Epidemiology and Mechanisms of Resistance of Extensively Drug Resistant Gram-Negative Bacteria
Abstract
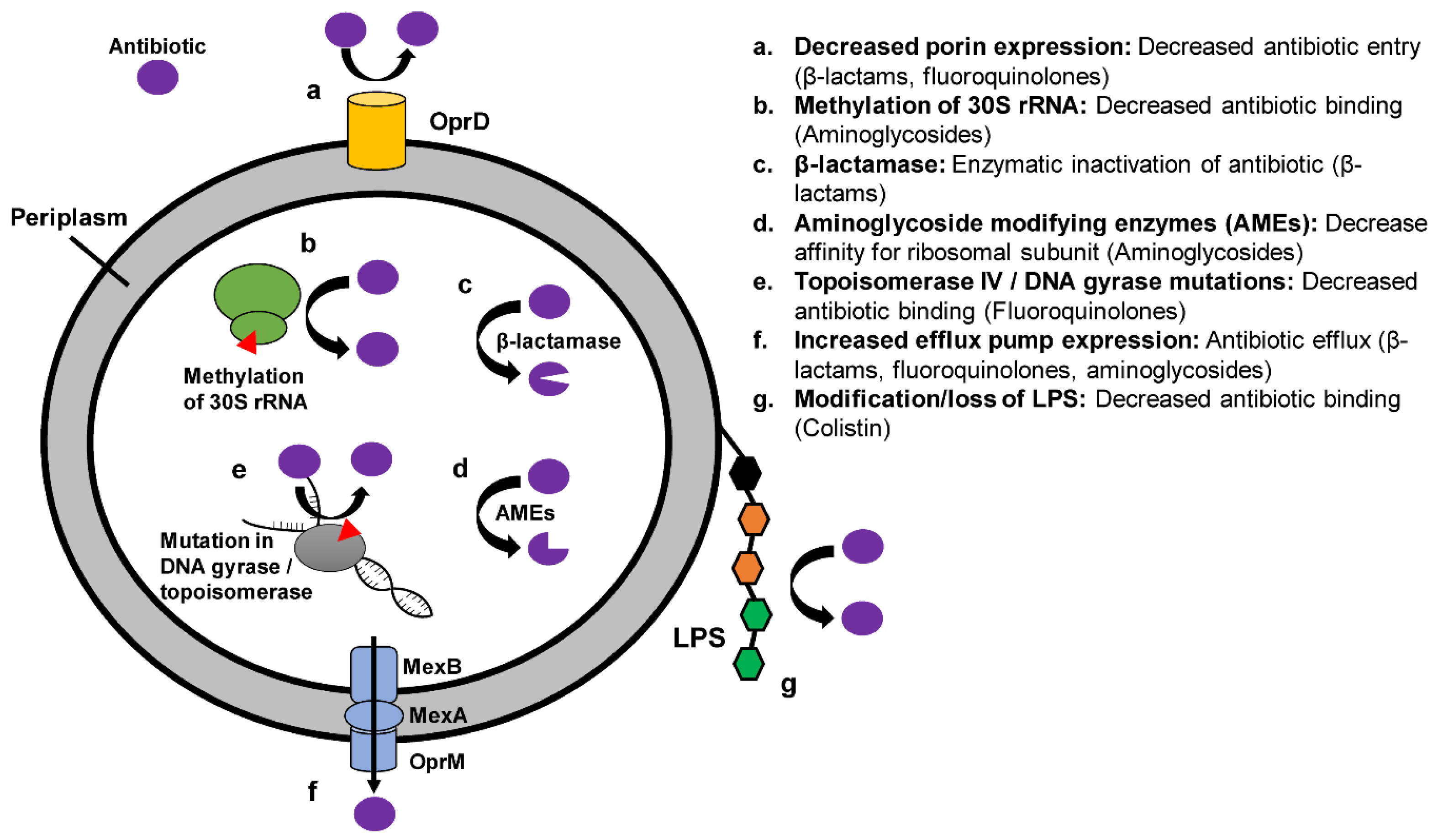
:1. Introduction
2. Mechanisms and Epidemiology of Antibiotic Resistance in Carbapenem Resistant Enterobacteriaceae (CRE)
2.1. Overview of Mechanisms of Resistance in CRE
2.2. Risk Factors for CRE Infection
2.3. Class A Carbapenemases
2.4. Class B Carbapenemases
2.5. Class D Carbapenemases
2.6. Efflux Pumps
2.7. Porin Mutations
2.8. Colistin Resistance
3. Mechanisms and Epidemiology of Antibiotic Resistance in XDR P. aeruginosa
3.1. Overview of Mechanisms of Resistance in XDR P. aeruginosa
3.2. Prevalence of and Risk Factors for XDR P. aeruginosa
3.3. Efflux Pumps
3.4. Porin Mutations
3.5. Modification of Antibiotic Target Site
3.6. Antibiotic Degradation
3.7. Clonal Structure of XDR P. aeruginosa
4. Mechanisms and Epidemiology of Antibiotic Resistance in XDR A. baumannii
4.1. Mechanisms of Resistance in XDR A. baumannii
4.2. Prevalence of and Risk Factors for XDR A. baumannii Infection
4.3. Outbreaks of XDR A. baumannii Infection
4.4. Efflux Pumps and Decreased Outer Membrane Permeability
4.5. Mutations in Antibiotic Target
4.6. Antibiotic Degradation
5. Conclusions
Author Contributions
Conflicts of Interest
References
- Tumbarello, M.; Viale, P.; Viscoli, C.; Trecarichi, E.M.; Tumietto, F.; Marchese, A.; Spanu, T.; Ambretti, S.; Ginocchio, F.; Cristini, F.; et al. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: Importance of combination therapy. Clin. Infect. Dis. 2012, 55, 943–950. [Google Scholar] [CrossRef]
- Pouch, S.M.; Kubin, C.J.; Satlin, M.J.; Tsapepas, D.S.; Lee, J.R.; Dube, G.; Pereira, M.R. Epidemiology and outcomes of carbapenem-resistant Klebsiella pneumoniae bacteriuria in kidney transplant recipients. Transpl. Infect. Dis. 2015, 17, 800–809. [Google Scholar] [CrossRef] [PubMed]
- Gasink, L.B.; Edelstein, P.H.; Lautenbach, E.; Synnestvedt, M.; Fishman, N.O. Risk factors and clinical impact of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. Infect. Control Hosp. Epidemiol. 2009, 30, 1180–1185. [Google Scholar] [CrossRef]
- Patel, G.; Huprikar, S.; Factor, S.H.; Jenkins, S.G.; Calfee, D.P. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect. Control Hosp. Epidemiol. 2008, 29, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Thaden, J.T.; Li, Y.; Ruffin, F.; Maskarinec, S.A.; Hill-Rorie, J.M.; Wanda, L.C.; Reed, S.D.; Fowler, V.G., Jr. Increased Costs Associated with Bloodstream Infections Caused by Multidrug-Resistant Gram-Negative Bacteria Are Due Primarily to Patients with Hospital-Acquired Infections. Antimicrob. Agents Chemother. 2017, 61, e01709-16. [Google Scholar] [CrossRef] [PubMed]
- Evans, H.L.; Lefrak, S.N.; Lyman, J.; Smith, R.L.; Chong, T.W.; McElearney, S.T.; Schulman, A.R.; Hughes, M.G.; Raymond, D.P.; Pruett, T.L.; et al. Cost of Gram-negative resistance. Crit. Care Med. 2007, 35, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Ye, H.; Xu, Y.; Ni, Y.; Hu, Y.; Yu, Y.; Huang, Z.; Ma, L. Clinical and economic outcomes associated with community-acquired intra-abdominal infections caused by extended spectrum beta-lactamase (ESBL) producing bacteria in China. Curr. Med. Res. Opin. 2010, 26, 1443–1449. [Google Scholar] [CrossRef]
- Lautenbach, E.; Patel, J.B.; Bilker, W.B.; Edelstein, P.H.; Fishman, N.O. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae: Risk factors for infection and impact of resistance on outcomes. Clin. Infect. Dis. 2001, 32, 1162–1171. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Dou, Y.; Zhou, K.; Chen, Y.; Han, L.; Guo, X.; Sun, J. Coexistence of blaOXA-48 and Truncated blaNDM-1 on Different Plasmids in a Klebsiella pneumoniae Isolate in China. Front. Microbiol 2017, 8, 133. [Google Scholar] [CrossRef]
- Kapmaz, M.; Erdem, F.; Abulaila, A.; Yeniaras, E.; Oncul, O.; Aktas, Z. First detection of NDM-1 with CTX-M-9, TEM, SHV and rmtC in Escherichia coli ST471 carrying IncI2, A/C and Y plasmids from clinical isolates in Turkey. J. Glob. Antimicrob. Resist. 2016, 7, 152–153. [Google Scholar] [CrossRef]
- Marshall, S.; Hujer, A.M.; Rojas, L.J.; Papp-Wallace, K.M.; Humphries, R.M.; Spellberg, B.; Hujer, K.M.; Marshall, E.K.; Rudin, S.D.; Perez, F.; et al. Can Ceftazidime-Avibactam and Aztreonam Overcome beta-Lactam Resistance Conferred by Metallo-beta-Lactamases in Enterobacteriaceae? Antimicrob. Agents Chemother. 2017, 61, e02243-16. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Meletis, G.; Chatzidimitriou, D.; Malisiovas, N. Double- and multi-carbapenemase-producers: The excessively armored bacilli of the current decade. Eur J. Clin. Microbiol. Infect. Dis. 2015, 34, 1487–1493. [Google Scholar] [CrossRef]
