Potential Inhibitory Effect of Apis mellifera’s Venom and of Its Two Main Components—Melittin and PLA2—on Escherichia coli F1F0-ATPase
Abstract
:1. Introduction
2. Results
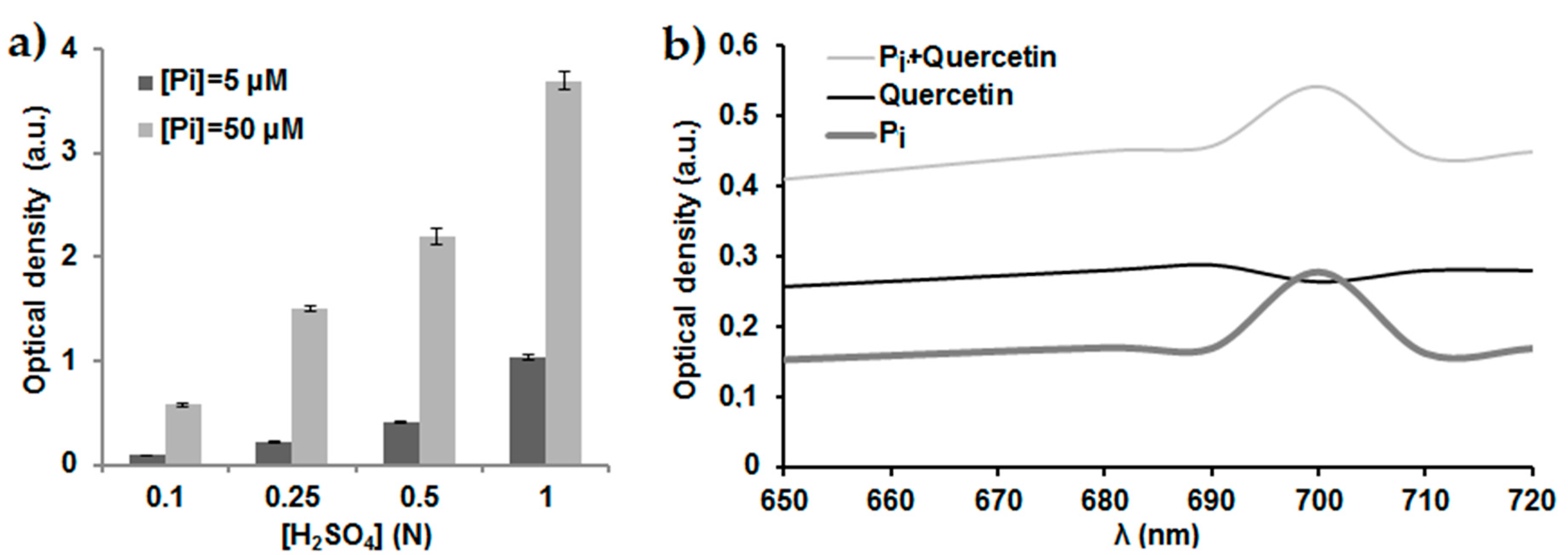
2.1. Optimization of the Phosphate Dosage Method
2.2. Screening of Natural Components as Potential Inhibitors of E. coli F1F0-ATPase
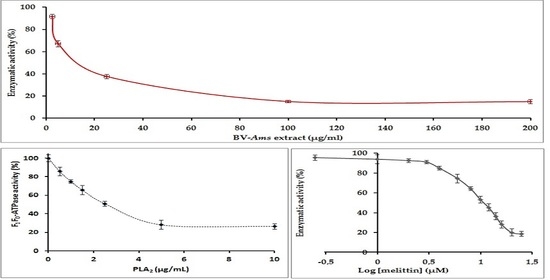
2.3. Study of the Effect of BV-Am on the Membrane-Bound E. coli. F1F0-ATPase
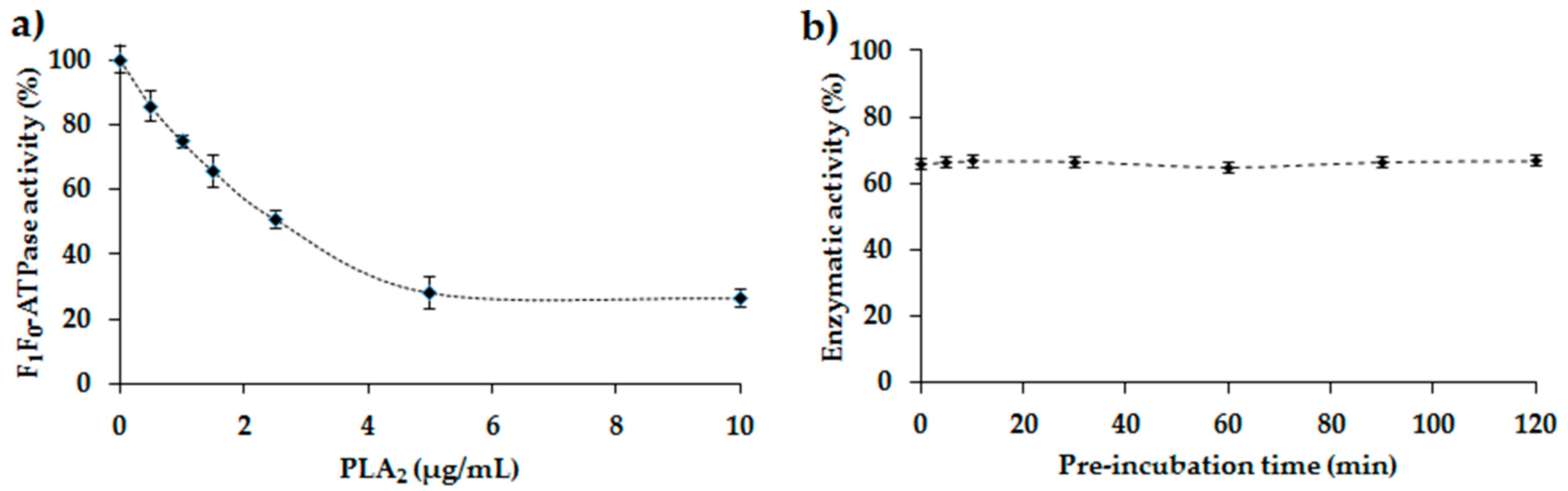
2.4. Study of the Inhibition of the E. coli F1F0-ATPase by PLA2
2.5. Inhibition of E. coli F1F0-ATPase by Melittin: Determination of the IC50 Value
2.6. Determination of the Kinetic Parameters of the Enzymatic Reaction Catalyzed by F1F0-ATPase in the Absence (Km and Vmax) and in the Presence of Melittin (K′m and V′max)
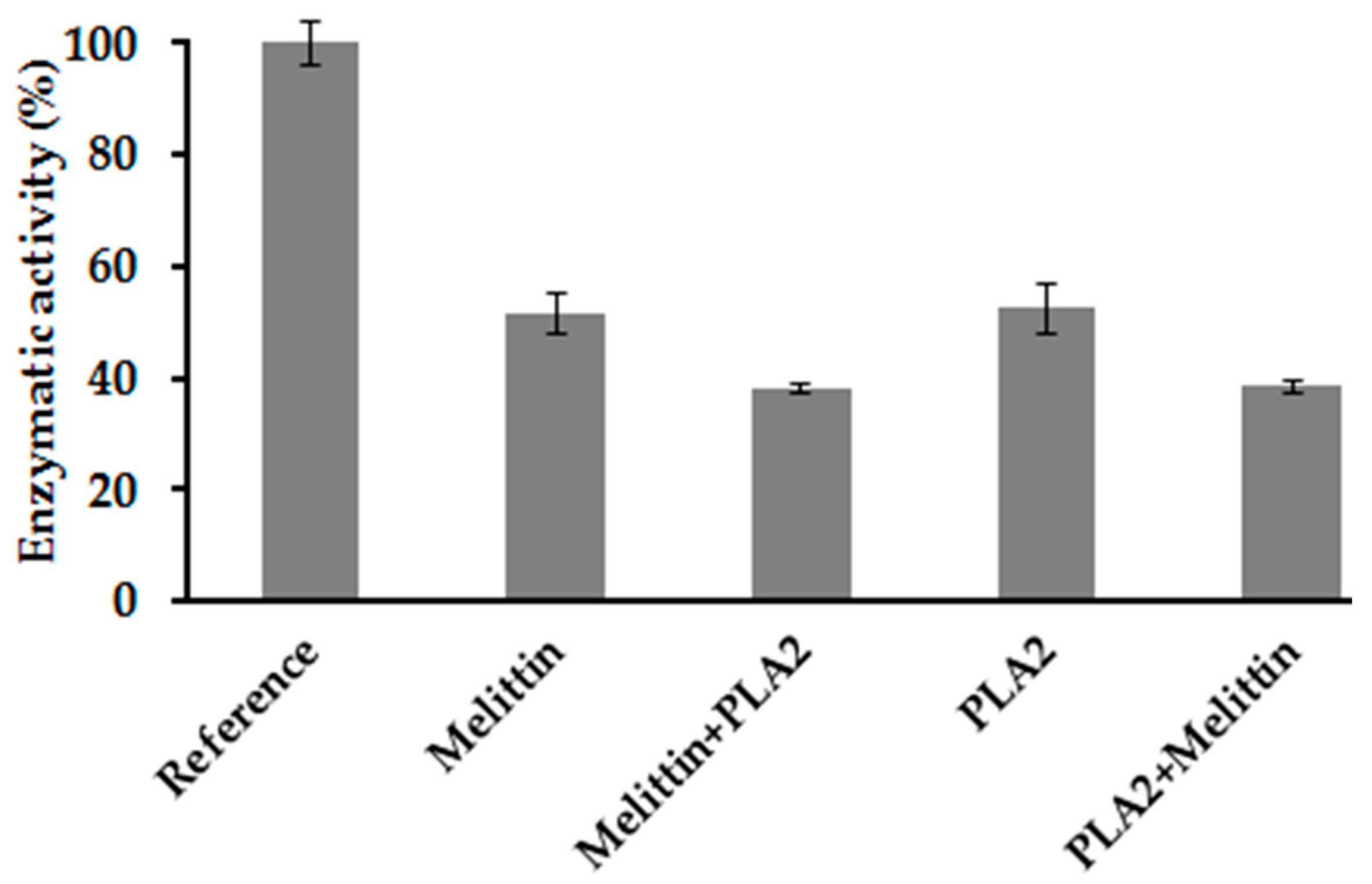
2.7. Combination Effect between Melittin and PLA2
3. Discussion
4. Materials and Methods
4.1. Venom
4.2. Reagents and Chemicals
4.3. Sources of Bacterial Strain and Culture Media
4.4. Bacterial Cultures
4.5. Preparation of the Bacterial Sample
4.6. Phosphate Quantification Protocol
4.7. Study of the Inhibition of E. coli F1F0-ATPase
4.8. Determination of IC50 Values of References and Potential Inhibitors of Membrane-Bound E. coli F1F0-ATPase
4.9. Study of the Potential Inhibitory Effect of BV-Am on the Membrane-Bound E. coli F1F0-ATPase
4.10. Study of the Potential Inhibitory Effect of PLA2 on the Membrane-Bound E. coli F1F0-ATPase
4.11. Study of the Potential Inhibitory Effect of Melittin on the Membrane-Bound E. coli F1F0-ATPase
4.12. Study of the Effect of Combination of PLA2 and Melittin on the Membrane-Bound E. coli F1F0-ATPase
