Emerging Resistance and Virulence Patterns in Salmonella enterica: Insights into Silver Nanoparticles as an Antimicrobial Strategy
Abstract
:1. Introduction
2. Results
2.1. Isolate Characterisation
2.2. Soluble Enzymatic VFs Detection
2.3. Biofilm Formation
2.4. AR and Virulence Genes
2.5. WGS Analysis of Salmonella enterica subsp. Enterica Isolates
2.6. Pangenome Analysis of S. enterica Strains
2.7. AMR, MGE and Phage Predictions in Context of Clustering
2.8. Conjugation Experiments
2.9. Antimicrobial Efficiency of AgNPs Against S. Typhimurium, S. Hadar and S. Infantis
2.9.1. Qualitative Screening of AgNP Against Selected S. enterica Isolates
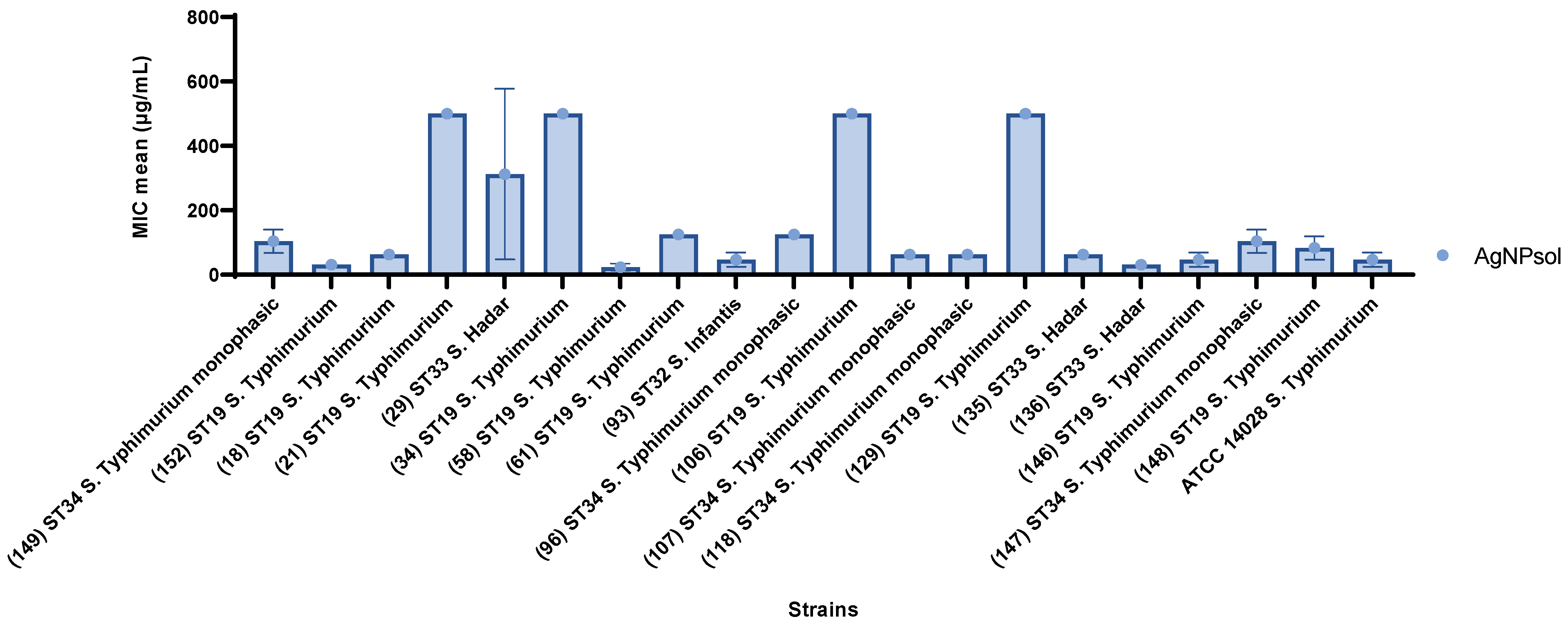
2.9.2. Quantitative Antibacterial Evaluation of AgNPsol Against Selected S. enterica Isolates
2.9.3. Anti-Adherence Activity of AgNPsol
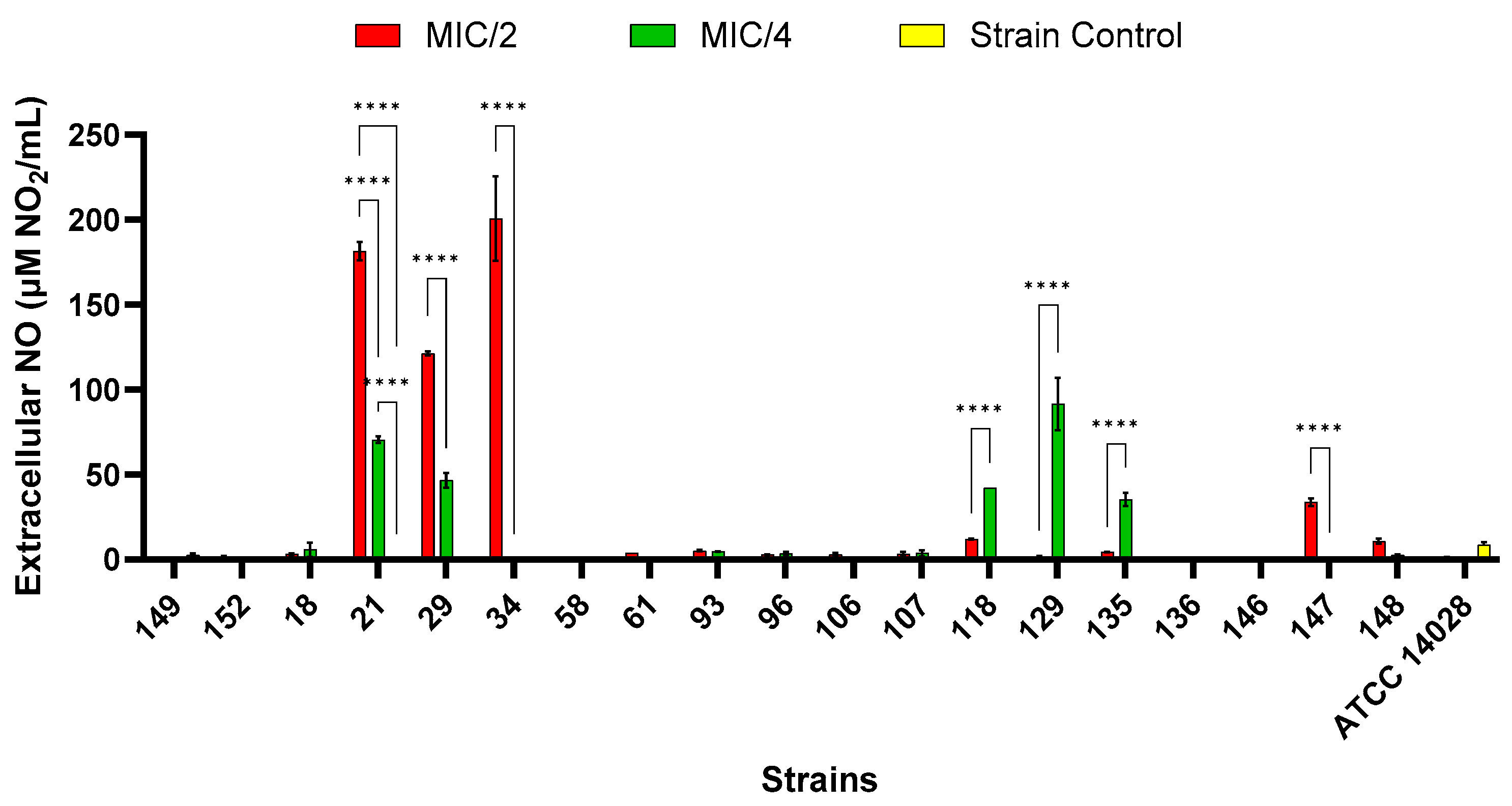
2.9.4. Extracellular Nitric Oxide Production
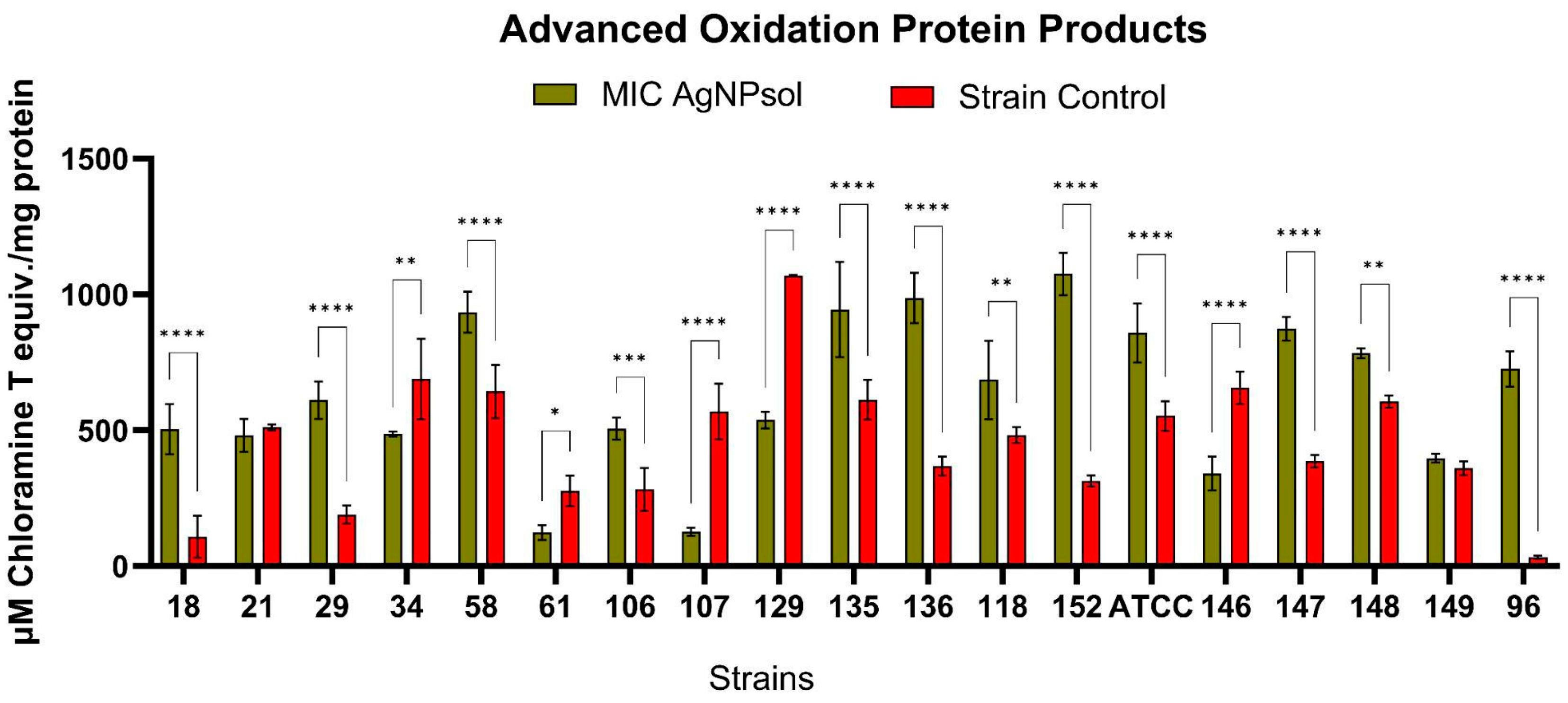
2.9.5. Advanced Oxidation Protein Products
2.10. Cell Viability
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Antibiotic Susceptibility Testing
4.2. Evaluation of the Soluble Enzymatic Virulence Factors
4.3. Biofilm Formation Assay
4.4. Molecular Characterization of Salmonella Strains
4.4.1. DNA Extraction
4.4.2. Polymerase Chain Reaction for ARGs and VFs Detection
4.4.3. Whole-Genome Sequencing (WGS) and Bioinformatics Analyses of Salmonella Isolates
4.4.4. Conjugation Assays Were Performed on Seven Representative Isolates
4.4.5. GenBank Accession Numbers
4.5. Antimicrobial, Anti-Biofilm Activity, and the Impact on Biochemical Processes of AgNP Solutions (AgNPs)
4.5.1. Qualitative Screening of AgNP Against Selected S. enterica Strains
4.5.2. Quantitative Evaluation of AgNPsol Efficacy
4.5.3. The Influence of AgNPs on Adherence Capacity
4.5.4. Extracellular Nitric Oxide Quantification
4.5.5. Advanced Oxidation Protein Products Quantification
4.6. Biocompatibility
4.7. Statistical Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ziyate, N.; Karraouan, B.; Kadiri, A.; Darkaoui, S.; Soulaymani, A.; Bouchrif, B. Prevalence and Antimicrobial Resistance of Salmonella Isolates in Moroccan Laying Hens Farms. J. Appl. Poult. Res. 2016, 25, 539–546. [Google Scholar] [CrossRef]
- Al-Rifai, R.H.; Chaabna, K.; Denagamage, T.; Alali, W.Q. Prevalence of Non-Typhoidal Salmonella enterica in Food Products in the Middle East and North Africa: A Systematic Review and Meta-Analysis. Food Control 2020, 109, 106908. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2017/2018. EFSA J. 2020, 18, e06007. [Google Scholar] [CrossRef]
- Randall, L.P.; Cooles, S.W.; Osborn, M.K.; Piddock, L.J.V.; Woodward, M.J. Antibiotic Resistance Genes, Integrons and Multiple Antibiotic Resistance in Thirty-Five Serotypes of Salmonella enterica Isolated from Humans and Animals in the UK. J. Antimicrob. Chemother. 2004, 53, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Thong, K.L.; Modarressi, S. Antimicrobial Resistant Genes Associated with Salmonella from Retail Meats and Street Foods. Food Res. Int. 2011, 44, 2641–2646. [Google Scholar] [CrossRef]
- Cuypers, W.L.; Jacobs, J.; Wong, V.; Klemm, E.J.; Deborggraeve, S.; Van Puyvelde, S. Fluoroquinolone Resistance in Salmonella: Insights by Whole-Genome Sequencing. Microb. Genom. 2018, 4, 000195. [Google Scholar] [CrossRef] [PubMed]
- Antunes, P.; Machado, J.; Sousa, J.C.; Peixe, L. Dissemination of Sulfonamide Resistance Genes (Sul1, Sul2, and Sul3) in Portuguese Salmonella enterica Strains and Relation with Integrons. Antimicrob. Agents Chemother. 2005, 49, 836–839. [Google Scholar] [CrossRef] [PubMed]
- Pezzella, C.; Ricci, A.; DiGiannatale, E.; Luzzi, I.; Carattoli, A. Tetracycline and Streptomycin Resistance Genes, Transposons, and Plasmids in Salmonella enterica Isolates from Animals in Italy. Antimicrob. Agents Chemother. 2004, 48, 903–908. [Google Scholar] [CrossRef]
- Sultan, I.; Rahman, S.; Jan, A.T.; Siddiqui, M.T.; Mondal, A.H.; Haq, Q.M.R. Antibiotics, Resistome and Resistance Mechanisms: A Bacterial Perspective. Front. Microbiol. 2018, 9, 2066. [Google Scholar] [CrossRef] [PubMed]
