Pharmacodynamic Profiling of Amoxicillin: Targeting Multidrug-Resistant Gram-Positive Pathogens Staphylococcus aureus and Staphylococcus pseudintermedius in Canine Clinical Isolates †
Abstract
:1. Introduction
2. Results
2.1. Evaluation of MIC MBC and ECOFF in Antimicrobial Studies
2.2. Mutation Prevention Concentration Analysis
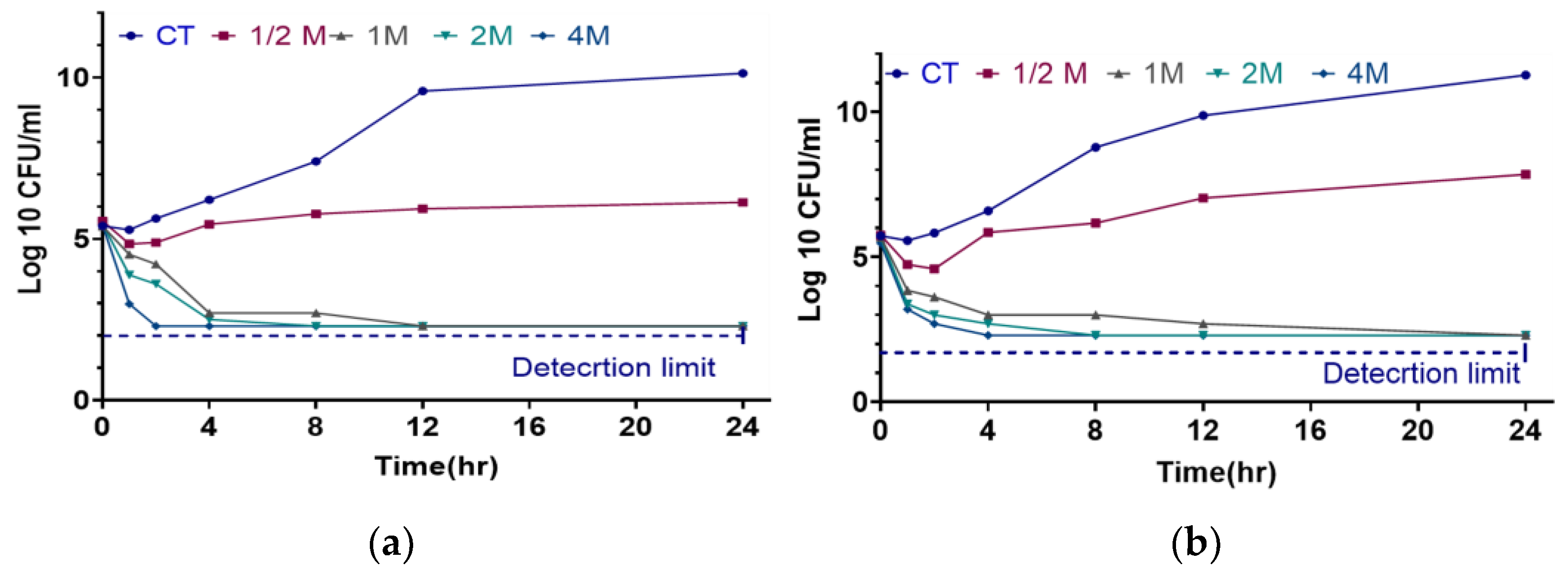
2.3. Time-Kill Assay
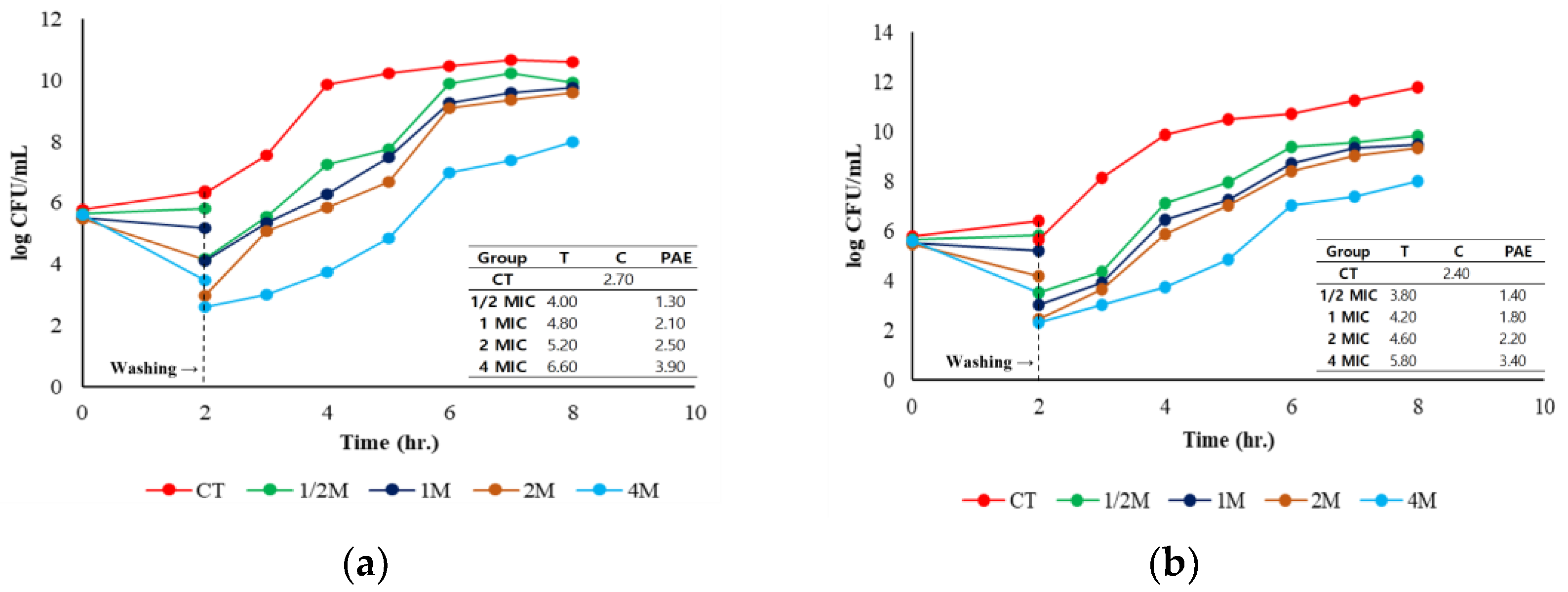
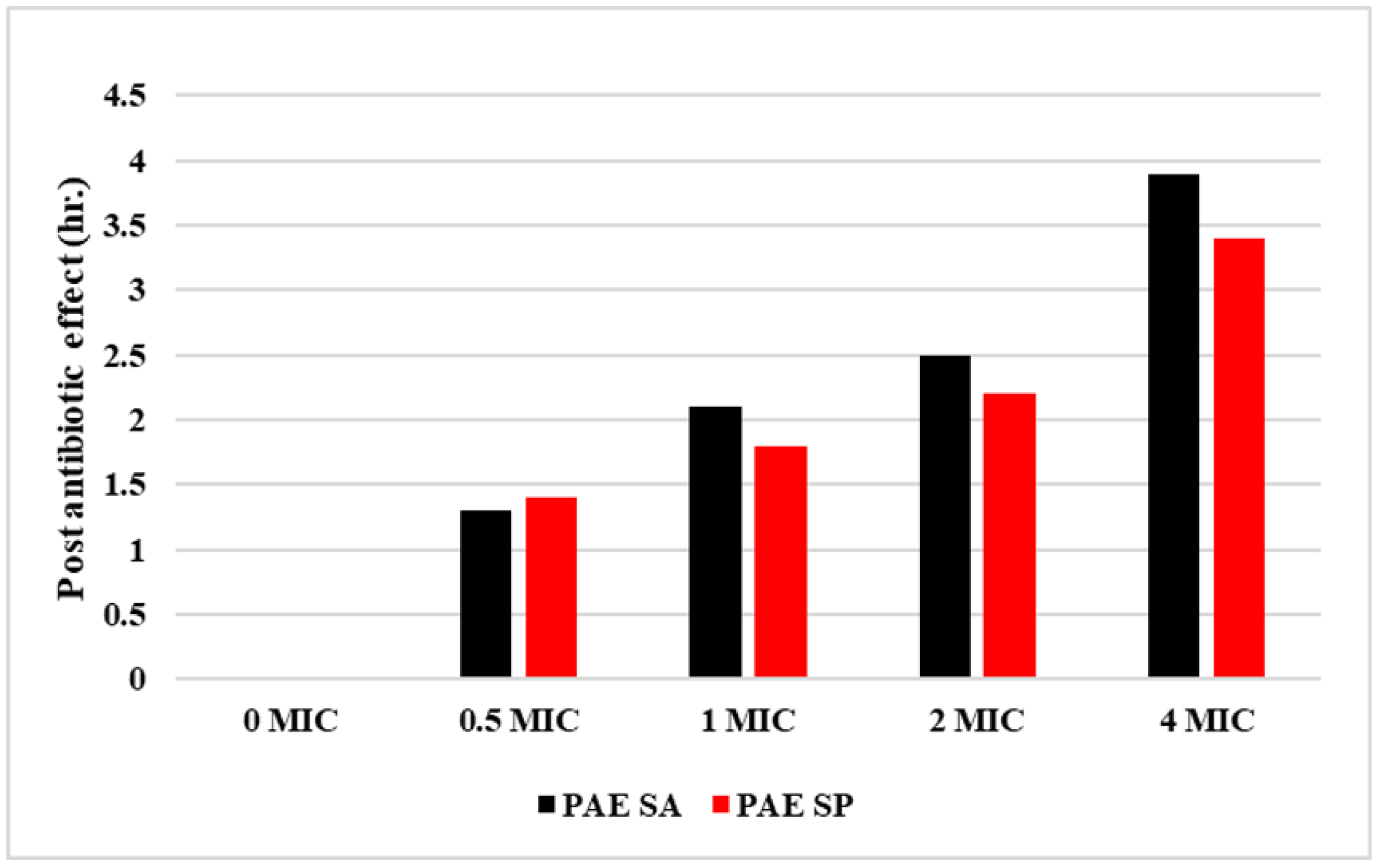
2.4. Post-Antibiotic Effect
3. Discussion
4. Materials and Methods
4.1. Chemicals, Reagents, and Bacterial Strains
4.2. S. aureus and S. pseudintermedius Strains, Culture Conditions, and Media
4.3. Minimum Inhibitory and Bactericidal Concentration
4.4. Mutation Prevention Concentration
4.5. Time-Kill Assay
4.6. Post-Antibiotic Effect
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chai, M.H.; Sukiman, M.Z.; Liew, Y.W.; Shapawi, M.S.; Roslan, F.S.; Hashim, S.N.; Mohamad, N.M.; Ariffin, S.M.Z.; Ghazali, M.F. Detection, molecular characterization, and antibiogram of multi-drug resistant and methicillin-resistant Staphylococcus aureus (MRSA) isolated from pets and pet owners in Malaysia. Iran. J. Vet. Res. 2021, 22, 277. [Google Scholar] [CrossRef] [PubMed]
- Grousd, J.A.; Rich, H.E.; Alcorn, J.F. Host-Pathogen Interactions in Gram-Positive Bacterial Pneumonia. Clin. Microbiol. Rev. 2019, 32, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Weese, J.S.; Prescott, J.F. Staphylococcal Infections. In Greene’s Infectious Diseases of the Dog and Cat, 5th ed.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 611–626. [Google Scholar] [CrossRef]
