Gut Bacteria of Water Monitor Lizard (Varanus salvator) Are a Potential Source of Antibacterial Compound(s)
Abstract
1. Introduction
2. Results
2.1. A Plethora of Bacteria Were Isolated from the Gut of Water Monitor Lizard
2.2. Bacterial Sensitivity Profile
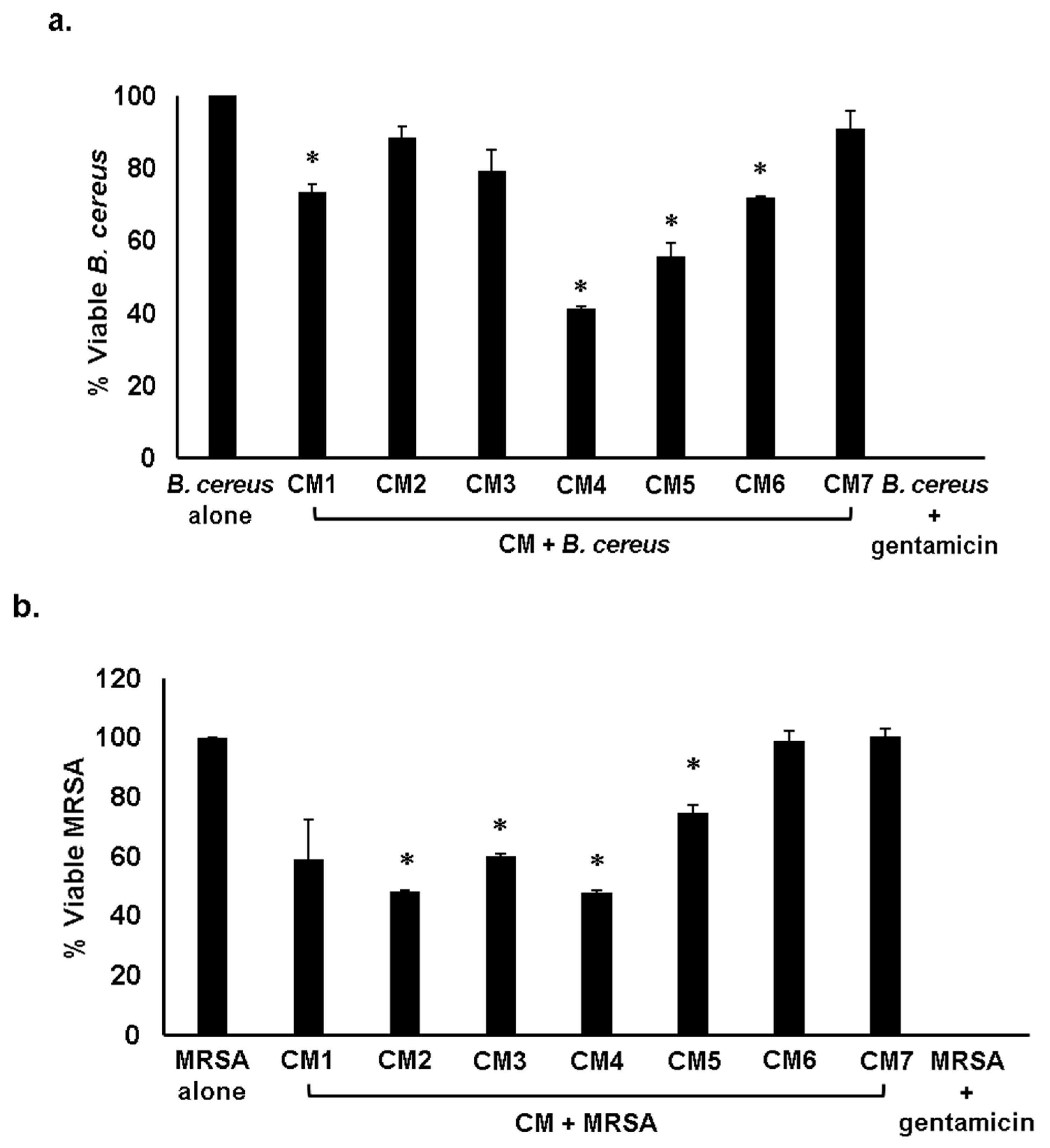
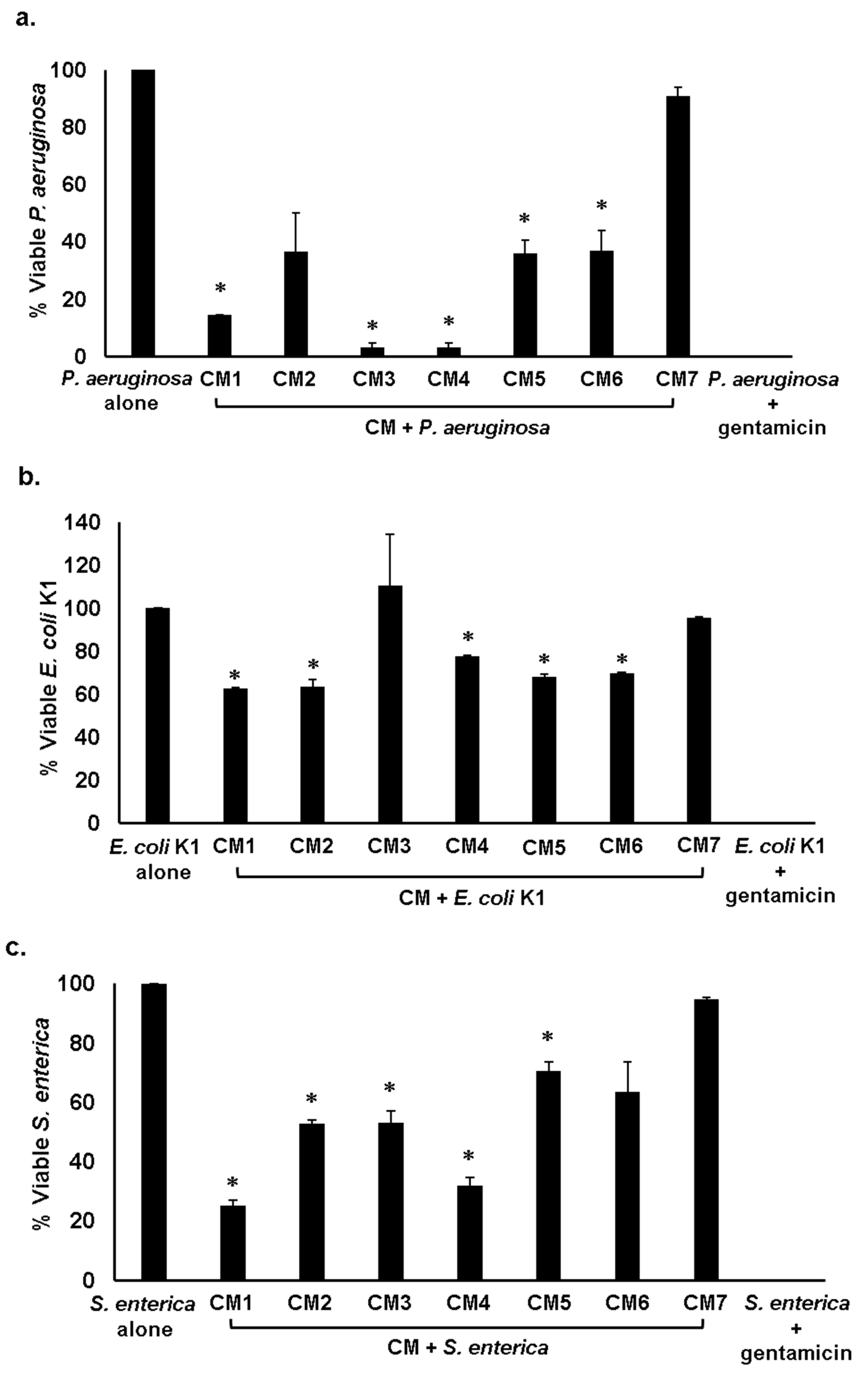
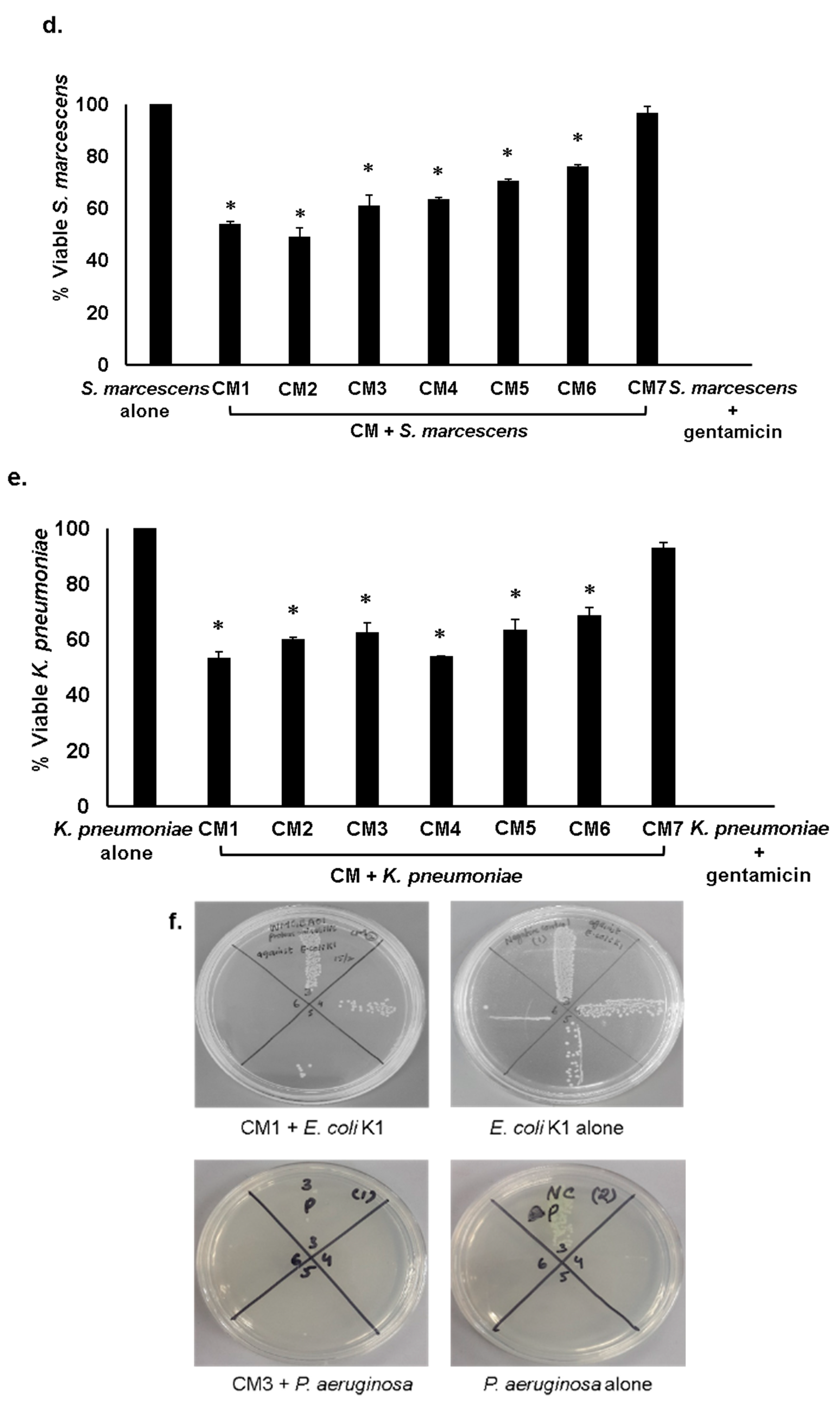
2.3. Gut Bacteria of Water Monitor Lizard Exhibited Bactericidal Properties Against Pathogenic Gram-Positive and Gram-Negative Bacteria
2.4. Antibacterial Properties of CM Were Heat-Resistant
2.5. Conditioned Medium Showed Limited Cytotoxic Effects on Human Cells
2.6. Liquid Chromatography-Mass Spectrometry Revealed a Plethora of Compound(s) from CM of Water Monitor Lizard Gut Microbiota
3. Discussion
4. Materials and Methods
4.1. Bacterial Cultures
4.2. Dissection of Water Monitor Lizard
4.3. Preparation of Bacterial Conditioned Medium
4.4. Antibacterial Assays
4.5. Host Cell Cytotoxicity Analyses
4.6. Mass Spectrometric Analysis of Culture Supernatant
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethical Approval and Consent to Participate
References
- Murray, C.J.; Lopez, A.D. Global and regional cause-of-death patterns in 1990. Bull. World Health Organ. 1994, 72, 447–480. [Google Scholar] [PubMed]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014; Available online: https://www.who.int/drugresistance/documents/surveillancereport/en/ (accessed on 29 December 2018).
- Lee, S.; Siddiqui, R.; Khan, N.A. Animals living in polluted environments are potential source of antimicrobials against infectious agents. Pathog. Glob. Health 2012, 106, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, J.; Siddiqui, R.; Khan, N.A. Acanthamoeba and bacteria produce antimicrobials to target their counterpart. Parasit. Vectors 2014, 7, e56. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, B.; Martin, J.; Ventura, S. United States Department of Health and Human Services, Centers for Disease Control and Prevention. National Center for Health Statistics. Natl. Vital. Stat. Rep. 2009, 57, e12. [Google Scholar]