- Diene, S.M.; Rolain, J.-M. Carbapenemase Genes and Genetic Platforms in Gram-Negative Bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter Species. Clin. Microbiol. Infect. 2014, 20, 831–838. [Google Scholar] [CrossRef]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus Biofilm: An Emerging Battleground in Microbial Communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Molin, S.; Tolker-Nielsen, T. Gene Transfer Occurs with Enhanced Efficiency in Biofilms and Induces Enhanced Stabilisation of the Biofilm Structure. Curr. Opin. Biotechnol. 2003, 14, 255–261. [Google Scholar] [CrossRef]
- Steenackers, H.; Hermans, K.; Vanderleyden, J.; De Keersmaecker, S.C.J. Salmonella Biofilms: An Overview on Occurrence, Structure, Regulation and Eradication. Food Res. Int. 2012, 45, 502–531. [Google Scholar] [CrossRef]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial Resistance and Virulence: A Successful or Deleterious Association in the Bacterial World? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Zeng, J.; Li, L.; Yang, J.; Tang, Z.; Xiong, W.; Li, Y.; Chen, S.; Zeng, Z. Coexistence of Antibiotic Resistance Genes and Virulence Factors Deciphered by Large-Scale Complete Genome Analysis. mSystems 2020, 5, 10–1128. [Google Scholar] [CrossRef]
- Escudeiro, P.; Pothier, J.; Dionisio, F.; Nogueira, T. Antibiotic Resistance Gene Diversity and Virulence Gene Diversity Are Correlated in Human Gut and Environmental Microbiomes. mSphere 2019, 4, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, M.; Brooks, B.D.; Brooks, A.E. The Complex Relationship between Virulence and Antibiotic Resistance. Genes 2017, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Eswarappa, S.M.; Panguluri, K.K.; Hensel, M.; Chakravortty, D. The YejABEF Operon of Salmonella Confers Resistance to Antimicrobial Peptides and Contributes to Its Virulence. Microbiology 2008, 154, 666–678. [Google Scholar] [CrossRef] [PubMed]
- Roux, D.; Danilchanka, O.; Guillard, T.; Cattoir, V.; Aschard, H.; Fu, Y.; Angoulvant, F.; Messika, J.; Ricard, J.-D.; Mekalanos, J.J.; et al. Fitness Cost of Antibiotic Susceptibility during Bacterial Infection. Sci. Transl. Med. 2015, 7, 297ra114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Dupont, A.; Torow, N.; Gohde, F.; Leschner, S.; Lienenklaus, S.; Weiss, S.; Brinkmann, M.M.; Kühnel, M.; Hensel, M.; et al. Age-Dependent Enterocyte Invasion and Microcolony Formation by Salmonella. PLoS Pathog. 2014, 10, e1004385. [Google Scholar] [CrossRef]
- Doorduyn, Y.; van Pelt, W.; Siezen, C.L.E.; van Der Horst, F.; van Duynhoven, Y.T.H.P.; Hoebee, B.; Janssen, R. Novel insight in the association between salmonellosis or campylobacteriosis and chronic illness, and the role of host genetics in susceptibility to these diseases. Epidemiol. Infect. 2008, 136, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- CDC. Salmonella Infection (Salmonellosis). Available online: https://www.cdc.gov/salmonella/spread/ (accessed on 10 December 2024).
- van Asten, A.J.A.M.; van Dijk, J.E. Distribution of “Classic” Virulence Factors among Salmonella spp. FEMS Immunol. Med. Microbiol. 2005, 44, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Eng, S.K.; Pusparajah, P.; Ab Mutalib, N.S.; Leng, S.H.; Chan, K.G.; Learn Han, L. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 2015, 8, 284–293. [Google Scholar] [CrossRef]
- Chaudhary, J.H.; Nayak, J.B.; Brahmbhatt, M.N.; Makwana, P.P. Virulence Genes Detection of Salmonella Serovars Isolated from Pork and Slaughterhouse Environment in Ahmedabad, Gujarat. Vet. World 2015, 8, 121–124. [Google Scholar] [CrossRef]
- Istivan, T.S.; Coloe, P.J. Phospholipase A in Gram-Negative Bacteria and Its Role in Pathogenesis. Microbiology 2006, 152, 1263–1274. [Google Scholar] [CrossRef] [PubMed]
- Marinescu, F.; Marutescu, L.; Chifiriuc, M.; Marutescu, L. Virulence Markers of Escherichia Coli Strains Isolated from Hospital and Poultry Wastewater. Biointerface Res. Appl. Chem. 2013, 3, 579–587. Available online: www.BiointerfaceResearch.com (accessed on 1 January 2025).
