstein, Lianne McLeod, and Cheryl L. Waldner. 2024. "Opportunities for Laboratory Testing to Inform Antimicrobial Use for Bovine Respiratory Disease: Application of Information Quality Value Stream Maps in Commercial Feedlots" Antibiotics 13, no. 9: 903. https://doi.org/10.3390/antibiotics13090903
APA StyleOtto, S. J. G., Pollock, C. M., Relf-Eckstein, J.-A., McLeod, L., & Waldner, C. L. (2024). Opportunities for Laboratory Testing to Inform Antimicrobial Use for Bovine Respiratory Disease: Application of Information Quality Value Stream Maps in Commercial Feedlots. Antibiotics, 13(9), 903. https://doi.org/10.3390/antibiotics13090903