- Hampton, T. Report reveals scope of US antibiotic resistance threat. JAMA 2013, 310, 1661–1663. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.C.; Fowler, T.; Watson, J.; Livermore, D.M.; Walker, D. Annual Report of the Chief Medical Officer: Infection and the rise of antimicrobial resistance. Lancet 2013, 381, 1606–1609. [Google Scholar] [CrossRef]
- Challinor, V.L.; Bode, H.B. Bioactive natural products from novel microbial sources. Ann. N. Y. Acad. Sci 2015, 1354, 82–97. [Google Scholar] [CrossRef]
- Sakoulas, G.; Nam, S.J.; Loesgen, S.; Fenical, W.; Jensen, P.R.; Nizet, V.; Hensler, M. Novel bacterial metabolite merochlorin A demonstrates in vitro activity against multi-drug resistant methicillin-resistant Staphylococcus aureus. PLoS ONE 2012, 7, e29439. [Google Scholar] [CrossRef]
- Akbar, N.; Siddiqui, R.; Iqbal, M.; Sagathevan, K.; Khan, N.A. Gut bacteria of cockroaches are a potential source of antibacterial compound(s). Lett. Appl. Microbiol. 2018, 66, 416–426. [Google Scholar] [CrossRef]
- Sagheer, M.; Siddiqui, R.; Iqbal, J.; Khan, N.A. Black cobra (Naja naja karachiensis) lysates exhibit broad-spectrum antimicrobial activities. Pathog. Glob. Health 2014, 108, 129–136. [Google Scholar] [CrossRef]
- Uyeda, L.; Iskandar, E.; Wirsing, A.; Kyes, R. Nocturnal activity of Varanus salvator on Tinjil Island, Indonesia. Biawak 2013, 7, 25–30. [Google Scholar]
- Uyeda, L.T. The Water Monitor Lizard Varanus salvator: Behavior, Ecology, and Human Dimensions in Banten, Indonesia. Ph.D. Thesis, University of Washington, Washington, DC, USA, 2015; p. 129. [Google Scholar]