4.13. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mainil, J. Escherichia coli virulence factors. Veter Immunol. Immunopathol. 2013, 152, 2–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanford, T.; Herbert, C.; Ronald, H. Theodor Escherich: The First Pediatric Infectious Diseases Physician. Clin. Infect. Dis. 2007, 45, 1025–1029. [Google Scholar]
- Wright, G.D. Antibiotic resistance: Where does it come from and what can we do about it? BMC Biol. 2010, 8, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, K.M.; Anselmi, C.; Wittig, I.; Faraldo-Gomez, J.D.; K’uhlbrandt, W. Stucture of the year F1F0-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc. Natl. Acad. Sci. USA 2012, 109, 13602–13607. [Google Scholar] [CrossRef] [Green Version]
- Sangjin, H.; Peter, L. ATP Synthase and the Actions of Inhibitors Utilized To Study Its Roles in Human Health, Disease, and Other Scientific Areas. Microbiol. Mol. Biol. Rev. 2008, 72, 590–641. [Google Scholar]
- Esparza-Moltó, P.B.; Nuevo-Tapioles, C.; Cuezva, J.M. Regulation of the H+-ATP synthase by IF1: A role in mitohormesis. Cell Mol. Life Sci. 2017, 74, 2151–2166. [Google Scholar] [CrossRef] [Green Version]
- Issa, D.; Najjar, A.; Greige-Gerges, H.; Nehme, H. Screening of Some Essential Oil Constituents as Potential Inhibitors of the ATP Synthase of Escherichia coli. J. Food Sci. 2018, 84, 138–146. [Google Scholar] [CrossRef] [Green Version]
- Laughlin, T.F.; Ahmad, Z. Inhibition of Escherichia coli ATP synthase by amphibian antimicrobial peptides. Int. J. Biol. Macromol. 2010, 46, 367–374. [Google Scholar] [CrossRef] [Green Version]
- Nesci, S.; Trombetti, F.; Algieri, C.; Pagliarani, A. A Therapeutic Role for the F1F0-ATP Synthase. SLAS Discov. 2019, 24, 893–903. [Google Scholar]