- Weese, J.; van Duijkeren, E. Methicillin-Resistant Staphylococcus aureus and Staphylococcus pseudintermedius in Veterinary Medicine. Vet. Microbiol. 2010, 140, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Hisata, K.; Kuwahara-Arai, K.; Yamanoto, M.; Ito, T.; Nakatomi, Y.; Cui, L.; Baba, T.; Terasawa, M.; Sotozono, C.; Kinoshita, S.; et al. Dissemination of methicillin-resistant staphylococci among healthy Japanese children. Am. Soc. Microbiol. 2005, 43, 3364–3372. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, K.; Shimokubo, N.; Sakagami, A.; Ueno, H.; Muramatsu, Y.; Kadosawa, T.; Yanagisawa, C.; Hanaki, H.; Nakajima, C.; Suzuki, Y.; et al. Occurrence and Molecular Characteristics of Methicillin-Resistant Staphylococcus aureus and Methicillin-Resistant Staphylococcus pseudintermedius in an Academic Veterinary Hospital. Appl. Environ. Microbiol. 2010, 76, 5165–5174. [Google Scholar] [CrossRef]
- Loeffler, A.; Boag, A.K.; Sung, J.; Lindsay, J.A.; Guardabassi, L.; Dalsgaard, A.; Smith, H.; Stevens, K.B.; Lloyd, D.H. Prevalence of methicillin-resistant Staphylococcus aureus among staff and pets in a small animal referral hospital in the UK. J. Antimicrob. Chemother. 2005, 56, 692–697. [Google Scholar] [CrossRef]
- Boost, M.; O’Donoghue, M.; Siu, K.H. Characterisation of methicillin-resistant Staphylococcus aureus isolates from dogs and their owners. Clin. Microbiol. Infect. 2007, 13, 731–733. [Google Scholar] [CrossRef]
- Sasaki, T.; Kikuchi, K.; Tanaka, Y.; Takahashi, N.; Kamata, S.; Hiramatsu, K. Methicillin-Resistant Staphylococcus pseudintermedius in a Veterinary Teaching Hospital. J. Clin. Microbiol. 2007, 45, 1118–1125. [Google Scholar] [CrossRef]
- Moodley, A.; Nightingale, E.C.; Stegger, M.; Nielsen, S.S.; Skov, R.L.; Guardabassi, L. High risk for nasal carriage of methicillin-resistant Staphylococcus aureus among Danish veterinary practitioners. Scand. J. Work. Environ. Health 2008, 34, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Ubukata, K.; Nonoguchi, R.; Matsuhashi, M.; Konno, M. Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J. Bacteriol. 1989, 171, 2882–2885. [Google Scholar] [CrossRef]