- Ahgilan, A.; Sabaratnam, V.; Periasamy, V. Antimicrobial properties of vitamin B2. Int. J. Food Prop. 2016, 19, 1173–1181. [Google Scholar] [CrossRef]
- Massaro, C.F.; Shelley, D.; Heard, T.A.; Brooks, P. In vitro antibacterial phenolic extracts from “sugarbag” pot-honeys of Australian stingless bees (Tetragonula carbonaria). J. Agric. Food Chem. 2014, 62, 12209–12217. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xie, G.; Yan, X. Encyclopedia of traditional Chinese medicines. Isolat. Compd. AB 2011, 1, 455. [Google Scholar]
- Tada, H.; Yasuda, F. Siphonodiol, a new polyacetylenic metabolite from the sponge Siphonochalina truncate. Chem. Lett. 1984, 13, 779–780. [Google Scholar] [CrossRef]
- Filho, AA.O.; Fernandes, H.M.; Sousa, J.P.; Gabriela, L.A.; Barbosa-Filho, J.M.; Oliveira, T.L. Antibacterial activity of flavonoid 5.7. 4′-trimethoxyflavone isolated from Praxelis clematides RM King & Robinson. Bol. Latinoam. Caribe Plantas Med. Aromát 2013, 12, 400–404. [Google Scholar]
- Gaire, B.P.; Kwon, O.W.; Park, S.H.; Chun, K.H.; Kim, S.Y.; Shin, D.Y.; Choi, J.W. Neuroprotective effect of 6-paradol in focal cerebral ischemia involves the attenuation of neuroinflammatory responses in activated microglia. PLoS ONE 2015, 10, e0120203. [Google Scholar] [CrossRef]
- Zaher, A.M.; Moharram, A.M.; Davis, R.; Panizzi, P.; Makboul, M.A.; Calderón, A.I. Characterisation of the metabolites of an antibacterial endophyte Botryodiplodia theobromae Pat. of Dracaena draco L. by LC–MS/MS. Nat. Prod. Res. 2015, 29, 2275–2281. [Google Scholar] [CrossRef]
- Feng, L.; Maddox, M.M.; Alam, M.Z.; Tsutsumi, L.S.; Narula, G.; Bruhn, D.F.; Wu, X.; Sandhaus, S.; Lee, R.B.; Simmons, C.J.; et al. Synthesis, structure–activity relationship studies, and antibacterial evaluation of 4-chromanones and chalcones, as well as olympicin A and derivatives. J. Med. Chem. 2014, 57, 8398–8420. [Google Scholar] [CrossRef]