- Zulfiqar, A.; Florence, O.; Sofiya, A.; Thomas, F.L. ATP Synthase: A Molecular Therapeutic Drug Target for Antimicrobial and Antitumor Peptides. Curr. Med. Chem. 2013, 20, 1956–1973. [Google Scholar]
- Boyer, P.D. The ATP synthase - A splendid molecular machine. Annu. Rev. Biochem. 1997, 66, 717–749. [Google Scholar] [CrossRef] [Green Version]
- Azim, S.; McDowell, D.; Cartagena, A.; Rodriguez, R.; Laughlin, T.F.; Ahmad, Z. Venom peptides cathelicidin and lycotoxin cause strong inhibition of Escherichia coli ATP synthase. Int. J. Biol. Macromol. 2016, 87, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Hulin, M.; Bemrah, N.; Nougadere, A.; Volatier, J.; Sirot, V.; Leblanc, J.-C. Assessment of infant exposure to food chemicals: The French Total Diet Study design. Food Addit. Contam. Part A 2014, 31, 1–14. [Google Scholar] [CrossRef]
- Seow, Y.X.; Yeo, C.R.; Chung, H.L.; Yuk, H.-G. Plant Essential Oils as Active Antimicrobial Agents. Crit. Rev. Food Sci. Nutr. 2013, 54, 625–644. [Google Scholar] [CrossRef] [PubMed]
- Frangieh, J.; Salma, Y.; Haddad, K.; Mattei, C.; Legros, C.; Fajloun, Z.; El Obeid, D. First Characterization of The Venom from Apis mellifera syriaca, A Honeybee from The Middle East Region. Toxins 2019, 11, 191. [Google Scholar] [CrossRef] [Green Version]
- Wehbe, R.; Frangieh, J.; Rima, M.; El Obeid, D.; Sabatier, J.-M.; Fajloun, Z. Bee Venom: Overview of Main Compounds and Bioactivities for Therapeutic Interests. Molecules 2019, 24, 2997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zolfagharian, H.; Mohajeri, M.; Babaie, M. Bee Venom (Apis Mellifera) an Effective Potential Alternative to Gentamicin for Specific Bacteria Strains. J. Pharmacopunct. 