- Kaur, J.; Jain, S.K. Role of Antigens and Virulence Factors of Salmonella enterica Serovar Typhi in Its Pathogenesis. Microbiol. Res. 2012, 167, 199–210. [Google Scholar] [CrossRef] [PubMed]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, 6406. [Google Scholar] [CrossRef]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). The European Union One Health 2022 Zoonoses Report. EFSA J. 2023, 21, e8442. [Google Scholar] [CrossRef]
- Trchounian, A.; Gabrielyan, L.; Mnatsakanyan, N. Nanoparticles of Various Transition Metals and Their Applications as Antimicrobial Agents. In Metal Nanoparticles: Properties, Synthesis and Applications; Nova Science Publishers: Hauppauge, NY, USA, 2018; pp. 161–211. [Google Scholar]
- Lee, S.H.; Jun, B.-H. Silver Nanoparticles: Synthesis and Application for Nanomedicine. Int. J. Mol. Sci. 2019, 20, 865. [Google Scholar] [CrossRef] [PubMed]
- Gabrielyan, L.; Trchounian, A. Antibacterial Activities of Transient Metals Nanoparticles and Membranous Mechanisms of Action. World J. Microbiol. Biotechnol. 2019, 35, 162. [Google Scholar] [CrossRef] [PubMed]
- Franci, G.; Falanga, A.; Galdiero, S.; Palomba, L.; Rai, M.; Morelli, G.; Galdiero, M. Silver Nanoparticles as Potential Antibacterial Agents. Molecules 2015, 20, 8856–8874. [Google Scholar] [CrossRef]
- Rai, M.; Yadav, A.; Gade, A. Silver Nanoparticles as a New Generation of Antimicrobials. Biotechnol. Adv. 2009, 27, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Seil, J.T.; Webster, T.J. Antimicrobial Applications of Nanotechnology: Methods and Literature. Int. J. Nanomed. 2012, 7, 2767–2781. [Google Scholar] [CrossRef]
- Mahmoud Hoseinnejad, S.M.J.; Katouzian, I. Inorganic and Metal Nanoparticles and Their Antimicrobial Activity in Food Packaging Applications. Crit. Rev. Microbiol. 2018, 44, 161–181. [Google Scholar] [CrossRef] [PubMed]
- Sanyasi, S.; Majhi, R.K.; Kumar, S.; Mishra, M.; Ghosh, A.; Suar, M.; Satyam, P.V.; Mohapatra, H.; Goswami, C.; Goswami, L. Polysaccharide-Capped Silver Nanoparticles Inhibit Biofilm Formation and Eliminate Multi-Drug-Resistant Bacteria by Disrupting Bacterial Cytoskeleton with Reduced Cytotoxicity towards Mammalian Cells. Sci. Rep. 2016, 6, 24929. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, Y.; Kadoya, A.; Suzuki, S. An Integrative and Conjugative Element (ICE) Found in Shewanella halifaxensis Isolated from Marine Fish Intestine May Connect Genetic Materials between Human and Marine Environments. Microbes Environ. 2022, 37, ME22038. [Google Scholar] [CrossRef]
- Mohd-Zain, Z.; Turner, S.L.; Cerdeño-Tárraga, A.M.; Lilley, A.K.; Inzana, T.J.; Duncan, A.J.; Harding, R.M.; Hood, D.W.; Peto, T.E.; Crook, D.W. Transferable Antibiotic Resistance Elements in Haemophilus influenzae Share a Common Evolutionary Origin with a Diverse Family of Syntenic Genomic Islands. J. Bacteriol. 2004, 186, 8114–8122. [Google Scholar] [CrossRef]
- Sim, E.M.; Wang, Q.; Howar, P.; Kim, R.; Lim, L.; Hope, K.; Sintchenko, V. Persistent Salmonella enterica Serovar Typhi Sub-Populations within Host Interrogated by Whole Genome Sequencing and Metagenomics. PLoS ONE 2023, 18, e0289070. [Google Scholar] [CrossRef] [PubMed]
- Tsui, C.K.-M.; Ben Abid, F.; McElheny, C.L.; Hamed, M.M.; Perez-Lopez, A.; Omrani, A.S.; Doi, Y. Characterization of blaNDM in Two Escherichia Coli ST1193 Clinical Isolates in the Gulf Region. JAC Antimicrob. Resist. 2024, 6, dlae166. [Google Scholar] [CrossRef] [PubMed]
- Barraud, N.; Hassett, D.J.; Hwang, S.-H.; Rice, S.A.; Kjelleberg, S.; Webb, J.S. Involvement of Nitric Oxide in Biofilm Dispersal of Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 7344–7353. [Google Scholar] [CrossRef] [PubMed]
- Bamal, D.; Singh, A.; Chaudhary, G.; Kumar, M.; Singh, M.; Rani, N.; Mundlia, P.; Sehrawat, A.R. Silver Nanoparticles Biosynthesis, Characterization, Antimicrobial Activities, Applications, Cytotoxicity and Safety Issues: An Updated Review. Nanomaterials 2021, 11, 2086. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, K.G.; Delattre, V.; Frost, V.J.; Buck, G.W.; Phu, J.V.; Fernandez, T.G.; Pavel, I.E. Nanosilver: An Old Antibacterial Agent with Great Promise in the Fight against Antibiotic Resistance. Antibiotics 2023, 12, 1264. [Google Scholar] [CrossRef] [PubMed]
- Páez, P.L.; Becerra, M.C.; Albesa, I. Comparison of Macromolecular Oxidation by Reactive Oxygen Species in Three Bacterial Genera Exposed to Different Antibiotics. Cell Biochem. Biophys. 2011, 61, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Marinas, I.C.; Ignat, L.; Maurușa, I.E.; Gaboreanu, M.D.; Adina, C.; Popa, M.; Chifiriuc, M.C.; Angheloiu, M.; Georgescu, M.; Iacobescu, A.; et al. Insights into the Physico-Chemical and Biological Characterization of Sodium Lignosulfonate-Silver Nanosystems Designed for Wound Management. Heliyon 2024, 10, e26047. [Google Scholar] [CrossRef]
- Quinteros, M.A.; Cano Aristizábal, V.; Dalmasso, P.R.; Paraje, M.G.; Páez, P.L. Oxidative Stress Generation of Silver Nanoparticles in Three Bacterial Genera and Its Relationship with the Antimicrobial Activity. Toxicol. In Vitro 2016, 36, 216–223. [Google Scholar] [CrossRef] [PubMed]
- More, P.R.; Pandit, S.; Filippis, A.D.e.; Franci, G.; Mijakovic, I.; Galdiero, M. Silver Nanoparticles: Bactericidal and Mechanistic Approach against Drug Resistant Pathogens. Microorganisms 2023, 11, 369. [Google Scholar] [CrossRef] [PubMed]
- Afzal, S.; Abdul Manap, A.S.; Attiq, A.; Albokhadaim, I.; Kandeel, M.; Alhojaily, S.M. From Imbalance to Impairment: The Central Role of Reactive Oxygen Species in Oxidative Stress-Induced Disorders and Therapeutic Exploration. Front. Pharmacol. 2023, 14, 1269581. [Google Scholar] [CrossRef] [PubMed]