- Chakravorty, S.; Rayner, M.K.; De Koning, C.B.; Van Vuuren, S.F.; Van Otterlo, W.A. Synthesis and antimicrobial activity of the essential oil compounds (E)-and (Z)-3-hexenyl nonanoate and two analogues. S. Afr. J. Chem. 2012, 65, 202–205. [Google Scholar]
- Lee, M.R.; Raman, N.; Gellman, S.H.; Lynn, D.M.; Palecek, S.P. Hydrophobicity and helicity regulate the antifungal activity of 14-helical β-peptides. ACS Chem. Biol. 2014, 9, 1613–1621. [Google Scholar] [CrossRef] [PubMed]
- Opdyke, D.L. Monographs on fragrance raw materials. Food Chem. Toxicol. 1973, 11, 1011–1081. [Google Scholar] [CrossRef]
- León, A.; Del-Ángel, M.; Ávila, J.L.; Delgado, G. Phthalides: Distribution in nature, chemical reactivity, synthesis, and biological activity. Prog. Chem. Org. Nat. Prod. 2017, 104, 127–246. [Google Scholar] [PubMed]
- Pereira, C.B.; de Sá, N.P.; Borelli, B.M.; Rosa, C.A.; Barbeira, P.J.; Cota, B.B.; Johann, S. Antifungal activity of eicosanoic acids isolated from the endophytic fungus Mycosphaerella sp. against Cryptococcus neoformans and C. gattii. Microb. Pathog. 2016, 100, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Pawar, B.; Shinde, V.; Chaskar, A. n-Dodecylbenzene Sulfonic Acid (DBSA) as a Novel Brønsted Acid Catalyst for the Synthesis of Bis (indolyl) methanes and Bis (4-hydroxycoumarin-3-yl) methanes in Water. Green. Sust. Chem. 2013, 3, 56. [Google Scholar] [CrossRef]
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, G.Z.; Chen, H.H.; Wang, H.B.; Qin, S.; Zhu, W.Y.; Xu, L.H.; Jiang, C.L.; Li, W.J. Antitumour and antimicrobial activities of endophytic streptomycetes from pharmaceutical plants in rainforest. Lett. Appl. Microbiol. 2008, 47, 574–580. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Kumar, A.; Singh, R.; Pandey, K.D. Endophytic bacteria: A new source of bioactive compounds. 3 Biotech. 2017, 7, 315. [Google Scholar] [CrossRef]
