Incidence of Vancomycin Resistant Phenotype of the Methicillin Resistant Staphylococcus aureus Isolated from a Tertiary Care Hospital in Lahore
Abstract
:1. Introduction
2. Results
2.1. Antibiotic Susceptibility Pattern of MRSA
2.2. Antibiotic Susceptibility Pattern of VRSA/VISA Isolated from MRSA
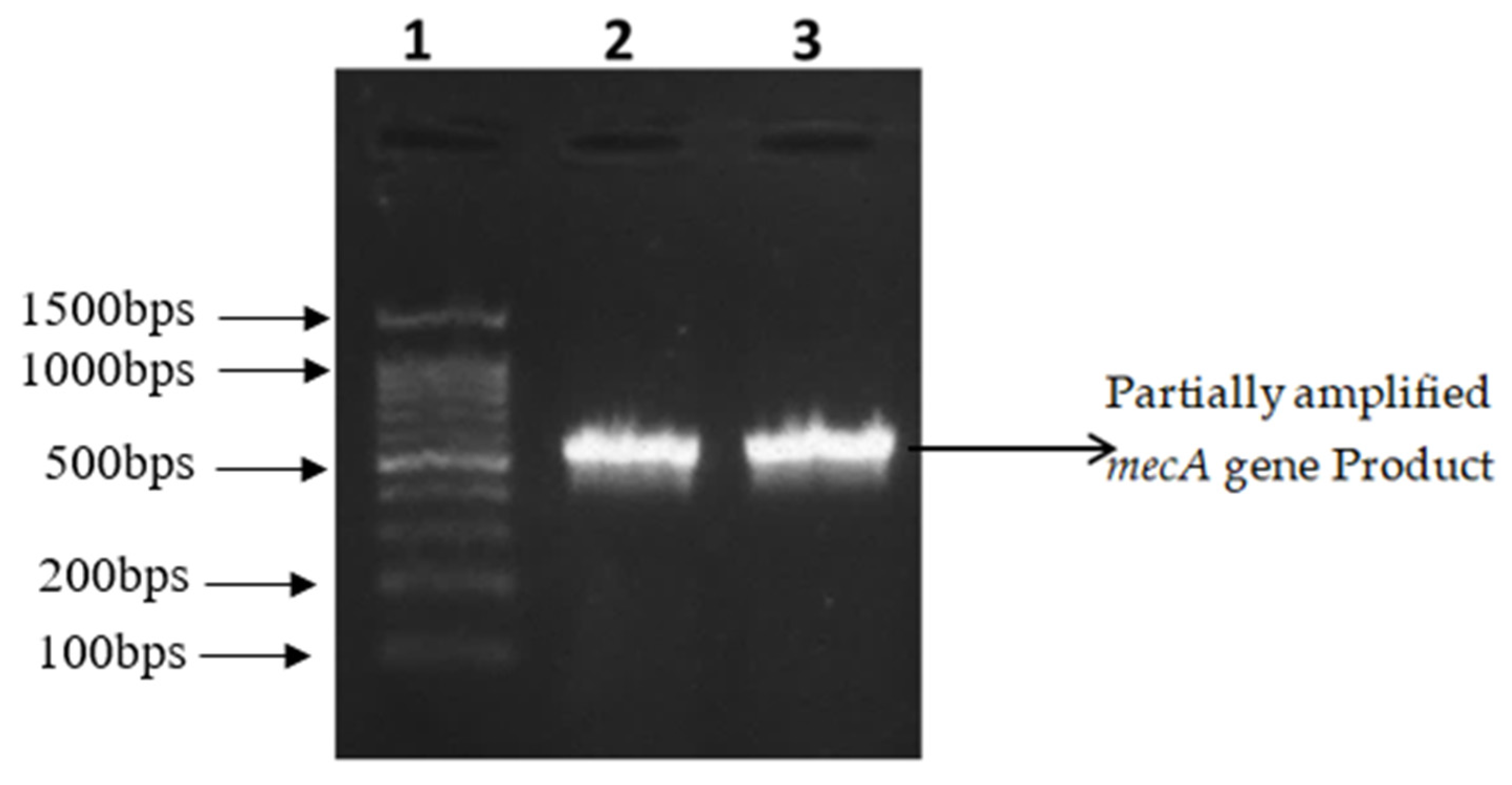
2.3. Amplification and Partial Sequencing of mecA and vanA
3. Discussion
4. Materials and Methods
4.1. Isolation and Identification of S. aureus
4.2. Antibiotic Susceptibility Testing
4.3. Phenotypic Detection of MRSA
4.4. MIC (Minimum Inhibitory Concentration) for the Confirmation of VRSA/VISA
4.5. Amplification of Methicillin and Vancomycin Resistance Genes
4.6. DNA Sequencing for mecA and vanA Gene Detection
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kuroda, M.; Ohta, T.; Uchiyama, I.; Baba, T.; Yuzawa, H.; Kobayashi, I.; Cui, L.; Oguchi, A.; Aoki, K.-I.; Nagai, Y. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 2001, 357, 1225–1240. [Google Scholar] [CrossRef]
- Saleem, N.; Nawaz, M.; Ghafoor, A.; Javeed, A.; Mustafa, A.; Yousuf, M.R.; Khan, I. Phenotypic and Molecular Analysis of Antibiotic Resistance in Lactobacilli of Poultry Origin from Lahore, Pakistan. Pak. Vet. J. 2018. [Google Scholar] [CrossRef]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Mcrobiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.J.; Huang, Y.C. New epidemiology of Staphylococcus aureus infection in Asia. Clin. Microbiol. Infect. 2014, 20, 605–623. [Google Scholar] [CrossRef] [Green Version]
- Kandemir, O.; Oztuna, V.; Colak, M.; Akdag, A.; Camdeviren, H. Comparison of the efficacy of tigecycline and teicoplanin in an experimental methicillin-resistant Staphylococcus aureus osteomyelitis model. J. Chemother. 2008, 20, 53–57. [Google Scholar] [CrossRef]
- Shamila-Syuhada, A.K.; Rusul, G.; Wan-Nadiah, W.A.; Chuah, L.-O. Prevalence and Antibiotics Resistance of Staphylococcus aureus Isolates Isolated from Raw Milk Obtained from Small-Scale Dairy Farms in Penang, Malaysia. Pak. Vet. J. 2016, 36, 98–102. [Google Scholar]
- Khanam, S.; Haq, J.A.; Shamsuzzaman, S.; Rahman, M.M.; Mamun, K.Z. Emergence of Vancomycin Resistant Staphylococcus aureus during Hospital Admission at a Tertiary Care Hospital in Bangladesh. Bangladesh J. Infec. Dis. 2016, 3, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Peacock, S.J.; Paterson, G.K. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef]