- Albiger, B.; Glasner, C.; Struelens, M.J.; Grundmann, H.; Monnet, D.L. Carbapenemase-producing Enterobacteriaceae in Europe: Assessment by national experts from 38 countries, May 2015. Euro Surveill. 2015, 20. [Google Scholar] [CrossRef] [PubMed]
- van Duin, D.; Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017, 8, 460–469. [Google Scholar] [CrossRef] [PubMed]
- van der Bij, A.K.; Pitout, J.D. The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J. Antimicrob. Chemother. 2012, 67, 2090–2100. [Google Scholar] [CrossRef]
- Arnold, R.S.; Thom, K.A.; Sharma, S.; Phillips, M.; Kristie Johnson, J.; Morgan, D.J. Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. South Med. J. 2011, 104, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Chiotos, K.; Tamma, P.D.; Flett, K.B.; Naumann, M.; Karandikar, M.V.; Bilker, W.B.; Zaoutis, T.; Han, J.H. Multicenter Study of the Risk Factors for Colonization or Infection with Carbapenem-Resistant Enterobacteriaceae in Children. Antimicrob. Agents Chemother. 2017, 61, e01440-17. [Google Scholar] [CrossRef] [PubMed]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile beta-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef]
- Sader, H.S.; Castanheira, M.; Flamm, R.K.; Farrell, D.J.; Jones, R.N. Antimicrobial activity of ceftazidime-avibactam against Gram-negative organisms collected from U.S. medical centers in 2012. Antimicrob. Agents Chemother. 2014, 58, 1684–1692. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Rhomberg, P.R.; Flamm, R.K.; Jones, R.N. Effect of the beta-Lactamase Inhibitor Vaborbactam Combined with Meropenem against Serine Carbapenemase-Producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2016, 60, 5454–5458. [Google Scholar] [CrossRef]
- Haidar, G.; Clancy, C.J.; Chen, L.; Samanta, P.; Shields, R.K.; Kreiswirth, B.N.; Nguyen, M.H. Identifying Spectra of Activity and Therapeutic Niches for Ceftazidime-Avibactam and Imipenem-Relebactam against Carbapenem-Resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 2017, 61, e00642-17. [Google Scholar] [CrossRef]
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef]
- Naas, T.; Dortet, L.; Iorga, B.I. Structural and Functional Aspects of Class A Carbapenemases. Curr Drug. Targets 2016, 17, 1006–1028. [Google Scholar] [CrossRef] [PubMed]
- Diene, S.M.; Rolain, J.M. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 2014, 20, 831–838. [Google Scholar] [CrossRef]
- Yigit, H.; Queenan, A.M.; Anderson, G.J.; Domenech-Sanchez, A.; Biddle, J.W.; Steward, C.D.; Alberti, S.; Bush, K.; Tenover, F.C. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001, 45, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Miriagou, V.; Tzouvelekis, L.S.; Rossiter, S.; Tzelepi, E.; Angulo, F.J.; Whichard, J.M. Imipenem resistance in a Salmonella clinical strain due to plasmid-mediated class A carbapenemase KPC-2. Antimicrob. Agents Chemother. 2003, 47, 1297–1300. [Google Scholar] [CrossRef] [PubMed]
- Yigit, H.; Queenan, A.M.; Rasheed, J.K.; Biddle, J.W.; Domenech-Sanchez, A.; Alberti, S.; Bush, K.; Tenover, F.C. Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing beta-lactamase KPC-2. Antimicrob. Agents Chemother. 2003, 47, 3881–3889. [Google Scholar] [CrossRef]
- Bradford, P.A.; Bratu, S.; Urban, C.; Visalli, M.; Mariano, N.; Landman, D.; Rahal, J.J.; Brooks, S.; Cebular, S.; Quale, J. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 beta-lactamases in New York City. Clin. Infect. Dis. 2004, 39, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Ferraro, M.J.; Pino, R.M.; Dew, R.B., 3rd; Moland, E.S.; Lockhart, T.J.; Thomson, K.S.; Goering, R.V.; Hanson, N.D. Plasmid-mediated carbapenem-hydrolyzing enzyme KPC-2 in an Enterobacter sp. Antimicrob. Agents Chemother. 2004, 48, 4438–4440. [Google Scholar] [CrossRef]
- Bratu, S.; Landman, D.; Alam, M.; Tolentino, E.; Quale, J. Detection of KPC carbapenem-hydrolyzing enzymes in Enterobacter spp. from Brooklyn, New York. Antimicrob. Agents Chemother. 2005, 49, 776–778. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention Website: Tracking CRE. Available online: https://www.cdc.gov/hai/organisms/cre/TrackingCRE.html (accessed on 22 March 2019).