- Martínez, S.R.; Aiassa, V.; Sola, C.; Becerra, M.C. Oxidative Stress Response in Reference and Clinical Staphylococcus aureus Strains under Linezolid Exposure. J. Glob. Antimicrob. Resist. 2020, 22, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Hassett, D.J.; Imlay, J.A. Bactericidal Antibiotics and Oxidative Stress: A Radical Proposal. ACS Chem. Biol. 2007, 2, 708–710. [Google Scholar] [CrossRef]
- Tan, E.; Chin, C.S.H.; Lim, Z.F.S.; Ng, S.K. HEK293 Cell Line as a Platform to Produce Recombinant Proteins and Viral Vectors. Front. Bioeng. Biotechnol. 2021, 9, 796991. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.S.; Ashitate, Y.; Lee, J.H.; Kim, S.H.; Matsui, A.; Insin, N.; Bawendi, M.G.; Semmler-Behnke, M.; Frangioni, J.V.; Tsuda, A. Rapid Translocation of Nanoparticles from the Lung Airspaces to the Body. Nat. Biotechnol. 2010, 28, 1300–1303. [Google Scholar] [CrossRef] [PubMed]
- ALAtawi, M.K.; AlAsmari, A.A.; AlAliany, A.D.; Almajed, M.M.; Sakran, M.I. Silver Nanoparticles Forensic Uses and Toxicity on Vital Organs and Different Body Systems. Adv. Toxicol. Toxic Eff. 2024, 8, 15–29. [Google Scholar] [CrossRef]
- Noga, M.; Milan, J.; Frydrych, A.; Jurowski, K. Toxicological Aspects, Safety Assessment, and Green Toxicology of Silver Nanoparticles (AgNPs)—Critical Review: State of the Art. Int. J. Mol. Sci. 2023, 24, 5133. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shan, K.; Shao, X.; Shi, X.; He, Y.; Liu, Z.; Jacob, J.A.; Deng, L. Nanotoxic Effects of Silver Nanoparticles on Normal HEK-293 Cells in Comparison to Cancerous Hela Cell Line. Int. J. Nanomed. 2021, 16, 753–761. [Google Scholar] [CrossRef]
- Kaur, R.; Singh, K.; Agarwal, S.; Masih, M.; Chauhan, A.; Gautam, P.K. Silver Nanoparticles Induces Apoptosis of Cancer Stem Cells in Head and Neck Cancer. Toxicol. Rep. 2024, 12, 10–17. [Google Scholar] [CrossRef]
- Guibourdenche, M.; Roggentin, P.; Mikoleit, M.; Fields, P.I.; Bockemühl, J.; Grimont, P.A.D.; Weill, F.-X. Supplement 2003–2007 (No. 47) to the White-Kauffmann-Le Minor Scheme. Res. Microbiol. 2010, 161, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Hasman, H.; Mevius, D.; Veldman, K.; Olesen, I.; Aarestrup, F.M. β-Lactamases among Extended-Spectrum β-Lactamase (ESBL)-Resistant Salmonella from Poultry, Poultry Products and Human Patients in The Netherlands. J. Antimicrob. Chemother. 2005, 56, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Srisanga, S.; Angkititrakul, S.; Sringam, P.; Le Ho, P.T.; Vo, A.T.T.; Chuanchuen, R. Phenotypic and Genotypic Antimicrobial Resistance and Virulence Genes of Salmonella enterica Isolated from Pet Dogs and Cats. J. Vet. Sci. 2017, 18, 273–281. [Google Scholar] [CrossRef]
- Robicsek, A.; Jacoby, G.A.; Hooper, D.C. The Worldwide Emergence of Plasmid-Mediated Quinolone Resistance. Lancet Infect. Dis. 2006, 6, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Bullen, J.J.; Rogers, H.J.; Spalding, P.B.; Ward, C.G. Iron and Infection: The Heart of the Matter. FEMS Immunol. Med. Microbiol. 2005, 43, 325–330. [Google Scholar] [CrossRef]
- Ramachandran, G. Gram-Positive and Gram-Negative Bacterial Toxins in Sepsis. Virulence 2014, 5, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Mogrovejo, D.C.; Perini, L.; Gostinčar, C.; Sepčić, K.; Turk, M.; Ambrožič-Avguštin, J.; Brill, F.H.H.; Gunde-Cimerman, N. Prevalence of Antimicrobial Resistance and Hemolytic Phenotypes in Culturable Arctic Bacteria. Front. Microbiol. 2020, 11, 570. [Google Scholar] [CrossRef]
- Beshiru, A.; Igbinosa, I.H.; Igbinosa, E.O. Biofilm Formation and Potential Virulence Factors of Salmonella Strains Isolated from Ready-to-Eat Shrimps. PLoS ONE 2018, 13, e0204345. [Google Scholar] [CrossRef]
- Di Rosa, R.; Creti, R.; Venditti, M.; D’Amelio, R.; Arciola, C.R.; Montanaro, L.; Baldassarri, L. Relationship between Biofilm Formation, the Enterococcal Surface Protein (Esp) and Gelatinase in Clinical Isolates of Enterococcus faecalis and Enterococcus faecium. FEMS Microbiol. Lett. 2006, 256, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Teng, F.; Murray, B.E. Gelatinase Is Important for Translocation of Enterococcus faecalis across Polarized Human Enterocyte-Like T84 Cells. Infect. Immun. 2005, 73, 1606–1612. [Google Scholar] [CrossRef] [PubMed]
- El-Sebay, N.A.; Shady, H.M.A.; El-Zeedy, S.A.E.R.; Samy, A.A. InvA Gene Sequencing of Salmonella Typhimurium Isolated from Egyptian Poultry. Asian J. Sci. Res. 2017, 10, 194–202. [Google Scholar] [CrossRef]
- Fluit, A.C. Towards More Virulent and Antibiotic-Resistant Salmonella? FEMS Immunol. Med. Microbiol. 2005, 43, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Abd-Elghany, S.M.; Sallam, K.I.; Abd-Elkhalek, A.; Tamura, T. Occurrence, Genetic Characterization and Antimicrobial Resistance of Salmonella Isolated from Chicken Meat and Giblets. Epidemiol. Infect. 2015, 143, 997–1003. [Google Scholar] [CrossRef]
- Lamas, A.; Fernandez-No, I.C.; Miranda, J.M.; Vázquez, B.; Cepeda, A.; Franco, C.M. Prevalence, Molecular Characterization and Antimicrobial Resistance of Salmonella Serovars Isolated from Northwestern Spanish Broiler Flocks (2011–2015). Poult. Sci. 2016, 95, 2097–2105. [Google Scholar] [CrossRef]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids Carrying Antimicrobial Resistance Genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef] [PubMed]