2016, 19, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Núñez, V.; Arce, V.; Gutiérrez, J.M.; Lomonte, B. Structural and functional characterization of myotoxin I, a Lys49 phospholipase A2 homologue from the venom of the snake Bothrops atrox. Toxicon 2004, 44, 91–101. [Google Scholar] [CrossRef]
- Samy, R.P.; Gopalakrishnakone, P.; Thwin, M.; Chow, T.; Bow, H.; Yap, E.; Thong, T. Antibacterial activity of snake, scorpion and bee venoms: A comparison with purified venom phospholipase A2 enzymes. J. Appl. Microbiol. 2007, 102, 650–659. [Google Scholar] [CrossRef]
- Lowry, O.H.; A Lopez, J. The determination of inorganic phosphate in the presence of labile phosphate esters. J. Biol. Chem. 1946, 162, 421–428. [Google Scholar]
- Rouessac, F.; Rouessac, A. Chemical Analysis: Modern Instrumentation Methods and Techniques, 2nd ed.; John Wiley & Sons Ltd.: Chichester, UK, 2007; pp. 198–199. [Google Scholar]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Jackson, N.; Czaplewski, L.; Piddock, L.J. Discovery and development of new antibacterial drugs: Learning from experience? J. Antimicrob. Chemother. 2018, 73, 1452–1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinaldi, A.C. Antimicrobial peptides from amphibian skin: An expanding scenario: Commentary. Curr. Opin. Chem. Biol. 2002, 6, 799–804. [Google Scholar] [CrossRef]
- Nasyrova, G.F.; Beliaeva, N.V.; E Simchuk, E.; A Palladina, T. Effect of phospholipase A2 on H+-ATPase in the plasma membrane of maize root cells. Ukr. Biokhimicheskiĭ Zhurnal 1996, 68, 38–44. [Google Scholar]
- Kim, B.; Araujo, R.; Howard, M.; Magni, R.; Liotta, L.A.; Luchini, A. Affinity enrichment for mass spectrometry: Improving the yield of low abundance biomarkers. Expert Rev. Proteom. 2018, 15, 353–366. [Google Scholar] [CrossRef]