- Guh, A.Y.; Bulens, S.N.; Mu, Y.; Jacob, J.T.; Reno, J.; Scott, J.; Wilson, L.E.; Vaeth, E.; Lynfield, R.; Shaw, K.M.; et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae in 7 US Communities, 2012-2013. JAMA 2015, 314, 1479–1487. [Google Scholar] [CrossRef]
- Satlin, M.J.; Chen, L.; Patel, G.; Gomez-Simmonds, A.; Weston, G.; Kim, A.C.; Seo, S.K.; Rosenthal, M.E.; Sperber, S.J.; Jenkins, S.G.; et al. Multicenter Clinical and Molecular Epidemiological Analysis of Bacteremia Due to Carbapenem-Resistant Enterobacteriaceae (CRE) in the CRE Epicenter of the United States. Antimicrob. Agents Chemother. 2017, 61, e02349-16. [Google Scholar] [CrossRef] [PubMed]
- Naas, T.; Nordmann, P.; Vedel, G.; Poyart, C. Plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob. Agents Chemother. 2005, 49, 4423–4424. [Google Scholar] [CrossRef] [PubMed]
- Spyropoulou, A.; Papadimitriou-Olivgeris, M.; Bartzavali, C.; Vamvakopoulou, S.; Marangos, M.; Spiliopoulou, I.; Anastassiou, E.D.; Christofidou, M. A ten-year surveillance study of carbapenemase-producing Klebsiella pneumoniae in a tertiary care Greek university hospital: Predominance of KPC- over VIM- or NDM-producing isolates. J. Med. Microbiol. 2016, 65, 240–246. [Google Scholar] [CrossRef]
- Parisi, S.G.; Bartolini, A.; Santacatterina, E.; Castellani, E.; Ghirardo, R.; Berto, A.; Franchin, E.; Menegotto, N.; De Canale, E.; Tommasini, T.; et al. Prevalence of Klebsiella pneumoniae strains producing carbapenemases and increase of resistance to colistin in an Italian teaching hospital from January 2012 To December 2014. BMC Infect. Dis. 2015, 15, 244. [Google Scholar] [CrossRef] [PubMed]
- Schwaber, M.J.; Carmeli, Y. An ongoing national intervention to contain the spread of carbapenem-resistant Enterobacteriaceae. Clin. Infect. Dis. 2014, 58, 697–703. [Google Scholar] [CrossRef]
- Chen, S.; Hu, F.; Xu, X.; Liu, Y.; Wu, W.; Zhu, D.; Wang, H. High prevalence of KPC-2-type carbapenemase coupled with CTX-M-type extended-spectrum beta-lactamases in carbapenem-resistant Klebsiella pneumoniae in a teaching hospital in China. Antimicrob. Agents Chemother. 2011, 55, 2493–2494. [Google Scholar] [CrossRef] [PubMed]
- Baraniak, A.; Izdebski, R.; Fiett, J.; Herda, M.; Derde, L.P.; Bonten, M.J.; Adler, A.; Carmeli, Y.; Goossens, H.; Hryniewicz, W.; et al. KPC-Like Carbapenemase-Producing Enterobacteriaceae Colonizing Patients in Europe and Israel. Antimicrob. Agents Chemother. 2015, 60, 1912–1917. [Google Scholar] [CrossRef]
- Kitchel, B.; Rasheed, J.K.; Patel, J.B.; Srinivasan, A.; Navon-Venezia, S.; Carmeli, Y.; Brolund, A.; Giske, C.G. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: Clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 2009, 53, 3365–3370. [Google Scholar] [CrossRef]
- Ocampo, A.M.; Chen, L.; Cienfuegos, A.V.; Roncancio, G.; Chavda, K.D.; Kreiswirth, B.N.; Jimenez, J.N. A Two-Year Surveillance in Five Colombian Tertiary Care Hospitals Reveals High Frequency of Non-CG258 Clones of Carbapenem-Resistant Klebsiella pneumoniae with Distinct Clinical Characteristics. Antimicrob. Agents Chemother. 2016, 60, 332–342. [Google Scholar] [CrossRef]
- Deleo, F.R.; Chen, L.; Porcella, S.F.; Martens, C.A.; Kobayashi, S.D.; Porter, A.R.; Chavda, K.D.; Jacobs, M.R.; Mathema, B.; Olsen, R.J.; et al. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc. Natl. Acad. Sci. USA 2014, 111, 4988–4993. [Google Scholar] [CrossRef]
- Chen, L.; Mathema, B.; Pitout, J.D.; DeLeo, F.R.; Kreiswirth, B.N. Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. MBio 2014, 5, e01355-14. [Google Scholar] [CrossRef]
- van Duin, D.; Perez, F.; Rudin, S.D.; Cober, E.; Hanrahan, J.; Ziegler, J.; Webber, R.; Fox, J.; Mason, P.; Richter, S.S.; et al. Surveillance of carbapenem-resistant Klebsiella pneumoniae: Tracking molecular epidemiology and outcomes through a regional network. Antimicrob. Agents Chemother. 2014, 58, 4035–4041. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.D.; Porter, A.R.; Freedman, B.; Pandey, R.; Chen, L.; Kreiswirth, B.N.; DeLeo, F.R. Antibody-Mediated Killing of Carbapenem-Resistant ST258 Klebsiella pneumoniae by Human Neutrophils. MBio 2018, 9, e00297-18. [Google Scholar] [CrossRef] [PubMed]
- Diago-Navarro, E.; Motley, M.P.; Ruiz-Perez, G.; Yu, W.; Austin, J.; Seco, B.M.S.; Xiao, G.; Chikhalya, A.; Seeberger, P.H.; Fries, B.C. Novel, Broadly Reactive Anticapsular Antibodies against Carbapenem-Resistant Klebsiella pneumoniae Protect from Infection. MBio 2018, 9, e00091-18. [Google Scholar] [CrossRef] [PubMed]
- Oteo, J.; Perez-Vazquez, M.; Bautista, V.; Ortega, A.; Zamarron, P.; Saez, D.; Fernandez-Romero, S.; Lara, N.; Ramiro, R.; Aracil, B.; et al. The spread of KPC-producing Enterobacteriaceae in Spain: WGS analysis of the emerging high-risk clones of Klebsiella pneumoniae ST11/KPC-2, ST101/KPC-2 and ST512/KPC-3. J. Antimicrob. Chemother. 2016, 71, 3392–3399. [Google Scholar] [CrossRef] [PubMed]
- Naas, T.; Vandel, L.; Sougakoff, W.; Livermore, D.M.; Nordmann, P. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A beta-lactamase, Sme-1, from Serratia marcescens S6. Antimicrob. Agents Chemother. 1994, 38, 1262–1270. [Google Scholar] [CrossRef]
- Walther-Rasmussen, J.; Hoiby, N. Class A carbapenemases. J. Antimicrob. Chemother. 2007, 60, 470–482. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, B.A.; Bush, K.; Keeney, D.; Yang, Y.; Hare, R.; O’Gara, C.; Medeiros, A.A. Characterization of IMI-1 beta-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob. Agents Chemother. 1996, 40, 2080–2086. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L. Emerging carbapenemases in Gram-negative aerobes. Clin. Microbiol. Infect. 2002, 8, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Meini, M.R.; Llarrull, L.I.; Vila, A.J. Overcoming differences: The catalytic mechanism of metallo-beta-lactamases. FEBS Lett. 2015, 589, 3419–3432. [Google Scholar] [CrossRef]