- Chambers, H.F. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 2001, 7, 178. [Google Scholar] [CrossRef]
- Tenover, F.C.; Biddle, J.W.; Lancaster, M.V. Increasing resistance to vancomycin and other glycopeptides in Staphylococcus aureus. Emerg. Infect. Dis. 2001, 7, 327. [Google Scholar] [CrossRef] [Green Version]
- Clark, N.C.; Weigel, L.M.; Patel, J.B.; Tenover, F.C. Comparison of Tn1546-like elements in vancomycin-resistant Staphylococcus aureus isolates from Michigan and Pennsylvania. Antimicrob. Agent Chemother. 2005, 49, 470–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perwaiz, S.; Barakzi, Q.; Farooqi, B.J.; Khursheed, N.; Sabir, N. Antimicrobial susceptibility pattern of clinical isolates of methicillin resistant Staphylococcus aureus. J. Pak. Med. Assoc. 2007, 57, 2. [Google Scholar] [PubMed]
- Akinkunmi, E.; Lamikanra, A. A study of the intestinal carriage of antibiotic resistant Staphylococcus aureus by Nigerian children. Afr. Health Sci. 2012, 12, 381–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kejela, T.; Bacha, K. Prevalence and antibiotic susceptibility pattern of methicillin-resistant Staphylococcus aureus (MRSA) among primary school children and prisoners in Jimma Town, Southwest Ethiopia. Ann. Clin. Microbiol. Antimicrob. 2013, 12, 11. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Tudela, J.; Barchiesi, F.; Bille, J.; Chryssanthou, E.; Cuenca-Estrella, M.; Denning, D.; Donnelly, J.; Dupont, B.; Fegeler, W.; Moore, C.; et al. Method for the determination of minimum inhibitory concentration (MIC) by broth dilution of fermentative yeasts. Clin. Microbiol. Infect. 2003, 9, i–viii. [Google Scholar] [CrossRef] [Green Version]
- Wikler, M.A. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically: Approved Standard; CLSI (NCCLS): Wayne, PA, USA, 2006. [Google Scholar]
- Altaf, M.; Ijaz, M.; Iqbal, M.K.; Rehman, A.; Avais, M.; Ghaffar, A.; Ayyub, R.M. Molecular Characterization of Methicillin Resistant Staphylococcus aureus (MRSA) and Associated Risk Factors with the Occurrence of Goat Mastitis. Pak. Vet. J. 2019. [Google Scholar] [CrossRef]
- Hussain, K.; Ijaz, M.; Farooqi, S.H.; Rizvi, B.; Nayab, S.; Ali, A.; Ghaffar, A.; Aqib, A.I.; Iqbal, M.K. Molecular Characterization of Clostridium perfringens Toxino-types and Type’D’Multidrug Resistance Profile in Diarrheic Sheep. Pak. Vet. J. 2018, 38, 271–275. [Google Scholar]