- Hanselman, B.; Kruth, S.; Weese, J.S. Methicillin-resistant staphylococcal colonization in dogs entering a veterinary teaching hospital. Vet. Microbiol. 2008, 126, 277–281. [Google Scholar] [CrossRef]
- Morris, D.O.; Rook, K.A.; Shofer, F.S.; Rankin, S.C. Screening of Staphylococcus aureus, Staphylococcus intermedius, and Staphylococcus schleiferi isolates obtained from small companion animals for antimicrobial resistance: A retrospective review of 749 isolates (2003–04). Veter. Dermatol. 2006, 17, 332–337. [Google Scholar] [CrossRef]
- Kitano, K.; Tomasz, A. Triggering of Autolytic Cell Wall Degradation in Escherichia coli by Beta-Lactam Antibiotics. Antimicrob. Agents Chkmotherapy 1979, 16, 8384848. [Google Scholar] [CrossRef]
- Dubée, V.; Chau, F.; Arthur, M.; Garry, L.; Benadda, S.; Mesnage, S.; Lefort, A.; Fantin, B. The In Vitro Contribution of Autolysins to Bacterial Killing Elicited by Amoxicillin Increases with Inoculum Size in Enterococcus faecalis. Antimicrob. Agents Chemother. 2011, 55, 910–912. [Google Scholar] [CrossRef]
- Lima, L.M.; da Silva, B.N.M.; Barbosa, G.; Barreiro, E.J. β-lactam antibiotics: An overview from a medicinal chemistry perspective. Eur. J. Med. Chem. 2020, 208, 112829. [Google Scholar] [CrossRef]
- Neu, H.C. Amoxicillin. Ann. Intern. Med. 1979, 90, 356–360. [Google Scholar] [CrossRef]
- Ahmad, I.; Huang, L.; Hao, H.; Sanders, P.; Yuan, Z. Application of PK/PD Modeling in Veterinary Field: Dose Optimization and Drug Resistance Prediction. Biomed. Res. Int. 2016, 2016, 5465678. [Google Scholar] [CrossRef] [PubMed]
- Elad, D. Increased Resistance of Staphylococcus pseudintermedius to the Commonly Used Antibiotics in Canine Dermatology. Isr. J. Vet. Med. 2011, 66, 143. [Google Scholar]