- Walsh, T.R.; Toleman, M.A.; Poirel, L.; Nordmann, P. Metallo-beta-lactamases: The quiet before the storm? Clin. Microbiol. Rev. 2005, 18, 306–325. [Google Scholar] [CrossRef]
- Kohira, N.; West, J.; Ito, A.; Ito-Horiyama, T.; Nakamura, R.; Sato, T.; Rittenhouse, S.; Tsuji, M.; Yamano, Y. In Vitro Antimicrobial Activity of a Siderophore Cephalosporin, S-649266, against Enterobacteriaceae Clinical Isolates, Including Carbapenem-Resistant Strains. Antimicrob. Agents Chemother. 2016, 60, 729–734. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, F.; Zhao, C.; Wang, Z.; Nichols, W.W.; Testa, R.; Li, H.; Chen, H.; He, W.; Wang, Q.; et al. In vitro activities of ceftazidime-avibactam and aztreonam-avibactam against 372 Gram-negative bacilli collected in 2011 and 2012 from 11 teaching hospitals in China. Antimicrob. Agents Chemother. 2014, 58, 1774–1778. [Google Scholar] [CrossRef] [PubMed]
- Kumarasamy, K.K.; Toleman, M.A.; Walsh, T.R.; Bagaria, J.; Butt, F.; Balakrishnan, R.; Chaudhary, U.; Doumith, M.; Giske, C.G.; Irfan, S.; et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef]
- Ranjan, A.; Shaik, S.; Mondal, A.; Nandanwar, N.; Hussain, A.; Semmler, T.; Kumar, N.; Tiwari, S.K.; Jadhav, S.; Wieler, L.H.; et al. Molecular Epidemiology and Genome Dynamics of New Delhi Metallo-beta-Lactamase-Producing Extraintestinal Pathogenic Escherichia coli Strains from India. Antimicrob. Agents Chemother. 2016, 60, 6795–6805. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Shukla, S.K.; Prasad, K.N.; Ovejero, C.M.; Pati, B.K.; Tripathi, A.; Singh, A.; Srivastava, A.K.; Gonzalez-Zorn, B. Prevalence and molecular characterisation of New Delhi metallo-beta-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int. J. Antimicrob. Agents 2014, 44, 30–37. [Google Scholar] [CrossRef]
- Nahid, F.; Khan, A.A.; Rehman, S.; Zahra, R. Prevalence of metallo-beta-lactamase NDM-1-producing multi-drug resistant bacteria at two Pakistani hospitals and implications for public health. J. Infect. Public Health 2013, 6, 487–493. [Google Scholar] [CrossRef]
- Sartor, A.L.; Raza, M.W.; Abbasi, S.A.; Day, K.M.; Perry, J.D.; Paterson, D.L.; Sidjabat, H.E. Molecular epidemiology of NDM-1-producing Enterobacteriaceae and Acinetobacter baumannii isolates from Pakistan. Antimicrob. Agents Chemother. 2014, 58, 5589–5593. [Google Scholar] [CrossRef]
- Walsh, T.R.; Weeks, J.; Livermore, D.M.; Toleman, M.A. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: An environmental point prevalence study. Lancet Infect. Dis. 2011, 11, 355–362. [Google Scholar] [CrossRef]
- Lauretti, L.; Riccio, M.L.; Mazzariol, A.; Cornaglia, G.; Amicosante, G.; Fontana, R.; Rossolini, G.M. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 1999, 43, 1584–1590. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Peirano, G.; Devinney, R.; Bradford, P.A.; Motyl, M.R.; Adams, M.D.; Chen, L.; Kreiswirth, B.; Pitout, J.D.D. Genomic epidemiology of global VIM-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2017, 72, 2249–2258. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Potron, A.; Nordmann, P. OXA-48-like carbapenemases: The phantom menace. J. Antimicrob. Chemother. 2012, 67, 1597–1606. [Google Scholar] [CrossRef]
- Poirel, L.; Heritier, C.; Tolun, V.; Nordmann, P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004, 48, 15–22. [Google Scholar] [CrossRef]
- Cizmeci, Z.; Aktas, E.; Otlu, B.; Acikgoz, O.; Ordekci, S. Molecular characterization of carbapenem-resistant Enterobacteriaceae yields increasing rates of NDM-1 carbapenemases and colistin resistance in an OXA-48-endemic area. J. Chemother. 2017, 29, 344–350. [Google Scholar] [CrossRef]
- Baran, I.; Aksu, N. Phenotypic and genotypic characteristics of carbapenem-resistant Enterobacteriaceae in a tertiary-level reference hospital in Turkey. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 20. [Google Scholar] [CrossRef]
- Li, X.Z.; Plesiat, P.; Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef]
- Goessens, W.H.; van der Bij, A.K.; van Boxtel, R.; Pitout, J.D.; van Ulsen, P.; Melles, D.C.; Tommassen, J. Antibiotic trapping by plasmid-encoded CMY-2 beta-lactamase combined with reduced outer membrane permeability as a mechanism of carbapenem resistance in Escherichia coli. Antimicrob. Agents Chemother. 2013, 57, 3941–3949. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Martinez, L.; Pascual, A.; Hernandez-Alles, S.; Alvarez-Diaz, D.; Suarez, A.I.; Tran, J.; Benedi, V.J.; Jacoby, G.A. Roles of beta-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob. Agents Chemother. 1999, 43, 1669–1673. [Google Scholar] [CrossRef]
- Livermore, D.M. Bacterial resistance to carbapenems. Adv. Exp. Med. Biol. 1995, 390, 25–47. [Google Scholar]
- Cornaglia, G.; Russell, K.; Satta, G.; Fontana, R. Relative importances of outer membrane permeability and group 1 beta-lactamase as determinants of meropenem and imipenem activities against Enterobacter cloacae. Antimicrob. Agents Chemother. 1995, 39, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Shields, R.K.; Nguyen, M.H.; Potoski, B.A.; Press, E.G.; Chen, L.; Kreiswirth, B.N.; Clarke, L.G.; Eschenauer, G.A.; Clancy, C.J. Doripenem MICs and ompK36 porin genotypes of sequence type 258, KPC-producing Klebsiella pneumoniae may predict responses to carbapenem-colistin combination therapy among patients with bacteremia. Antimicrob. Agents Chemother. 2015, 59, 1797–1801. [Google Scholar] [CrossRef]