- Salmonella Hadar. Available online: https://confluence.cornell.edu/display/foodsafety/salmonella+hadar (accessed on 2 November 2024).
- Dhanani, A.S.; Block, G.; Dewar, K.; Forgetta, V.; Topp, E.; Beiko, R.G.; Diarra, S.M. Genomic Comparison of Non-Typhoidal Salmonella enterica Serovars Typhimurium, Enteritidis, Heidelberg, Hadar and Kentucky Isolates from Broiler Chickens. PLoS ONE 2015, 10, e0137697. [Google Scholar] [CrossRef] [PubMed]
- Brandenburg, J.M.; Stapleton, G.S.; Kline, K.E.; Khoury, J.; Mallory, K.; Machesky, K.D.; Ladd-Wilson, S.G.; Scholz, R.; Freiman, J.; Schwensohn, C.; et al. Salmonella Hadar Linked to Two Distinct Transmission Vehicles Highlights Challenges to Enteric Disease Outbreak Investigations. Epidemiol. Infect. 2024, 152, e86. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, T.; Petridou, E.; Zdragas, A.; Nair, S.; Peters, T.; de Pinna, E.; Mandilara, G.; Passiotou, M.; Vatopoulos, A. Phenotypic and Molecular Characterization of Multidrug-Resistant Salmonella enterica Serovar Hadar in Greece, from 2007 to 2010. Clin. Microbiol. Infect. 2015, 21, 149.e1–149.e4. [Google Scholar] [CrossRef] [PubMed]
- Cernela, N.; Nüesch-Inderbinen, M.; Hächler, H.; Stephan, R. Antimicrobial Resistance Patterns and Genotypes of Salmonella enterica Serovar Hadar Strains Associated with Human Infections in Switzerland, 2005–2010. Epidemiol. Infect. 2014, 142, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Abgottspon, H.; Zurfluh, K.; Nüesch-Inderbinen, M.; Hächler, H.; Stephan, R. Quinolone Resistance Mechanisms in Salmonella enterica Serovars Hadar, Kentucky, Virchow, Schwarzengrund, and 4, 5, 12: I:−, isolated from Humans in Switzerland, and Identification of a Novel QnrD Variant, QnrD2, in S. Hadar. Antimicrob. Agents Chemother. 2014, 58, 3560–3563. [Google Scholar] [CrossRef]
- Nógrády, N.; Király, M.; Davies, R.; Nagy, B. Multidrug Resistant Clones of Salmonella Infantis of Broiler Origin in Europe. Int. J. Food Microbiol. 2012, 157, 108–112. [Google Scholar] [CrossRef]
- Cadel-Six, S.; Cherchame, E.; Douarre, P.-E.; Tang, Y.; Felten, A.; Barbet, P.; Litrup, E.; Banerji, S.; Simon, S.; Pasquali, F.; et al. The Spatiotemporal Dynamics and Microevolution Events That Favored the Success of the Highly Clonal Multidrug-Resistant Monophasic Salmonella Typhimurium Circulating in Europe. Front. Microbiol. 2021, 12, 651124. [Google Scholar] [CrossRef]
- Franco, A.; Leekitcharoenphon, P.; Feltrin, F.; Alba, P.; Cordaro, G.; Iurescia, M.; Tolii, R.; D’Iancu, M.; Staffolani, M.; Di Giannatale, E.; et al. Emergence of a Clonal Lineage of Multidrug-Resistant ESBL-Producing Salmonella Infantis Transmitted from Broilers and Broiler Meat to Humans in Italy between 2011 and 2014. PLoS ONE 2016, 10, e0144802. [Google Scholar] [CrossRef]
- Hindermann, D.; Gopinath, G.; Chase, H.; Negrete, F.; Althaus, D.; Zurfluh, K.; Tall, B.D.; Stephan, R.; Nüesch-Inderbinen, M. Salmonella enterica Serovar Infantis from Food and Human Infections, Switzerland, 2010–2015: Poultry-Related Multidrug Resistant Clones and an Emerging ESBL Producing Clonal Lineage. Front. Microbiol. 2017, 8, 1322. [Google Scholar] [CrossRef] [PubMed]
- Hauser, E.; Tietze, E.; Helmuth, R.; Junker, E.; Prager, R.; Schroeter, A.; Rabsch, W.; Fruth, A.; Toboldt, A.; Malorny, B. Clonal Dissemination of Salmonella enterica Serovar Infantis in Germany. Foodborne Pathog. Dis. 2012, 9, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Papić, B.; Kušar, D.; Mićunović, J.; Pirš, M.; Ocepek, M.; Avberšek, J. Clonal Spread of PESI-Positive Multidrug-Resistant ST32 Salmonella enterica Serovar Infantis Isolates among Broilers and Humans in Slovenia. Microbiol. Spectr. 2022, 10, e02481-22. [Google Scholar] [CrossRef]
- Tîrziu, E.; Lazăr, R.; Sala, C.; Nichita, I.; Morar, A.; Şereş, M.; Imre, K. Salmonella in Raw Chicken Meat from the Romanian Seaside: Frequency of Isolation and Antibiotic Resistance. J. Food Prot. 2015, 78, 1003–1006. [Google Scholar] [CrossRef]
- Mihaiu, L.; Lapusan, A.; Tanasuica, R.; Sobolu, R.; Mihaiu, R.; Oniga, O.; Mihaiu, M. First Study of Salmonella in Meat in Romania. J. Infect. Dev. Ctries. 2014, 8, 50–58. [Google Scholar] [CrossRef]
- Usein, C.-R.; Oprea, M.; Ciontea, A.S.; Dinu, S.; Cristea, D.; Zota, L.C.; Kotila, S. A Snapshot of the Genetic Diversity of Salmonella Enteritidis Population Involved in Human Infections in Romania Taken in the European Epidemiological Context. Pathogens 2021, 10, 1490. [Google Scholar] [CrossRef] [PubMed]
- Petrin, S.; Wijnands, L.; Benincà, E.; Mughini-Gras, L.; Delfgou-van Asch, E.H.M.; Villa, L.; Orsini, M.; Losasso, C.; Olsen, J.E.; Barco, L. Assessing Phenotypic Virulence of Salmonella enterica across Serovars and Sources. Front. Microbiol. 2023, 14, 1184387. [Google Scholar] [CrossRef] [PubMed]
- Teklemariam, A.D.; Al-Hindi, R.R.; Albiheyri, R.S.; Alharbi, M.G.; Alghamdi, M.A.; Filimban, A.A.R.; Al Mutiri, A.S.; Al-Alyani, A.M.; Alseghayer, M.S.; Almaneea, A.M.; et al. Human Salmonellosis: A Continuous Global Threat in the Farm-to-Fork Food Safety Continuum. Foods 2023, 12, 1756. [Google Scholar] [CrossRef]
- Mattock, J.; Chattaway, M.A.; Hartman, H.; Dallman, T.; Smith, A.; Keddy, K.; Petrovska, L.; Manners, E.; Duze, S.; Smouse, S.; et al. A One Health Perspective on Salmonella enterica Serovar Infantis, an Emerging Human Multidrug-Resistant Pathogen. Emerg. Infect. Dis. J. 2024, 30, 701. [Google Scholar] [CrossRef] [PubMed]