- Lee, E.B.; Abbas, M.A.; Park, J.; Tassew, D.D.; Park, S.C. Optimizing tylosin dosage for co-infection of Actinobacillus pleuropneumoniae and Pasteurella multocida in pigs using pharmacokinetic/pharmacodynamic modeling. Front Pharmacol. 2023, 14, 1258403. [Google Scholar] [CrossRef] [PubMed]
- Diekema, D.J.; Pfaller, M.A.; Schmitz, F.J.; Smayevsky, J.; Bell, J.; Jones, R.N.; Beach, M. Survey of Infections Due to Staphylococcus Species: Frequency of Occurrence and Antimicrobial Susceptibility of Isolates Collected in the United States, Canada. Clin. Infect. Dis. 2001, 32, 114–146. [Google Scholar] [CrossRef] [PubMed]
- Perreten, V.; Kadlec, K.; Schwarz, S.; Andersson, U.G.; Finn, M.; Greko, C.; Moodley, A.; Kania, S.A.; Frank, L.A.; Bemis, D.A.; et al. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: An international multicentre study. J. Antimicrob. Chemother. 2010, 65, 1145–1154. [Google Scholar] [CrossRef]
- Rubin, J.E.; Ball, K.R.; Chirino-Trejo, M. Antimicrobial susceptibility of Staphylococcus aureus and Staphylococcus pseudintermedius isolated from various animals. Can. Vet. J. 2011, 52, 153. [Google Scholar] [PubMed]
- Kırmusaoğlu, S.; Kırmusaoğlu, S. MRSA and MSSA: The Mechanism of Methicillin Resistance and the Influence of Methicillin Resistance on Biofilm Phenotype of Staphylococcus aureus. In The Rise of Virulence and Antibiotic Resistance in Staphylococcus aureus; BoD—Books on Demand: Norderstedt, Germany, 2017. [Google Scholar] [CrossRef]
- Rodríguez-Melcón, C.; Alonso-Calleja, C.; García-Fernández, C.; Carballo, J.; Capita, R. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) for Twelve Antimicrobials (Biocides and Antibiotics) in Eight Strains of Listeria monocytogenes. Biology 2022, 11, 46. [Google Scholar] [CrossRef]