- Clancy, C.J.; Chen, L.; Hong, J.H.; Cheng, S.; Hao, B.; Shields, R.K.; Farrell, A.N.; Doi, Y.; Zhao, Y.; Perlin, D.S.; et al. Mutations of the ompK36 porin gene and promoter impact responses of sequence type 258, KPC-2-producing Klebsiella pneumoniae strains to doripenem and doripenem-colistin. Antimicrob. Agents Chemother. 2013, 57, 5258–5265. [Google Scholar] [CrossRef]
- Clancy, C.J.; Hao, B.; Shields, R.K.; Chen, L.; Perlin, D.S.; Kreiswirth, B.N.; Nguyen, M.H. Doripenem, gentamicin, and colistin, alone and in combinations, against gentamicin-susceptible, KPC-producing Klebsiella pneumoniae strains with various ompK36 genotypes. Antimicrob. Agents Chemother. 2014, 58, 3521–3525. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, L.M.; Rhomberg, P.R.; Sader, H.S.; Jones, R.N. Emergence of serine carbapenemases (KPC and SME) among clinical strains of Enterobacteriaceae isolated in the United States Medical Centers: Report from the MYSTIC Program (1999-2005). Diagn. Microbiol. Infect. Dis. 2006, 56, 367–372. [Google Scholar] [CrossRef]
- Evans, M.E.; Feola, D.J.; Rapp, R.P. Polymyxin B sulfate and colistin: Old antibiotics for emerging multiresistant gram-negative bacteria. Ann. Pharmacother. 1999, 33, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, H.; Liu, Y.H.; Feng, Y. Towards Understanding MCR-like Colistin Resistance. Trends Microbiol. 2018, 26, 794–808. [Google Scholar] [CrossRef]
- Rossolini, G.M.; Mantengoli, E. Treatment and control of severe infections caused by multiresistant Pseudomonas aeruginosa. Clin. Microbiol. Infect. 2005, 11 (Suppl. 4), 17–32. [Google Scholar] [CrossRef]
- Falagas, M.E.; Rafailidis, P.I.; Matthaiou, D.K.; Virtzili, S.; Nikita, D.; Michalopoulos, A. Pandrug-resistant Klebsiella pneumoniae, Pseudomonas aeruginosa and Acinetobacter baumannii infections: Characteristics and outcome in a series of 28 patients. Int. J. Antimicrob. Agents 2008, 32, 450–454. [Google Scholar] [CrossRef]
- Palavutitotai, N.; Jitmuang, A.; Tongsai, S.; Kiratisin, P.; Angkasekwinai, N. Epidemiology and risk factors of extensively drug-resistant Pseudomonas aeruginosa infections. PLoS ONE 2018, 13, e0193431. [Google Scholar] [CrossRef]
- Zavascki, A.P.; Carvalhaes, C.G.; Picao, R.C.; Gales, A.C. Multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: Resistance mechanisms and implications for therapy. Expert Rev. Anti Infect. Ther. 2010, 8, 71–93. [Google Scholar] [CrossRef]
- Cabot, G.; Ocampo-Sosa, A.A.; Dominguez, M.A.; Gago, J.F.; Juan, C.; Tubau, F.; Rodriguez, C.; Moya, B.; Pena, C.; Martinez-Martinez, L.; et al. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob. Agents Chemother. 2012, 56, 6349–6357. [Google Scholar] [CrossRef] [PubMed]
- Samonis, G.; Vardakas, K.Z.; Kofteridis, D.P.; Dimopoulou, D.; Andrianaki, A.M.; Chatzinikolaou, I.; Katsanevaki, E.; Maraki, S.; Falagas, M.E. Characteristics, risk factors and outcomes of adult cancer patients with extensively drug-resistant Pseudomonas aeruginosa infections. Infection 2014, 42, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Saderi, H.; Owlia, P. Detection of Multidrug Resistant (MDR) and Extremely Drug Resistant (XDR) P. aeruginosa Isolated from Patients in Tehran, Iran. Iran J. Pathol. 2015, 10, 265–271. [Google Scholar]
- Safaei, H.G.; Moghim, S.; Isfahani, B.N.; Fazeli, H.; Poursina, F.; Yadegari, S.; Nasirmoghadas, P.; Hosseininassab Nodoushan, S.A. Distribution of the Strains of Multidrug-resistant, Extensively Drug-resistant, and Pandrug-resistant Pseudomonas aeruginosa Isolates from Burn Patients. Adv. Biomed. Res. 2017, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Bodro, M.; Sabe, N.; Tubau, F.; Llado, L.; Baliellas, C.; Gonzalez-Costello, J.; Cruzado, J.M.; Carratala, J. Extensively drug-resistant Pseudomonas aeruginosa bacteremia in solid organ transplant recipients. Transplantation 2015, 99, 616–622. [Google Scholar] [CrossRef]
- Park, Y.S.; Lee, H.; Chin, B.S.; Han, S.H.; Hong, S.G.; Hong, S.K.; Kim, H.Y.; Uh, Y.; Shin, H.B.; Choo, E.J.; et al. Acquisition of extensive drug-resistant Pseudomonas aeruginosa among hospitalized patients: Risk factors and resistance mechanisms to carbapenems. J. Hosp. Infect. 2011, 79, 54–58. [Google Scholar] [CrossRef]
- Ziha-Zarifi, I.; Llanes, C.; Kohler, T.; Pechere, J.C.; Plesiat, P. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 1999, 43, 287–291. [Google Scholar] [CrossRef]
- Livermore, D.M. Of Pseudomonas, porins, pumps and carbapenems. J. Antimicrob. Chemother. 2001, 47, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Fruci, M.; Poole, K. Aminoglycoside-inducible expression of the mexAB-oprM multidrug efflux operon in Pseudomonas aeruginosa: Involvement of the envelope stress-responsive AmgRS two-component system. PLoS ONE 2018, 13, e0205036. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.T.; Brinkman, F.S.; Hancock, R.E. Aminoglycoside efflux in Pseudomonas aeruginosa: Involvement of novel outer membrane proteins. Antimicrob. Agents Chemother. 2003, 47, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Masuda, N.; Sakagawa, E.; Ohya, S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1995, 39, 645–649. [Google Scholar] [CrossRef]
- Masuda, N.; Ohya, S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1992, 36, 1847–1851. [Google Scholar] [CrossRef]
- Akasaka, T.; Tanaka, M.; Yamaguchi, A.; Sato, K. Type II topoisomerase mutations in fluoroquinolone-resistant clinical strains of Pseudomonas aeruginosa isolated in 1998 and 1999: Role of target enzyme in mechanism of fluoroquinolone resistance. Antimicrob. Agents Chemother. 2001, 45, 2263–2268. [Google Scholar] [CrossRef] [PubMed]