- Gavrish, E.; Sit, C.S.; Clarissa, S.; Cao, S.; Kandror, O.; Spoering, A.; Peoples, A.; Ling, L.; Fetterman, A.; Hughes, D.; et al. Lassomycin, a Ribosomally Synthesized Cyclic Peptide, Kills Mycobacterium tuberculosis by Targeting the ATP-Dependent Protease ClpC1P1P2. Chem. Biol. 2014, 21, 509–518. [Google Scholar] [CrossRef]
- Atta, H.M. Biochemical studies on antibiotic production from Streptomyces sp.: Taxonomy, fermentation, isolation and biological properties. J. Saudi Chem. Soc. 2015, 19, 12–22. [Google Scholar] [CrossRef]
- Galindo, C.; Gutierrez, J.C.; Chopra, A. Potential involvement of galectin-3 and SNAP23 in Aeromonas hydrophila cytotoxic enterotoxin-induced host cell apoptosis. Microb. Pathog. 2006, 40, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Chopra, A.; Xu, X.J.; Ribardo, D.; Gonzalez, M.; Kuhl, K.; Peterson, J.; Houston, C. The cytotoxic enterotoxin of Aeromonas hydrophila induces proinflammatory cytokine production and activates arachidonic acid metabolism in macrophages. Infect. Immun. 2000, 68, 2808–2818. [Google Scholar] [CrossRef] [PubMed]
- Krzymińska, S.; Mokracka, J.; Koczura, R.; Cwiertnia, A.; Kaznowski, A. Aeromonas spp.-mediated cell-contact cytotoxicity is associated with the presence of type III secretion system. Antonie Van Leeuwenhoek 2012, 101, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Birch, A.; Robertson, W.; Geoghegan, I.; Mcgavin, W.; Alphey, T.; Phillips, M.; Fellows, L.; Watson, A.; Simmonds, M.; Porter, E. DMDP-a plant-derived sugar analogue with systemic activity against plant parasitic nematodes. Nematologica 1993, 39, 521–535. [Google Scholar]
- Niranjan, A.; Prakash, D. Chemical constituents and biological activities of turmeric (Curcuma longa L.)—A review. Int. J. Food Sci. Technol. 2008, 45, 109. [Google Scholar]