- Gonzalez, N.; Sevillano, D.; Alou, L.; Cafini, F.; Gimenez, M.J.; Gomez-Lus, M.L.; Prieto, J.; Aguilar, L. Influence of the MBC/MIC ratio on the antibacterial activity of vancomycin versus linezolid against methicillin-resistant Staphylococcus aureus isolates in a pharmacodynamic model simulating serum and soft tissue interstitial fluid concentrations reported in diabetic patients. J. Antimicrob. Chemother. 2013, 68, 2291–2295. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-B.; Lee, K. A Pharmacodynamic Study of Aminoglycosides against Pathogenic E. coli through Monte Carlo Simulation. Pharmaceuticals 2024, 17, 27. [Google Scholar] [CrossRef] [PubMed]
- Standard Operating Procedure. MIC Distributions and the Setting of Epidemiological Cut-Off (ECOFF) Values. 2021. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/EUCAST_SOPs/2021/EUCAST_SOP_10.2_MIC_distributions_and_epidemiological_cut-off_value__ECOFF__setting_20211202.pdf (accessed on 12 October 2024).
- Kahlmeter, G.; Turnidge, J. How to: ECOFFs—The why, the how, and the don’ts of EUCAST epidemiological cutoff values. Clin. Microbiol. Infect. 2022, 28, 952–954. [Google Scholar] [CrossRef]
- Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018.
- Firsov, A.A.; Vostrov, S.N.; Lubenko, I.Y.; Drlica, K.; Portnoy, Y.A.; Zinner, S.H. In Vitro Pharmacodynamic Evaluation of the Mutant Selection Window Hypothesis Using Four Fluoroquinolones against Staphylococcus aureus. Am. Soc. Microbiol. 2003, 47, 1604–1613. [Google Scholar] [CrossRef] [PubMed]
- Birhanu, B.T.; Lee, E.B.; Lee, S.J.; Park, S.C. Targeting Salmonella Typhimurium Invasion and Intracellular Survival Using Pyrogallol. Front. Microbiol. 2021, 12, 631426. [Google Scholar] [CrossRef]
- MacKenzie, F.; Gould, I.M. The post-antibiotic effect. J. Antimicrob. Chemother. 1993, 32, 519–537. [Google Scholar] [CrossRef]
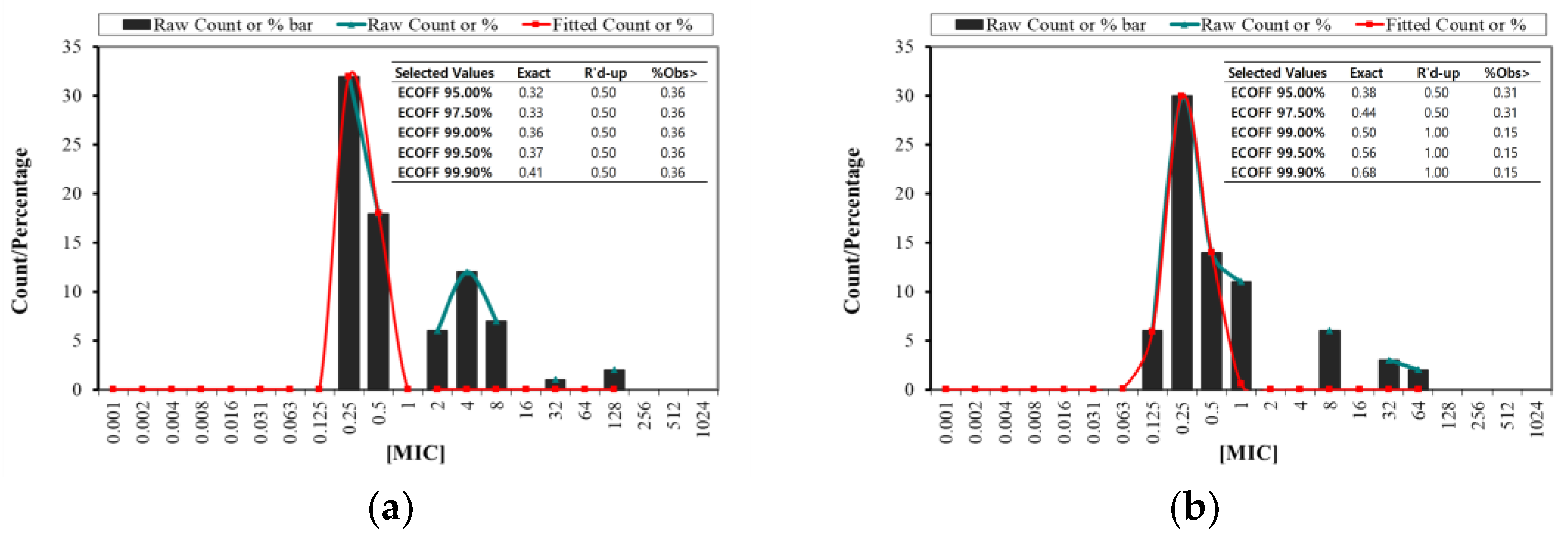

| Species | S | I | R | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC (μg/mL) | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 |