- Morais Cabral, J.H.; Jackson, A.P.; Smith, C.V.; Shikotra, N.; Maxwell, A.; Liddington, R.C. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 1997, 388, 903–906. [Google Scholar] [CrossRef] [PubMed]
- Kato, J.; Suzuki, H.; Ikeda, H. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J. Biol. Chem. 1992, 267, 25676–25684. [Google Scholar] [PubMed]
- Yokoyama, K.; Doi, Y.; Yamane, K.; Kurokawa, H.; Shibata, N.; Shibayama, K.; Yagi, T.; Kato, H.; Arakawa, Y. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 2003, 362, 1888–1893. [Google Scholar] [CrossRef]
- Doi, Y.; de Oliveira Garcia, D.; Adams, J.; Paterson, D.L. Coproduction of novel 16S rRNA methylase RmtD and metallo-beta-lactamase SPM-1 in a panresistant Pseudomonas aeruginosa isolate from Brazil. Antimicrob. Agents Chemother. 2007, 51, 852–856. [Google Scholar] [CrossRef]
- Gurung, M.; Moon, D.C.; Tamang, M.D.; Kim, J.; Lee, Y.C.; Seol, S.Y.; Cho, D.T.; Lee, J.C. Emergence of 16S rRNA methylase gene armA and cocarriage of bla(IMP-1) in Pseudomonas aeruginosa isolates from South Korea. Diagn. Microbiol. Infect. Dis. 2010, 68, 468–470. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef]
- Bush, K. Alarming beta-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 2010, 13, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Poole, K. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005, 49, 479–487. [Google Scholar] [CrossRef]
- Aggen, J.B.; Armstrong, E.S.; Goldblum, A.A.; Dozzo, P.; Linsell, M.S.; Gliedt, M.J.; Hildebrandt, D.J.; Feeney, L.A.; Kubo, A.; Matias, R.D.; et al. Synthesis and spectrum of the neoglycoside ACHN-490. Antimicrob. Agents Chemother. 2010, 54, 4636–4642. [Google Scholar] [CrossRef]
- Mulet, X.; Cabot, G.; Ocampo-Sosa, A.A.; Dominguez, M.A.; Zamorano, L.; Juan, C.; Tubau, F.; Rodriguez, C.; Moya, B.; Pena, C.; et al. Biological markers of Pseudomonas aeruginosa epidemic high-risk clones. Antimicrob. Agents Chemother. 2013, 57, 5527–5535. [Google Scholar] [CrossRef]
- Pena, C.; Cabot, G.; Gomez-Zorrilla, S.; Zamorano, L.; Ocampo-Sosa, A.; Murillas, J.; Almirante, B.; Pomar, V.; Aguilar, M.; Granados, A.; et al. Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin. Infect. Dis. 2015, 60, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.; Mulet, X.; Lopez-Causape, C.; Juan, C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist. Updat. 2015, 21–22, 41–59. [Google Scholar] [CrossRef] [PubMed]
- Kos, V.N.; Deraspe, M.; McLaughlin, R.E.; Whiteaker, J.D.; Roy, P.H.; Alm, R.A.; Corbeil, J.; Gardner, H. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob. Agents Chemother. 2015, 59, 427–436. [Google Scholar] [CrossRef]
- Weldhagen, G.F.; Poirel, L.; Nordmann, P. Ambler class A extended-spectrum beta-lactamases in Pseudomonas aeruginosa: Novel developments and clinical impact. Antimicrob. Agents Chemother. 2003, 47, 2385–2392. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Ronco, E.; Naas, T.; Duport, C.; Michel-Briand, Y.; Labia, R. Characterization of a novel extended-spectrum beta-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1993, 37, 962–969. [Google Scholar] [CrossRef]
- Vanegas, J.M.; Cienfuegos, A.V.; Ocampo, A.M.; Lopez, L.; del Corral, H.; Roncancio, G.; Sierra, P.; Echeverri-Toro, L.; Ospina, S.; Maldonado, N.; et al. Similar frequencies of Pseudomonas aeruginosa isolates producing KPC and VIM carbapenemases in diverse genetic clones at tertiary-care hospitals in Medellin, Colombia. J. Clin. Microbiol. 2014, 52, 3978–3986. [Google Scholar] [CrossRef]
- Correa, A.; Del Campo, R.; Perenguez, M.; Blanco, V.M.; Rodriguez-Banos, M.; Perez, F.; Maya, J.J.; Rojas, L.; Canton, R.; Arias, C.A.; et al. Dissemination of high-risk clones of extensively drug-resistant Pseudomonas aeruginosa in Colombia. Antimicrob. Agents Chemother. 2015, 59, 2421–2425. [Google Scholar] [CrossRef]
- Guzvinec, M.; Izdebski, R.; Butic, I.; Jelic, M.; Abram, M.; Koscak, I.; Baraniak, A.; Hryniewicz, W.; Gniadkowski, M.; Tambic Andrasevic, A. Sequence types 235, 111, and 132 predominate among multidrug-resistant Pseudomonas aeruginosa clinical isolates in Croatia. Antimicrob. Agents Chemother. 2014, 58, 6277–6283. [Google Scholar] [CrossRef]
- Garcia-Castillo, M.; Del Campo, R.; Morosini, M.I.; Riera, E.; Cabot, G.; Willems, R.; van Mansfeld, R.; Oliver, A.; Canton, R. Wide dispersion of ST175 clone despite high genetic diversity of carbapenem-nonsusceptible Pseudomonas aeruginosa clinical strains in 16 Spanish hospitals. J. Clin. Microbiol. 2011, 49, 2905–2910. [Google Scholar] [CrossRef] [PubMed]
- Viedma, E.; Juan, C.; Villa, J.; Barrado, L.; Orellana, M.A.; Sanz, F.; Otero, J.R.; Oliver, A.; Chaves, F. VIM-2-producing multidrug-resistant Pseudomonas aeruginosa ST175 clone, Spain. Emerg. Infect. Dis. 2012, 18, 1235–1241. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.; Nielsen, T.B.; Bonomo, R.A.; Pantapalangkoor, P.; Luna, B.; Spellberg, B. Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin. Microbiol. Rev. 2017, 30, 409–447. [Google Scholar] [CrossRef] [PubMed]
- Weiner, L.M.; Webb, A.K.; Limbago, B.; Dudeck, M.A.; Patel, J.; Kallen, A.J.; Edwards, J.R.; Sievert, D.M. Antimicrobial-Resistant Pathogens Associated With Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011-2014. Infect. Control Hosp. Epidemiol. 2016, 37, 1288–1301. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Surveillance of Antimicrobial Resistance in Europe 2017. European Center for Disease Prevention: Sona, Sweden. Available online: https://ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2017. (accessed on 1 January 2019).