- Littlefield-Wyer, J.G.; Brooks, P.; Katouli, M. Application of biochemical fingerprinting and fatty acid methyl ester profiling to assess the effect of the pesticide Atradex on aquatic microbial communities. Environ. Pollut. 2008, 153, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Fadlallah, S.M.; Shehab, M.; Cheaito, K.; Saleh, M.; Ghosn, N.; Ammar, W.; El Hajj, R.; Matar, G.M. Molecular epidemiology and antimicrobial resistance of Salmonella species from clinical specimens and food Items in Lebanon. J. Infect. Dev. Ctries 2017, 11, 19–27. [Google Scholar] [CrossRef]
- Ali, S.M.; Siddiqui, R.; Ong, S.K.; Shah, M.R.; Anwar, A.; Heard, P.J.; Khan, N.A. Identification and characterization of antibacterial compound (s) of cockroaches (Periplaneta americana). Appl. Microbiol. Biotechnol. 2017, 101, 253–286. [Google Scholar] [CrossRef]
- Siddiqui, R.; Saleem, S.; Khan, N.A. The effect of peptidic and non-peptidic proteasome inhibitors on the biological properties of Acanthamoeba castellanii belonging to the T4 genotype. Exp. Parasitol. 2016, 168, 16–24. [Google Scholar] [CrossRef]
- Yasmin, S.; Hafeez, F.Y.; Mirza, M.S.; Rasul, M.; Arshad, H.M.; Zubair, M.; Iqbal, M. Biocontrol of bacterial leaf blight of rice and profiling of secondary metabolites produced by rhizospheric Pseudomonas aeruginosa BRp3. Front. Microbiol. 2017, 8, 1895. [Google Scholar] [CrossRef]




| Bacterial Source | Conditioned Medium |
|---|---|
| Proteus mirabilis | CM1 |
| Aeromonas hydrophila | CM2 |
| Citrobacter ferundii | CM3 |
| Escherichia coli | CM4 |
| Staphylococcus sp. | CM5 |
| Staphylococcus aureus | CM6 |
| Conditioned Media | Antibacterial Activities against Gram-Positive Bacteria | Antibacterial Activities against Gram-Negative Bacteria | ||||||
|---|---|---|---|---|---|---|---|---|
| Bacillus cereus | MRSA | Streptococcus pyogenes | Escherichia coli K1 | Klebsiella pneumoniae | Pseudomonas aeruginosa | Serratia marcescens | Salmonella enterica | |