- Seong, M.; Lee, D.G. Silver Nanoparticles Against Salmonella enterica Serotype Typhimurium: Role of Inner Membrane Dysfunction. Curr. Microbiol. 2017, 74, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Alfei, S.; Schito, G.C.; Schito, A.M.; Zuccari, G. Reactive Oxygen Species (ROS)-Mediated Antibacterial Oxidative Therapies: Available Methods to Generate ROS and a Novel Option Proposal. Int. J. Mol. Sci. 2024, 25, 7182. [Google Scholar] [CrossRef]
- da Cruz Nizer, W.S.; Adams, M.E.; Allison, K.N.; Montgomery, M.C.; Mosher, H.; Cassol, E.; Overhage, J. Oxidative Stress Responses in Biofilms. Biofilm 2024, 7, 100203. [Google Scholar] [CrossRef]
- Seixas, A.F.; Quendera, A.P.; Sousa, J.P.; Silva, A.F.Q.; Arraiano, C.M.; Andrade, J.M. Bacterial Response to Oxidative Stress and RNA Oxidation. Front. Genet. 2022, 12, 821535. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, L.; Wang, W.; Gao, H. Nitric Oxide, Nitric Oxide Formers and Their Physiological Impacts in Bacteria. Int. J. Mol. Sci. 2022, 23, 10778. [Google Scholar] [CrossRef] [PubMed]
- Do, T.; Vaculciakova, S.; Kluska, K.; Peris-Díaz, M.D.; Priborsky, J.; Guran, R.; Krężel, A.; Adam, V.; Zitka, O. Antioxidant-Related Enzymes and Peptides as Biomarkers of Metallic Nanoparticles (Eco)Toxicity in the Aquatic Environment. Chemosphere 2024, 364, 142988. [Google Scholar] [CrossRef] [PubMed]
- Kędziora, A.; Speruda, M.; Krzyżewska, E.; Rybka, J.; Łukowiak, A.; Bugla-Płoskońska, G. Similarities and Differences between Silver Ions and Silver in Nanoforms as Antibacterial Agents. Int. J. Mol. Sci. 2018, 19, 444. [Google Scholar] [CrossRef]
- Dias de Emery, B.; Zottis Chitolina, G.; Qadir, M.I.; Quedi Furian, T.; Apellanis Borges, K.; de Souza Moraes, H.L.; Pippi Salle, C.T.; Pinheiro do Nascimento, V. Antimicrobial and Antibiofilm Activity of Silver Nanoparticles against Salmonella Enteritidis. Braz. J. Microbiol. 2023, 54, 285–292. [Google Scholar] [CrossRef]
- Abd-Elhakeem, M.A.; Badawy, I.; Raafat, A. Efficacy of Silver Nanoparticles as Antimicrobial Agent against Salmonella Infection and Accompanied Biochemical. Immunol. Histopathol. Chang. Rats 2016, 54, 13–19. [Google Scholar]
- Sanguiñedo, P.; Fratila, R.; Estevez, M.B.; Fuente, J.; Grazu, V.; Alborés, S. Extracellular Biosynthesis of Silver Nanoparticles Using Fungi and Their Antibacterial Activity. Nano Biomed. Eng. 2018, 10, 165–173. [Google Scholar] [CrossRef]
- Chiao, S.H.; Lin, S.H.; Shen, C.I.; Liao, J.W.; Bau, I.J.; Wei, J.C.; Tseng, L.P.; Hsu, S.; Lai, P.S.; Lin, S.Z.; et al. Efficacy and Safety of Nanohybrids Comprising Silver Nanoparticles and Silicate Clay for Controlling Salmonella Infection. Int. J. Nanomed. 2012, 7, 2421–2432. [Google Scholar] [CrossRef]
- Yakoup, A.Y.; Kamel, A.G.; Elbermawy, Y.; Abdelsattar, A.S.; El-Shibiny, A. Characterization, Antibacterial, and Cytotoxic Activities of Silver Nanoparticles Using the Whole Biofilm Layer as a Macromolecule in Biosynthesis. Sci. Rep. 2024, 14, 364. [Google Scholar] [CrossRef]
- Shumbula, N.P.; Nkabinde, S.S.; Ndala, Z.B.; Mpelane, S.; Shumbula, M.P.; Mdluli, P.S.; Njengele-Tetyana, Z.; Tetyana, P.; Hlatshwayo, T.; Mlambo, M.; et al. Evaluating the Antimicrobial Activity and Cytotoxicity of Polydopamine Capped Silver and Silver/Polydopamine Core-Shell Nanocomposites. Arab. J. Chem. 2022, 15, 103798. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Sriram, B.; Sathiyaseelan, A.; Mariadoss, A.V.A.; Hu, X.; Han, K.-S.; Vishnupriya, V.; MubarakAli, D.; Wang, M.-H. Synthesis, Characterization, and Cytotoxicity of Starch-Encapsulated Biogenic Silver Nanoparticle and Its Improved Anti-Bacterial Activity. Int. J. Biol. Macromol. 2021, 182, 1409–1418. [Google Scholar] [CrossRef] [PubMed]
- Sathiyaseelan, A.; Saravanakumar, K.; Mariadoss, A.V.A.; Wang, M.-H. Biocompatible Fungal Chitosan Encapsulated Phytogenic Silver Nanoparticles Enhanced Antidiabetic, Antioxidant and Antibacterial Activity. Int. J. Biol. Macromol. 2020, 153, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Abdelmoneim, D.; Porter, G.; Duncan, W.; Lim, K.; Easingwood, R.; Woodfield, T.; Coates, D. Three-Dimensional Evaluation of the Cytotoxicity and Antibacterial Properties of Alpha Lipoic Acid-Capped Silver Nanoparticle Constructs for Oral Applications. Nanomaterials 2023, 13, 052007. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Chen, L.; Qiu, X. Rapid Synthesis of Ferulic Acid-Derived Lignin Coated Silver Nanoparticles with Low Cytotoxicity and High Antibacterial Activity. Int. J. Biol. Macromol. 2024, 277, 134471. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, H.M.; Mosleh, A.M.; Elghany, A.A.; Shams-Eldin, E.; Abu Serea, E.S.; Ali, S.A.; Shalan, A.E. Coated Silver Nanoparticles: Synthesis, Cytotoxicity, and Optical Properties. RSC Adv. 2019, 9, 20118–20136. [Google Scholar] [CrossRef] [PubMed]
- Roy, N.; Sengupta, R.; Bhowmick, A.K. Modifications of Carbon for Polymer Composites and Nanocomposites. Prog. Polym. Sci. 2012, 37, 781–819. [Google Scholar] [CrossRef]
- Ferdous, Z.; Nemmar, A. Health Impact of Silver Nanoparticles: A Review of the Biodistribution and Toxicity Following Various Routes of Exposure. Int. J. Mol. Sci. 2020, 21, 2375. [Google Scholar] [CrossRef]
- Hashim, M.; Mujahid, H.; Hassan, S.; Bukhari, S.; Anjum, I.; Hano, C.; Abbasi, B.H.; Anjum, S. Implication of Nanoparticles to Combat Chronic Liver and Kidney Diseases: Progress and Perspectives. Biomolecules 2022, 12, 1337. [Google Scholar] [CrossRef]
- Corbu, V.M.; Dumbravă, A.Ş.; Marinescu, L.; Motelica, L.; Chircov, C.; Surdu, A.V.; Gheorghe-Barbu, I.; Pecete, I.; Balotescu, I.; Popa, M.; et al. Alternative Mitigating Solutions Based on Inorganic Nanoparticles for the Preservation of Cultural Heritage. Front. Mater. 2023, 10, 1272869. [Google Scholar] [CrossRef]