- Hsu, L.Y.; Apisarnthanarak, A.; Khan, E.; Suwantarat, N.; Ghafur, A.; Tambyah, P.A. Carbapenem-Resistant Acinetobacter baumannii and Enterobacteriaceae in South and Southeast Asia. Clin. Microbiol. Rev. 2017, 30, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, C.H.; Nastro, M.; Famiglietti, A. Carbapenemases in Acinetobacter baumannii. Review of their dissemination in Latin America. Rev. Argent. Microbiol. 2018, 50, 327–333. [Google Scholar] [CrossRef]
- Mahgoub, S.; Ahmed, J.; Glatt, A.E. Underlying characteristics of patients harboring highly resistant Acinetobacter baumannii. Am. J. Infect. Control 2002, 30, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Manikal, V.M.; Landman, D.; Saurina, G.; Oydna, E.; Lal, H.; Quale, J. Endemic carbapenem-resistant Acinetobacter species in Brooklyn, New York: Citywide prevalence, interinstitutional spread, and relation to antibiotic usage. Clin. Infect. Dis. 2000, 31, 101–106. [Google Scholar] [CrossRef]
- Catalano, M.; Quelle, L.S.; Jeric, P.E.; Di Martino, A.; Maimone, S.M. Survival of Acinetobacter baumannii on bed rails during an outbreak and during sporadic cases. J. Hosp. Infect. 1999, 42, 27–35. [Google Scholar] [CrossRef]
- Villegas, M.V.; Hartstein, A.I. Acinetobacter outbreaks, 1977–2000. Infect. Control Hosp. Epidemiol. 2003, 24, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Bianco, A.; Quirino, A.; Giordano, M.; Marano, V.; Rizzo, C.; Liberto, M.C.; Foca, A.; Pavia, M. Control of carbapenem-resistant Acinetobacter baumannii outbreak in an intensive care unit of a teaching hospital in Southern Italy. BMC Infect. Dis. 2016, 16, 747. [Google Scholar] [CrossRef] [PubMed]
- Ansaldi, F.; Canepa, P.; Bassetti, M.; Zancolli, M.; Molinari, M.P.; Talamini, A.; Ginocchio, F.; Durando, P.; Mussap, M.; Orengo, G.; et al. Sequential outbreaks of multidrug-resistant Acinetobacter baumannii in intensive care units of a tertiary referral hospital in Italy: Combined molecular approach for epidemiological investigation. J. Hosp. Infect. 2011, 79, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Tanguy, M.; Kouatchet, A.; Tanguy, B.; Pichard, E.; Fanello, S.; Joly-Guillou, M.L. Management of an Acinetobacter baumannii outbreak in an intensive care unit. Med. Mal. Infect. 2017, 47, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Lolans, K.; Rice, T.W.; Munoz-Price, L.S.; Quinn, J.P. Multicity outbreak of carbapenem-resistant Acinetobacter baumannii isolates producing the carbapenemase OXA-40. Antimicrob. Agents Chemother. 2006, 50, 2941–2945. [Google Scholar] [CrossRef] [PubMed]
- Naas, T.; Bogaerts, P.; Bauraing, C.; Degheldre, Y.; Glupczynski, Y.; Nordmann, P. Emergence of PER and VEB extended-spectrum beta-lactamases in Acinetobacter baumannii in Belgium. J. Antimicrob. Chemother. 2006, 58, 178–182. [Google Scholar] [CrossRef]
- Coelho, J.M.; Turton, J.F.; Kaufmann, M.E.; Glover, J.; Woodford, N.; Warner, M.; Palepou, M.F.; Pike, R.; Pitt, T.L.; Patel, B.C.; et al. Occurrence of carbapenem-resistant Acinetobacter baumannii clones at multiple hospitals in London and Southeast England. J. Clin. Microbiol. 2006, 44, 3623–3627. [Google Scholar] [CrossRef]
- Chen, Y.; Ai, L.; Guo, P.; Huang, H.; Wu, Z.; Liang, X.; Liao, K. Molecular characterization of multidrug resistant strains of Acinetobacter baumannii isolated from pediatric intensive care unit in a Chinese tertiary hospital. BMC Infect. Dis. 2018, 18, 614. [Google Scholar] [CrossRef]
- Saharman, Y.R.; Karuniawati, A.; Sedono, R.; Aditianingsih, D.; Sudarmono, P.; Goessens, W.H.F.; Klaassen, C.H.W.; Verbrugh, H.A.; Severin, J.A. Endemic carbapenem-nonsusceptible Acinetobacter baumannii-calcoaceticus complex in intensive care units of the national referral hospital in Jakarta, Indonesia. Antimicrob. Resist. Infect. Control 2018, 7, 5. [Google Scholar] [CrossRef]
- Landelle, C.; Legrand, P.; Lesprit, P.; Cizeau, F.; Ducellier, D.; Gouot, C.; Brehaut, P.; Soing-Altrach, S.; Girou, E.; Brun-Buisson, C. Protracted outbreak of multidrug-resistant Acinetobacter baumannii after intercontinental transfer of colonized patients. Infect. Control Hosp. Epidemiol. 2013, 34, 119–124. [Google Scholar] [CrossRef]
- Wybo, I.; Blommaert, L.; De Beer, T.; Soetens, O.; De Regt, J.; Lacor, P.; Pierard, D.; Lauwers, S. Outbreak of multidrug-resistant Acinetobacter baumannii in a Belgian university hospital after transfer of patients from Greece. J. Hosp. Infect. 2007, 67, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.Z.; Hsueh, P.R.; Lee, L.N.; Yu, C.J.; Yang, P.C.; Luh, K.T. Severe community-acquired pneumonia due to Acinetobacter baumannii. Chest 2001, 120, 1072–1077. [Google Scholar] [CrossRef]
- Leung, W.S.; Chu, C.M.; Tsang, K.Y.; Lo, F.H.; Lo, K.F.; Ho, P.L. Fulminant community-acquired Acinetobacter baumannii pneumonia as a distinct clinical syndrome. Chest 2006, 129, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Magnet, S.; Courvalin, P.; Lambert, T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 2001, 45, 3375–3380. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Yum, J.H.; Kim, C.K.; Yong, D.; Jeon, E.H.; Jeong, S.H.; Ahn, J.Y.; Lee, K. Role of OXA-23 and AdeABC efflux pump for acquiring carbapenem resistance in an Acinetobacter baumannii strain carrying the blaOXA-66 gene. Ann. Clin. Lab. Sci. 2010, 40, 43–48. [Google Scholar] [PubMed]
- Damier-Piolle, L.; Magnet, S.; Bremont, S.; Lambert, T.; Courvalin, P. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2008, 52, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Huys, G.; Cnockaert, M.; Vaneechoutte, M.; Woodford, N.; Nemec, A.; Dijkshoorn, L.; Swings, J. Distribution of tetracycline resistance genes in genotypically related and unrelated multiresistant Acinetobacter baumannii strains from different European hospitals. Res. Microbiol. 2005, 156, 348–355. [Google Scholar] [CrossRef]
- Smani, Y.; Fabrega, A.; Roca, I.; Sanchez-Encinales, V.; Vila, J.; Pachon, J. Role of OmpA in the multidrug resistance phenotype of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2014, 58, 1806–1808. [Google Scholar] [CrossRef]
- Vila, J.; Ruiz, J.; Goni, P.; Jimenez de Anta, T. Quinolone-resistance mutations in the topoisomerase IV parC gene of Acinetobacter baumannii. J. Antimicrob. Chemother. 1997, 39, 757–762. [Google Scholar] [CrossRef]
- Vila, J.; Ruiz, J.; Goni, P.; Marcos, A.; Jimenez de Anta, T. Mutation in the gyrA gene of quinolone-resistant clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 1995, 39, 1201–1203. [Google Scholar] [CrossRef]
- Seward, R.J.; Towner, K.J. Molecular epidemiology of quinolone resistance in Acinetobacter spp. Clin. Microbiol. Infect. 1998, 4, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.D.; Nickel, G.C.; Bajaksouzian, S.; Lavender, H.; Murthy, A.R.; Jacobs, M.R.; Bonomo, R.A. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 2009, 53, 3628–3634. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Harrison, P.; Hale, J.D.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; St. Michael, F.; Cox, A.D.; et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [Google Scholar] [CrossRef]