| SA | 32 | 18 | 6 | 12 | 7 | 1 | 2 | ||||
| SP | 6 | 30 | 14 | 11 | 6 | 3 | 2 | ||||
| S. aureus | S. pseudintermedius | |
|---|---|---|
| Observations | 78 | 72 |
| Distributions | 7 | 7 |
| % Resistance | 35.89 | 15.27 |
| % Sensitive | 64.10 | 69.44 |
| MIC range | 0.25–128 μg/mL | 0.125–64 μg/mL |
| MBC range | 0.25–512 μg/mL | 0.125–128 μg/mL |
| MIC50 | 0.50 μg/mL | 0.25 μg/mL |
| MIC90 | 8 | 8 |
| MBC50 | 0.50 | 1 |
| MBC90 | 8 | 8 |
| MIC/MBC50 | 1 | 0.25 |
| MIC/MBC90 | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sayem, S.A.J.; Lee, G.-Y.; Abbas, M.A.; Park, S.-C.; Lee, S.-J. Pharmacodynamic Profiling of Amoxicillin: Targeting Multidrug-Resistant Gram-Positive Pathogens Staphylococcus aureus and Staphylococcus pseudintermedius in Canine Clinical Isolates. Antibiotics 2025, 14, 99. https://doi.org/10.3390/antibiotics14010099
Sayem SAJ, Lee G-Y, Abbas MA, Park S-C, Lee S-J. Pharmacodynamic Profiling of Amoxicillin: Targeting Multidrug-Resistant Gram-Positive Pathogens Staphylococcus aureus and Staphylococcus pseudintermedius in Canine Clinical Isolates. Antibiotics. 2025; 14(1):99. https://doi.org/10.3390/antibiotics14010099
Chicago/Turabian StyleSayem, Syed Al Jawad, Ga-Yeong Lee, Muhammad Aleem Abbas, Seung-Chun Park, and Seung-Jin Lee. 2025. "Pharmacodynamic Profiling of Amoxicillin: Targeting Multidrug-Resistant Gram-Positive Pathogens Staphylococcus aureus and Staphylococcus pseudintermedius in Canine Clinical Isolates" Antibiotics 14, no. 1: 99. https://doi.org/10.3390/antibiotics14010099
APA StyleSayem, S. A. J., Lee, G.-Y., Abbas, M. A., Park, S.-C., & Lee, S.-J. (2025). Pharmacodynamic Profiling of Amoxicillin: Targeting Multidrug-Resistant Gram-Positive Pathogens Staphylococcus aureus and Staphylococcus pseudintermedius in Canine Clinical Isolates. Antibiotics, 14(1), 99. https://doi.org/10.3390/antibiotics14010099