- Xuan, L.; Ju, Z.; Skonieczna, M.; Zhou, P.-K.; Huang, R. Nanoparticles-Induced Potential Toxicity on Human Health: Applications, Toxicity Mechanisms, and Evaluation Models. MedComm 2023, 4, e327. [Google Scholar] [CrossRef] [PubMed]
- Awashra, M.; Młynarz, P. The Toxicity of Nanoparticles and Their Interaction with Cells: An in Vitro Metabolomic Perspective. Nanoscale Adv. 2023, 5, 2674–2723. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Truşcă, B.S.; Gheorghe-Barbu, I.; Manea, M.; Ianculescu, E.; Barbu, I.C.; Măruțescu, L.G.; Dițu, L.-M.; Chifiriuc, M.-C.; Lazăr, V. Snapshot of Phenotypic and Molecular Virulence and Resistance Profiles in Multidrug-Resistant Strains Isolated in a Tertiary Hospital in Romania. Pathogens 2023, 12, 609. [Google Scholar] [CrossRef] [PubMed]
- Lazar, V.; Herlea, V.; Cernat, R.; Bulai, D.; Balotescu, C.M.; Moraru, A. Microbiologie General-Manual De Lucrări Practice; Editura Universitatii din Bucuresti: București, Romania, 2004; p. 320. [Google Scholar]
- Stepanović, S.; Vuković, D.; Hola, V.; Bonaventura, G.D.I.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of Biofilm in Microtiter Plates: Overview of Testing Conditions and Practical Recommendations for Assessment of Biofilm Production by Staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Gheorghe, I.; Cristea, V.C.; Marutescu, L.; Popa, M.; Murariu, C.; Trusca, B.S.; Borcan, E.; Ghita, C.; Lazar, V.; Chifiriuc, M.C. Resistance and Virulence Features in Carbapenem-Resistant Acinetobacter baumannii Community Acquired and Nosocomial Isolates in Romania. Rev. Chim. 2019, 70, 3502–3507. [Google Scholar] [CrossRef]
- Eftekhar, F.; Hosseini-Mazinani, S.M.; Ghandili, S.; Hamraz, M.; Zamani, S. PCR Detection of Plasmid Mediated TEM, SHV and AmpC? —Lactamases in Community and Nosocomialurinary Isolates of Escherichia coli. Iran. J. Biotechnol. 2005, 3, 48–54. [Google Scholar]
- Bali, E.; Açık, L.; Sultan, N. Phenotypic and Molecular Characterization of SHV, TEM, CTX-M and Extended-Spectrum β-Lactamase Produced by Escherichia coli, Acinobacter baumannii and Klebsiella Isolates in a Turkish Hospital. Afr. J. Microbiol. Res. 2010, 4, 650–654. [Google Scholar]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for Detection of Acquired Carbapenemase Genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I.; Chee-Sanford, J.C.; Garrigues, N.; Teferedegne, B.; Krapac, I.J.; White, B.A.; Mackie, R.I. Development, Validation, and Application of PCR Primers for Detection of Tetracycline Efflux Genes of Gram-Negative Bacteria. Appl. Environ. Microbiol. 2002, 68, 1786–1793. [Google Scholar] [CrossRef] [PubMed]
- Frank, T.; Gautier, V.; Talarmin, A.; Bercion, R.; Arlet, G. Characterization of Sulphonamide Resistance Genes and Class 1 Integron Gene Cassettes in Enterobacteriaceae, Central African Republic (CAR). J. Antimicrob. Chemother. 2007, 59, 742–745. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.-J.; Nordmann, P. Multiplex PCR for Detection of Plasmid-Mediated Quinolone Resistance Qnr Genes in ESBL-Producing Enterobacterial Isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.H.; Ou, J.T. Rapid Identification of Salmonella Serovars in Feces by Specific Detection of Virulence Genes, InvA and SpvC, by an Enrichment Broth Culture-Multiplex PCR Combination Assay. J. Clin. Microbiol. 1996, 34, 2619–2622. [Google Scholar] [CrossRef] [PubMed]
- Rahn, K.; De Grandis, S.A.; Clarke, R.C.; McEwen, S.A.; Galán, J.E.; Ginocchio, C.; Curtiss, R.; Gyles, C.L. Amplification of an InvA Gene Sequence of Salmonella Typhimurium by Polymerase Chain Reaction as a Specific Method of Detection of Salmonella. Mol. Cell. Probes 1992, 6, 271–279. [Google Scholar] [CrossRef]
- Seemann, T. Shovill. Available online: https://github.com/tseemann/shovill (accessed on 1 November 2024).
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and Standardized Annotation of Bacterial Genomes via Alignment-Free Sequence Identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef]
- Seemann, T. ABRicate. Available online: https://github.com/tseemann/abricate (accessed on 1 November 2024).
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and Refined Dataset for Big Data Analysis—10 Years On. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef] [PubMed]
- Gheorghe-Barbu, I.; Surleac, M.; Barbu, I.C.; Paraschiv, S.; Bănică, L.M.; Rotaru, L.-I.; Vrâncianu, C.O.; Niță Lazăr, M.; Oțelea, D.; Chifiriuc, M.C. Decoding the Resistome, Virulome and Mobilome of Clinical versus Aquatic Acinetobacter Baumannii in Southern Romania. Heliyon 2024, 10, e33372. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. MLST. Available online: https://github.com/tseemann/mlst (accessed on 1 November 2024).
- Yoshida, C.E.; Kruczkiewicz, P.; Laing, C.R.; Lingohr, E.J.; Gannon, V.P.; Nash, J.H.; Taboada, E.N. The Salmonella In Silico Typing Resource (SISTR): An Open Web-Accessible Tool for Rapidly Typing and Subtyping Draft Salmonella Genome Assemblies. PLoS ONE 2016, 11, e0147101. [Google Scholar] [CrossRef]
- Pathogen Watch. PathogenWatch. Available online: https://pathogen.watch/ (accessed on 1 November 2024).
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; Abudahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An Interactive Viewer for Bacterial Population Genomics. Bioinformatics 2018, 34, 292–293. [Google Scholar] [CrossRef] [PubMed]