- Nemec, A.; Dolzani, L.; Brisse, S.; van den Broek, P.; Dijkshoorn, L. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J. Med. Microbiol. 2004, 53, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Seward, R.J.; Lambert, T.; Towner, K.J. Molecular epidemiology of aminoglycoside resistance in Acinetobacter spp. J. Med. Microbiol. 1998, 47, 455–462. [Google Scholar] [CrossRef]
- Doi, Y.; Adams, J.M.; Yamane, K.; Paterson, D.L. Identification of 16S rRNA methylase-producing Acinetobacter baumannii clinical strains in North America. Antimicrob. Agents Chemother. 2007, 51, 4209–4210. [Google Scholar] [CrossRef]
- Zhou, H.; Du, X.X.; Yang, Q.; Zhou, J.Y.; Yu, Y.S.; Li, L.J. Study on carbapenemase and 16S rRNA methylase of imipenem-resistant Acinetobacter baumannii. Zhonghua Liu Xing Bing Xue Za Zhi 2009, 30, 269–272. [Google Scholar] [PubMed]
- Brigante, G.; Migliavacca, R.; Bramati, S.; Motta, E.; Nucleo, E.; Manenti, M.; Migliorino, G.; Pagani, L.; Luzzaro, F.; Vigano, F.E. Emergence and spread of a multidrug-resistant Acinetobacter baumannii clone producing both the carbapenemase OXA-23 and the 16S rRNA methylase ArmA. J. Med. Microbiol. 2012, 61, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Aghazadeh, M.; Rezaee, M.A.; Nahaei, M.R.; Mahdian, R.; Pajand, O.; Saffari, F.; Hassan, M.; Hojabri, Z. Dissemination of aminoglycoside-modifying enzymes and 16S rRNA methylases among Acinetobacter baumannii and Pseudomonas aeruginosa isolates. Microb. Drug Resist. 2013, 19, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Bakour, S.; Touati, A.; Bachiri, T.; Sahli, F.; Tiouit, D.; Naim, M.; Azouaou, M.; Rolain, J.M. First report of 16S rRNA methylase ArmA-producing Acinetobacter baumannii and rapid spread of metallo-beta-lactamase NDM-1 in Algerian hospitals. J. Infect. Chemother. 2014, 20, 696–701. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Nordmann, P. Carbapenem resistance in Acinetobacter baumannii: Mechanisms and epidemiology. Clin. Microbiol. Infect. 2006, 12, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Corvec, S.; Caroff, N.; Espaze, E.; Giraudeau, C.; Drugeon, H.; Reynaud, A. AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J. Antimicrob. Chemother. 2003, 52, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Turton, J.F.; Woodford, N.; Glover, J.; Yarde, S.; Kaufmann, M.E.; Pitt, T.L. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 2006, 44, 2974–2976. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Berrazeg, M.; Fagade, O.E.; Adelowo, O.O.; Alli, J.A.; Rolain, J.M. Emergence of multidrug-resistant Acinetobacter baumannii producing OXA-23 carbapenemase, Nigeria. Int. J. Infect. Dis. 2013, 17, e469–e470. [Google Scholar] [CrossRef]
- Poirel, L.; Naas, T.; Nordmann, P. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Corvec, S.; Poirel, L.; Naas, T.; Drugeon, H.; Nordmann, P. Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007, 51, 1530–1533. [Google Scholar] [CrossRef] [PubMed]
- Merino, M.; Poza, M.; Roca, I.; Barba, M.J.; Sousa, M.D.; Vila, J.; Bou, G. Nosocomial outbreak of a multiresistant Acinetobacter baumannii expressing OXA-23 carbapenemase in Spain. Microb. Drug Resist. 2014, 20, 259–263. [Google Scholar] [CrossRef]
- Afzal-Shah, M.; Woodford, N.; Livermore, D.M. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D beta-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2001, 45, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Heritier, C.; Poirel, L.; Aubert, D.; Nordmann, P. Genetic and Functional Analysis of the Chromosome-Encoded Carbapenem-Hydrolyzing Oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2003, 47, 268–273. [Google Scholar] [CrossRef]
- Tada, T.; Miyoshi-Akiyama, T.; Shimada, K.; Shimojima, M.; Kirikae, T. Dissemination of 16S rRNA methylase ArmA-producing Acinetobacter baumannii and emergence of OXA-72 carbapenemase coproducers in Japan. Antimicrob. Agents Chemother. 2014, 58, 2916–2920. [Google Scholar] [CrossRef]
- de Sa Cavalcanti, F.L.; Almeida, A.C.; Vilela, M.A.; de Morais Junior, M.A.; de Morais, M.M.; Leal-Balbino, T.C. Emergence of extensively drug-resistant OXA-72-producing Acinetobacter baumannii in Recife, Brazil: Risk of clonal dissemination? Diagn. Microbiol. Infect. Dis. 2013, 77, 250–251. [Google Scholar] [CrossRef]
- Povilonis, J.; Seputiene, V.; Krasauskas, R.; Juskaite, R.; Miskinyte, M.; Suziedelis, K.; Suziedeliene, E. Spread of carbapenem-resistant Acinetobacter baumannii carrying a plasmid with two genes encoding OXA-72 carbapenemase in Lithuanian hospitals. J. Antimicrob. Chemother. 2013, 68, 1000–1006. [Google Scholar] [CrossRef] [PubMed]
- Goic-Barisic, I.; Towner, K.J.; Kovacic, A.; Sisko-Kraljevic, K.; Tonkic, M.; Novak, A.; Punda-Polic, V. Outbreak in Croatia caused by a new carbapenem-resistant clone of Acinetobacter baumannii producing OXA-72 carbapenemase. J. Hosp. Infect. 2011, 77, 368–369. [Google Scholar] [CrossRef] [PubMed]
- Robledo, I.E.; Aquino, E.E.; Sante, M.I.; Santana, J.L.; Otero, D.M.; Leon, C.F.; Vazquez, G.J. Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob. Agents Chemother. 2010, 54, 1354–1357. [Google Scholar] [CrossRef]
- Potron, A.; Poirel, L.; Nordmann, P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int. J. Antimicrob. Agents 2015, 45, 568–585. [Google Scholar] [CrossRef]


| Ambler Class | β-Lactamases | Active Site Agent | Examples | Substrates |
|---|---|---|---|---|
| A | Penicillinases | Serine | PSE TEM, SHV, CTX-M, VEB, PER, GES KPC, SME, IMI/NMC-A | Penicillins Penicillins, 3rd generation cephalosporins All β-lactams |
| B | Metallo-β-lactamases | Zinc | IMP, VIM, NDM, SPM, GIM | All β-lactams, except monobactams |
| C | Cephalosporinases | Serine | AmpC | Cephamycins, 3rd generation cephalosporins |
| D | Oxacillinases | Serine | OXA | All β-lactams, though class D enzymes have highly variable spectra of activity |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eichenberger, E.M.; Thaden, J.T. Epidemiology and Mechanisms of Resistance of Extensively Drug Resistant Gram-Negative Bacteria. Antibiotics 2019, 8, 37. https://doi.org/10.3390/antibiotics8020037
Eichenberger EM, Thaden JT. Epidemiology and Mechanisms of Resistance of Extensively Drug Resistant Gram-Negative Bacteria. Antibiotics. 2019; 8(2):37. https://doi.org/10.3390/antibiotics8020037
Chicago/Turabian StyleEichenberger, Emily M., and Joshua T. Thaden. 2019. "Epidemiology and Mechanisms of Resistance of Extensively Drug Resistant Gram-Negative Bacteria" Antibiotics 8, no. 2: 37. https://doi.org/10.3390/antibiotics8020037
APA StyleEichenberger, E. M., & Thaden, J. T. (2019). Epidemiology and Mechanisms of Resistance of Extensively Drug Resistant Gram-Negative Bacteria. Antibiotics, 8(2), 37. https://doi.org/10.3390/antibiotics8020037