- Ji, G.; Chen, Q.; Gong, X.; Zheng, F.; Li, S.; Liu, Y. Topoisomerase Mutations are Associated with High-Level Ciprofloxacin Resistance in Staphylococcus saprophyticus, Enterococcus faecalis and Escherichia coli Isolated from Ducks. Pak. Vet. J. 2018, 38, 39–45. [Google Scholar] [CrossRef]
- Hasan, R.; Acharjee, M.; Noor, R. Prevalence of vancomycin resistant Staphylococcus aureus (VRSA) in methicillin resistant S. aureus (MRSA) strains isolated from burn wound infections. Tzu Chi Med. J. 2016, 28, 49–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thati, V.; Shivannavar, C.T.; Gaddad, S.M. Vancomycin resistance among methicillin resistant Staphylococcus aureus isolates from intensive care units of tertiary care hospitals in Hyderabad. Indian J. Med. Res. 2011, 134, 704. [Google Scholar] [CrossRef] [PubMed]
- Anupurba, S.; Sen, M.; Nath, G.; Sharma, B.; Gulati, A.; Mohapatra, T. Prevalence of methicillin resistant Staphylococcus aureus in a tertiary referral hospital in eastern Uttar Pradesh. Indian J. Med. Microbiol. 2003, 21, 49. [Google Scholar] [PubMed]
- Perveen, I.; Majid, A.; Knawal, S.; Naz, I.; Sehar, S.; Ahmed, S.; Raza, M.A. Prevalence and antimicrobial susceptibility pattern of methicillin-resistant Staphylococcus aureus and coagulase-negative Staphylococci in Rawalpindi, Pakistan. J. Adv. Med. Med. Res. 2013, 3, 198–209. [Google Scholar] [CrossRef] [Green Version]
- Hussain, M.S.; Naqvi, A.; Sharaz, M. Methicillin resistant Staphylococcus aureus (MRSA); Prevalence and Susceptibility Pattern of (MRSA) Isolated from Pus in Tertiary care of district hospital of Rahim Yar Khan. Prof. Med. J. 2019, 26. [Google Scholar] [CrossRef]
- Afroz, S. Detection of MRSA in Patients and Carriers by Evaluating Different Methods of Identification, Its Typing and Susceptibility to Vancomycin. Ph.D. Thesis, DMCH, Department of Microbiology, University of Dhaka, Dhaka, Bangladesh, 2005. [Google Scholar]
- Ghias, W.; Sharif, M.; Yazdani, F.A.; Rabbani, M. Isolation and identification of Methicillin and Vancomycin resistance Staphylococcus aureus from pus samples of injured skin patients in Lahore, Pakistan. Biomed. Lett. 2016, 2, 103–112. [Google Scholar]
- Maalik, A.; Ali, S.; Anam Iftikhar, M.R.; Ahmad, H.; Khan, I. Prevalence and Antibiotic Resistance of Staphylococcus aureus and Risk Factors for Bovine Subclinical Mastitis in District Kasur, Punjab, Pakistan. Pak. J. Zool 2019, 51, 1123–1130. [Google Scholar] [CrossRef]