- Corbu, V.M.; Georgescu, A.M.; Marinas, I.C.; Pericleanu, R.; Mogos, D.V.; Dumbravă, A.Ș.; Marinescu, L.; Pecete, I.; Vassu-Dimov, T.; Czobor Barbu, I.; et al. Phenotypic and Genotypic Characterization of Resistance and Virulence Markers in Candida Spp. Isolated from Community-Acquired Infections in Bucharest, and the Impact of AgNPs on the Highly Resistant Isolates. J. Fungi 2024, 10, 563. [Google Scholar] [CrossRef]
- Braik, A.; Lahouel, M.; Merabet, R.; Djebar, M.R.; Morin, D. Myocardial Protection by Propolis during Prolonged Hypothermic Preservation. Cryobiology 2019, 88, 29–37. [Google Scholar] [CrossRef] [PubMed]
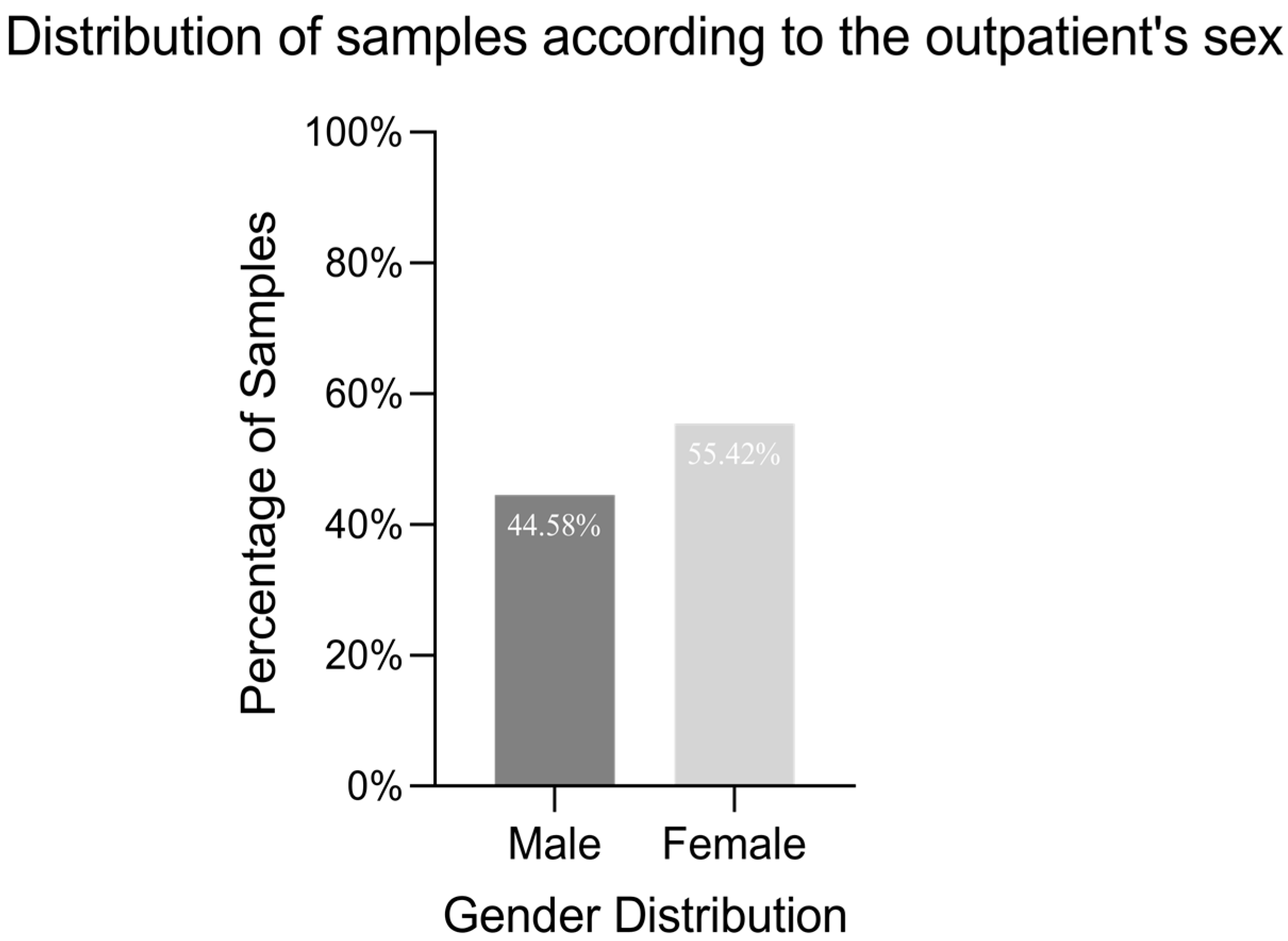
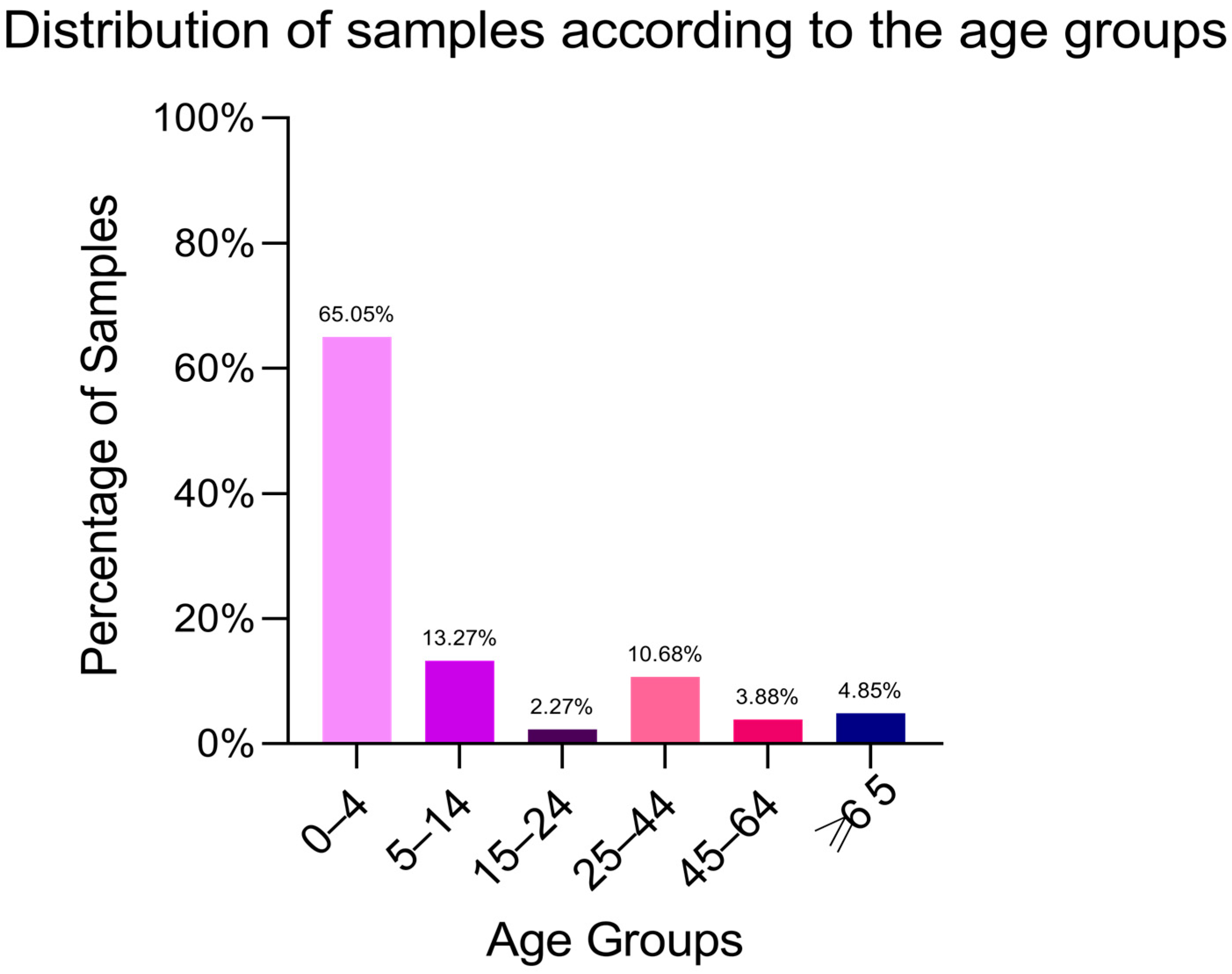



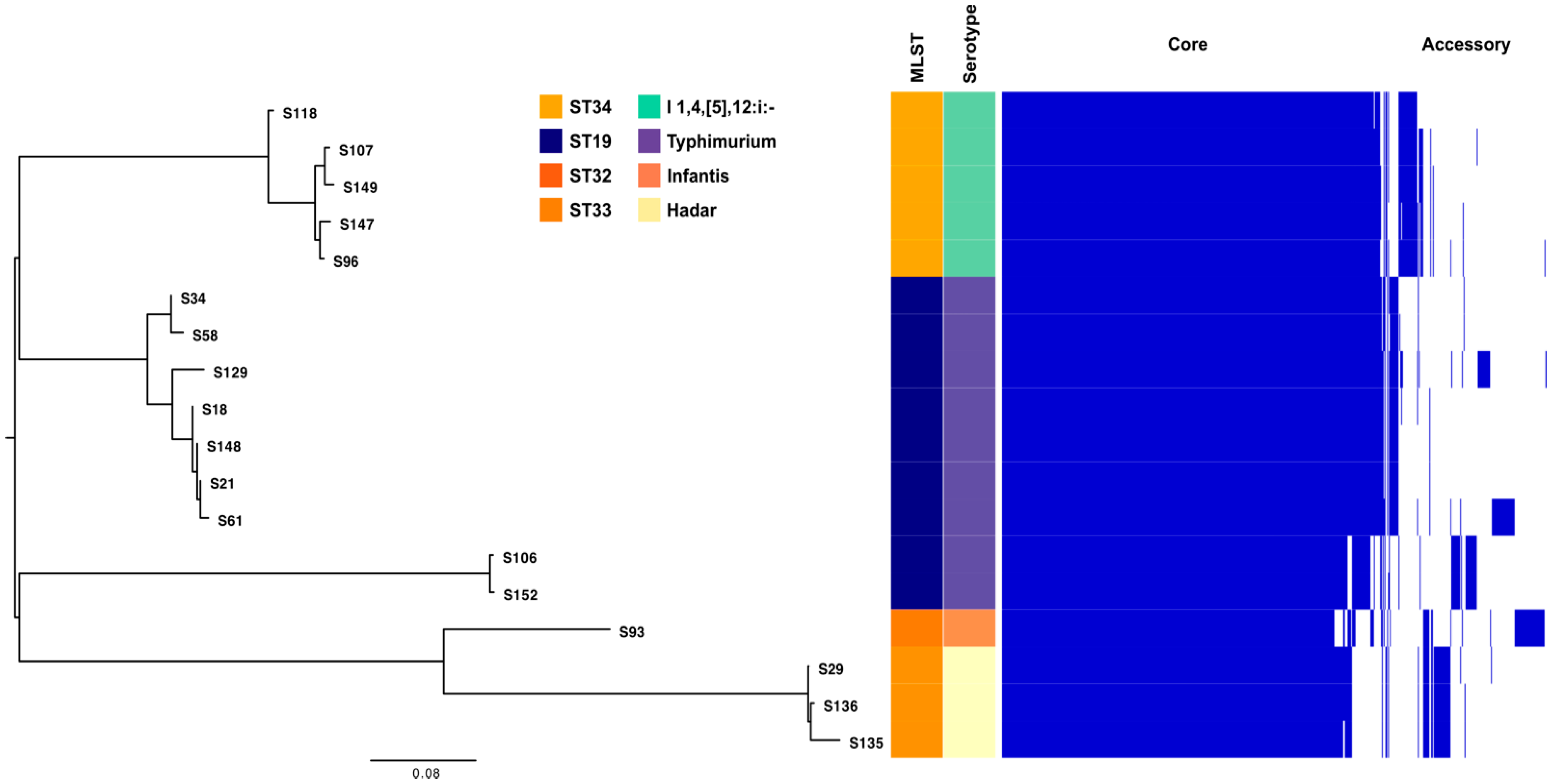
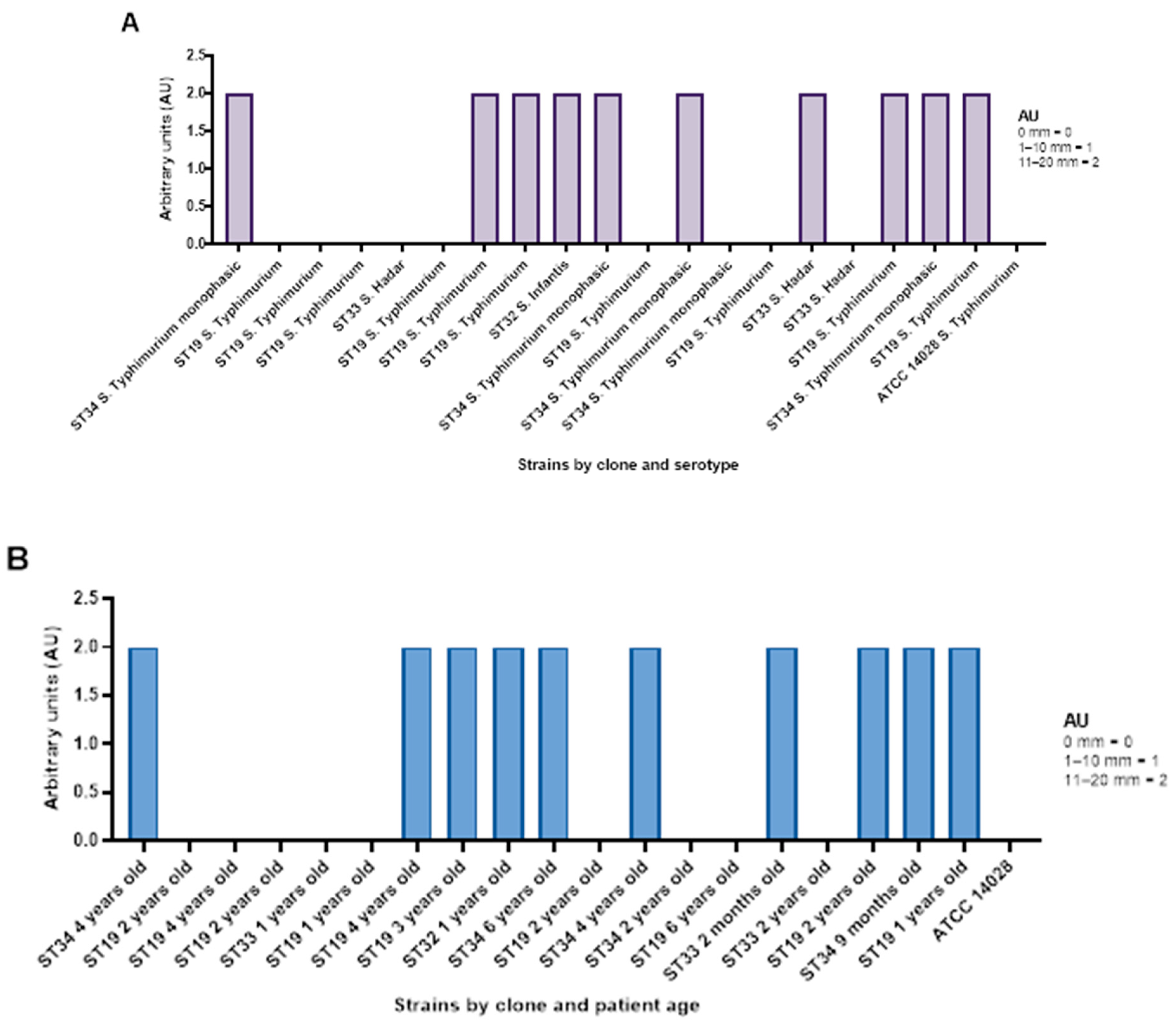


| Bacterial Strain | Age Group | Number of Strains (%) | Isolation Source | β- and α-Haemolysins (%) | Lecitinase (%) | Amylase (%) | Lipase (%) | Caseinase (%) | Gelatinase (%) | Esculin Hydrolysis (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Salmonella spp. | 0–4 | 201 (65.05%) | Stool samples | 201 (100%, β) | 38 (19%) | 45 (22%) | 108 (54%) | 80 (40%) | 189 (94%) | 201 (100%) |
| 5–14 | 41 (13.27%) | 41 (100%, β) | 2 (5%) | 7 (17%) | 33 (80%) | 5 (12%) | 37 (90%) | 41 (100%) | ||
| 15–24 | 7 (2.27%) | 5 (71%, α) | 0 | 1 (14%) | 5 (71%) | 5 (71%) | 2 (29%) | 5 (71%) | ||
| 25–44 | 33 (10.68%) | 15 (45%, α) | 1 (3%) | 0 | 2 (6%) | 0 | 12 (36%) | 21 (64%) | ||
| 45–64 | 12 (3.88%) | 8 (66%, α) | 0 | 0 | 2 (17%) | 0 | 11 (91%) | 4 (33%) | ||
| >65 | 15 (4.85%) | 3 (20%, α) | 0 | 0 | 1 (7%) | 0 | 5 (33%) | 3 (20%) |
| Antibiotic Classes | ARGs | 0–4 Years (%) | Total No of Strains-17 | 6 Years (%) | Total No of Strains-2 | STs | |||
|---|---|---|---|---|---|---|---|---|---|
| 34 | 19 | 33 | 32 | ||||||
| β-lactams | blaTEM-1 | 70.58 | 12 | 100 | 2 | ||||
| Aminoglycosides | aadA1 | 0 | 50 | 1 | |||||
| aadA2 | 0 | 5.88 | 1 | ||||||
| aadA5 | 5.88 | 1 | 0 | ||||||
| ant(2″)-Ia | 0 | 50 | 1 | ||||||
| aph(3″)-Ib | 35.29 | 6 | 50 | 1 | |||||
| aph(6)-Id | 35.29 | 6 | 50 | 1 | |||||
| Quinolones | qnrA1 | 35.29 | 6 | 50 | 1 | ||||
| qnrB19 | 17.64 | 3 | 0 | ||||||
| Sulphonamides | sul1 | 47.05 | 8 | 0 | |||||
| sul2 | 23.52 | 4 | 50 | 1 | |||||
| sul3 | 0 | 50 | 1 | ||||||
| Tetracyclines | tet(B) | 11.76 | 2 | 50 | 1 | ||||
| tet(A) | 17.64 | 3 | 0 | ||||||
| Macrolides | mph(A) | 11.76 | 2 | 0 | |||||
| Thrimethoprim | dfrA5 | 47.05 | 8 | 0 | |||||
| dfrA12 | 0 | 50 | 1 | ||||||
| drfA17 | 5.88 | 1 | 0 | ||||||
| Chloramphenicol | cmlA1 | 0 | 50 | 1 | |||||
| cmlA5 | 35.29 | 6 | 0 | ||||||
| Serotype | Antigen Structure | Number of Isolates | ST |
|---|---|---|---|
| Salmonella Typhimurium | 1,4,[5],12:i:1,2 | 10 | 19 |
| Salmonella Typhimurium monophasic | 1,4,[5],12:i:- | 5 | 34 |
| Salmonella Hadar | 6,8:z10:e,n,x | 3 | 33 |
| Salmonella Infantis | 6,7,14:r:1,5 | 1 | 32 |
| Receptor | Donors | ARG | Conjugation Frequency | Transferred ARGs |
|---|---|---|---|---|
| Escherichia coli J53 | S135 S. Hadar | tet(A) | 3.5 × 10−10 | - |
| S136 S. Hadar | tet(A) | 2.36 × 10−10 | - | |
| S29 S. Hadar | tet(A) | 3.3 × 10−6 | - | |
| S96 S. 1,4,[5],12:i:- | tet(B) | 4.66 × 10−1 | - | |
| S107 S. 1,4,[5],12:i:- | tet (B) | 3.33 × 10−1 | - | |
| S149 S. 1,4,[5],12:i:- | tet(B) | 3.5 | - |
| Class | Antibiotic(s) Tested |
|---|---|
| β-lactams inhibitors | Amoxicillin-clavulanate |
| Ampicillin-sulbactam | |
| Piperacillin-tazobactam | |
| Monobactams | Aztreonam |
| Imipenem | |
| Carbapenems | Ertapenem |
| Meropenem | |
| Doripenem | |
| Folate Pathway Antagonists | Trimethoprim-sulfamethoxazole |
| Ampicillin | |
| Penicillins | Piperacillin |
| Tetracyclines | Tetracycline |
| Quinolones and Fluoroquinolones | Ciprofloxacin Ofloxacin Levofloxacin Moxifloxacin |
| Cefepime | |
| Cephalosporins | Cefotaxime |
| Ceftazidime | |
| Ceftriaxone | |
| Cefpodoxime | |
| Other | Tigecycline |
| Chloramphenicol | |
| Fosfomycin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gheorghe-Barbu, I.; Czobor Barbu, I.; Dragomir, R.-I.; Marinaș, I.C.; Stan, M.S.; Pericleanu, R.; Dumbravă, A.Ș.; Rotaru, L.-I.; Paraschiv, S.; Bănică, L.M.; et al. Emerging Resistance and Virulence Patterns in Salmonella enterica: Insights into Silver Nanoparticles as an Antimicrobial Strategy. Antibiotics 2025, 14, 46. https://doi.org/10.3390/antibiotics14010046
Gheorghe-Barbu I, Czobor Barbu I, Dragomir R-I, Marinaș IC, Stan MS, Pericleanu R, Dumbravă AȘ, Rotaru L-I, Paraschiv S, Bănică LM, et al. Emerging Resistance and Virulence Patterns in Salmonella enterica: Insights into Silver Nanoparticles as an Antimicrobial Strategy. Antibiotics. 2025; 14(1):46. https://doi.org/10.3390/antibiotics14010046
Chicago/Turabian StyleGheorghe-Barbu, Irina, Ilda Czobor Barbu, Rareș-Ionuț Dragomir, Ioana Cristina Marinaș, Miruna Silvia Stan, Radu Pericleanu, Andreea Ștefania Dumbravă, Liviu-Iulian Rotaru, Simona Paraschiv, Leontina Mirela Bănică, and et al. 2025. "Emerging Resistance and Virulence Patterns in Salmonella enterica: Insights into Silver Nanoparticles as an Antimicrobial Strategy" Antibiotics 14, no. 1: 46. https://doi.org/10.3390/antibiotics14010046
APA StyleGheorghe-Barbu, I., Czobor Barbu, I., Dragomir, R.-I., Marinaș, I. C., Stan, M. S., Pericleanu, R., Dumbravă, A. Ș., Rotaru, L.-I., Paraschiv, S., Bănică, L. M., Pecete, I., Oțelea, D., Cristea, V. C., Popa, M. I., Țânțu, M. M., & Surleac, M. (2025). Emerging Resistance and Virulence Patterns in Salmonella enterica: Insights into Silver Nanoparticles as an Antimicrobial Strategy. Antibiotics, 14(1), 46. https://doi.org/10.3390/antibiotics14010046