| CM1 | + | − | + | + | + | + | + | + |
| CM2 | − | + | − | + | + | − | + | + |
| CM3 | − | + | + | − | + | + | + | + |
| CM4 | + | + | + | + | + | + | + | + |
| CM5 | + | + | + | + | + | + | + | + |
| CM6 | + | − | + | + | + | + | + | − |
| CM7 | − | − | − | − | − | − | − | − |
| Bacteria | Strain |
|---|---|
| Methicillin-resistant Staphylococcus aureus | MTCC 381123 (clinical isolate) |
| Escherichia coli K1 | MTCC 710859 (clinical isolate) |
| Streptococcus pyogenes | ATCC 49399 (clinical isolate) |
| Bacillus cereus | MTCC 131621 (clinical isolate) |
| Pseudomonas aeruginosa | ATCC 10145 (clinical isolate) |
| Klebsiella pneumoniae | ATCC 13883 (clinical isolate) |
| Escherichia coli K-12 | MTCC 817356 (non-clinical isolate) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akbar, N.; Siddiqui, R.; Sagathevan, K.; Iqbal, M.; Khan, N.A. Gut Bacteria of Water Monitor Lizard (Varanus salvator) Are a Potential Source of Antibacterial Compound(s). Antibiotics 2019, 8, 164. https://doi.org/10.3390/antibiotics8040164
Akbar N, Siddiqui R, Sagathevan K, Iqbal M, Khan NA. Gut Bacteria of Water Monitor Lizard (Varanus salvator) Are a Potential Source of Antibacterial Compound(s). Antibiotics. 2019; 8(4):164. https://doi.org/10.3390/antibiotics8040164
Chicago/Turabian StyleAkbar, Noor, Ruqaiyyah Siddiqui, K Sagathevan, Mazhar Iqbal, and Naveed Ahmed Khan. 2019. "Gut Bacteria of Water Monitor Lizard (Varanus salvator) Are a Potential Source of Antibacterial Compound(s)" Antibiotics 8, no. 4: 164. https://doi.org/10.3390/antibiotics8040164
APA StyleAkbar, N., Siddiqui, R., Sagathevan, K., Iqbal, M., & Khan, N. A. (2019). Gut Bacteria of Water Monitor Lizard (Varanus salvator) Are a Potential Source of Antibacterial Compound(s). Antibiotics, 8(4), 164. https://doi.org/10.3390/antibiotics8040164