- Elhassan, M.M.; Ozbak, H.A.; Hemeg, H.A.; Elmekki, M.A.; Ahmed, L.M. Absence of the mecA gene in methicillin resistant Staphylococcus aureus isolated from different clinical specimens in shendi city, Sudan. Biomed. Res. Int. 2015. [Google Scholar] [CrossRef] [Green Version]
- Saadat, S.; Solhjoo, K.; Norooz-Nejad, M.J.; Kazemi, A. VanA and vanB positive vancomycin-resistant Staphylococcus aureus among clinical isolates in Shiraz, South of Iran. Oman Med. J. 2015, 29, 335. [Google Scholar] [CrossRef]
- Schweiger, E.; Scheinfeld, N.; Tischler, H.; Weinberg, J. Linezolid and quinupristin/dalfopristin: Novel antibiotics for gram-positive infections of the skin. J. Drug Dermtol. 2003, 2, 378–383. [Google Scholar]
- Hudzicki, J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol; American Society for Microbiology: Washington, WA, USA, 2009. [Google Scholar]
- Rasheed, M.; Ahmed, Z. Phenotypic methods of greater accuracy to detect the mecA gene product for the recognition of MRSA in resource constraint settings. Asian Paci. J. Trop. Med. 2010, 3, 741–744. [Google Scholar] [CrossRef] [Green Version]
- Azimian, A.; Havaei, S.A.; Fazeli, H.; Naderi, M.; Ghazvini, K.; Samiee, S.M.; Soleimani, M.; Peerayeh, S.N. Genetic characterization of a vancomycin-resistant Staphylococcus aureus isolated from respiratory tract of a hospitalized patient in a university hospital in north east of Iran. J. Clin. Microbiol. 2012. [Google Scholar] [CrossRef] [Green Version]
- Patel, H.; Vaghasiya, Y.; Vyas, B.; Chanda, S. Antibiotic-resistant Staphylococcus aureus: A challenge to researchers and clinicians. Bacteriol. J. 2012. [Google Scholar] [CrossRef] [Green Version]

| Antibiotic | Quantity (µg) | Antibiotic Sensitivity Pattern of S. aureus (n = 100) | ||
|---|---|---|---|---|
| Resistant | Intermediate | Sensitive | ||
| Methicillin * | 5 | 76 | 11 | 13 |
| Ampicillin * | 10 | 81 | 12 | 7 |
| Aztreonam * | 30 | 85 | 6 | 9 |
| Tobramycin * | 10 | 78 | 10 | 12 |
| Tetracycline * | 30 | 74 | 17 | 9 |
| Moxifloxacin * | 5 | 76 | 9 | 15 |
| Cefoxitin * | 30 | 76 | 4 | 20 |
| Ertapenem * | 10 | 83 | 4 | 13 |
| Tigecycline * | 30 | 2 | 0 | 98 |
| Vancomycin ** | 0.25–256 µg/mL | 14 | 30 | 56 |
| Phenotype | Distribution of SA, MRSA, and VRSA from Different Type of Specimens | Chi-Square Test | |||
|---|---|---|---|---|---|
| Blood n (%) | Pus n (%) | Sputum n (%) | Body Fluid n (%) | ||
| SA | 25 | 36 | 24 | 15 | p > 0.05 * |
| MRSA | 19 (76) | 29 (80.55) | 19 (79.16) | 9 (60) | |
| MRSA+ + VRSA+ | 12 (63.16) | 14 (48.28) | 13 (68.42) | 5 (55.56) | |
| MRSA+ + VRSA− | 7 (36.84) | 15 (51.72) | 6 (31.58) | 4 (44.44) | |
| Background | Status | No of Individual |
|---|---|---|
| Education of Patients | Illiterate | 12 |
| Less than Matric | 48 | |
| Matric to Graduation | 40 | |
| Economic Status in terms of Income Per month (PKR) | <25,000 | 53 |
| 25,000–50,000 | 38 | |
| >50,000 | 09 | |
| Knowledge about Antibiotics | Yes | 11 |
| No | 89 | |
| Use of Medication | Yes | 34 |
| No | 66 | |
| Use of Antibiotics for suggested number of days | Yes | 05 |
| No | 95 |
| Antibiotic | MRSA Isolates Showing Susceptibility to Other Antibiotics | ||
|---|---|---|---|
| Resistant n (%) | Intermediate n (%) | Sensitive n (%) | |
| Methicillin * | 76 (100) | - | - |
| Ampicillin * | 76 (100) | - | - |
| Ertapenem * | 76 (100) | - | - |
| Tobramycin * | 72 (94.7) | 1 (1.3) | 3 (3.9) |
| Tetracycline * | 70 (92.1) | 3 (3.9) | 3 (3.9) |
| Cefoxitin * | 76 (100) | - | - |
| Moxifloxacin * | 68 (89.4) | 4 (5.26) | 3 (3.9) |
| Vancomycin ** | 14 (18.42) | - | 32 (42.1) |
| Tigecycline * | 1 (1.3) | - | 75 (98.7) |
| Antibiotics | No. of Isolates Showing Antibiotic Susceptibility * | ||
|---|---|---|---|
| Resistant n (%) | Intermediate n (%) | Sensitive n (%) | |
| Methicillin | 44 (100) | - | - |
| Ampicillin | 44 (100) | - | - |
| Cefoxitin | 44 (100) | - | - |
| Tobramycin | 43 (97.7) | 1 (2.3) | - |
| Tetracycline | 42 (95.5) | 1 (2.3) | 1 (2.3) |
| Ertapenem | 42 (95.5) | 2 (4.5) | - |
| Moxifloxacin | 41 (93.2) | 1 (2.3) | 2 (4.5) |
| Tigecycline | 1 (2.3) | - | 43 (97.7) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saeed, A.; Ahsan, F.; Nawaz, M.; Iqbal, K.; Rehman, K.U.; Ijaz, T. Incidence of Vancomycin Resistant Phenotype of the Methicillin Resistant Staphylococcus aureus Isolated from a Tertiary Care Hospital in Lahore. Antibiotics 2020, 9, 3. https://doi.org/10.3390/antibiotics9010003
Saeed A, Ahsan F, Nawaz M, Iqbal K, Rehman KU, Ijaz T. Incidence of Vancomycin Resistant Phenotype of the Methicillin Resistant Staphylococcus aureus Isolated from a Tertiary Care Hospital in Lahore. Antibiotics. 2020; 9(1):3. https://doi.org/10.3390/antibiotics9010003
Chicago/Turabian StyleSaeed, Aqib, Fatima Ahsan, Muhammad Nawaz, Khadeja Iqbal, Kashif Ur Rehman, and Tayyaba Ijaz. 2020. "Incidence of Vancomycin Resistant Phenotype of the Methicillin Resistant Staphylococcus aureus Isolated from a Tertiary Care Hospital in Lahore" Antibiotics 9, no. 1: 3. https://doi.org/10.3390/antibiotics9010003