- Amini, A.; Raheem, S.; Steiner, A.; Deeba, F.; Ahmad, Z. Insect venom peptides as potent inhibitors of Escherichia coli ATP synthase. Int. J. Biol. Macromol. 2020, 150, 23–30. [Google Scholar] [CrossRef]
- Bowie, J.H.; Separovic, F.; Tyler, M.J. Host-defense peptides of Australian anurans. Part 2. Structure, activity, mechanism of action, and evolutionary significance. Peptides 2012, 37, 174–188. [Google Scholar] [CrossRef]
- Mann, A. Chapter 17—Conformational Restriction and/or Steric Hindrance in Medicinal Chemistry. In The Practice of Medicinal Chemistry, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2008; pp. 363–379. [Google Scholar]
- Pagliarani, A.; Nesci, S.; Ventrella, V. Novel Drugs Targeting the c-Ring of the F1F0-ATP Synthase. Mini Rev. Med. Chem. 2016, 16, 815–824. [Google Scholar] [CrossRef]
- Gledhill, J.R.; Walker, J.H. Inhibition sites in F1-ATPase from bovine heart mitochondria. Biochem. J. 2005, 386, 591–598. [Google Scholar] [CrossRef]
- Guo, H.; Suzuki, T.; Rubinstein, J.L. Structure of a bacterial ATP synthase. eLife 2019, 8, e43128. [Google Scholar] [CrossRef]
- Mowery, Y.M.; Pizzo, S.V. Targeting cell surface F1F0 ATP synthase in cancer therapy. Cancer Biol. Ther. 2008, 7, 1836–1838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esparza-Moltó, P.B.; Cuezva, J.M. The role of mitochondrial H+-ATP synthase in cancer. Front. Oncol. 2018, 8, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, Z.; Hassan, S.S.; Azim, S. A therapeutic connection between dietary phytochemicals and ATP synthase. Curr. Med. Chem. 2017, 24, 3894–3906. [Google Scholar] [CrossRef]
- Salomon, A.R.; Voehringer, D.W.; Herzenberg, L.A.; Khosla, C. Understanding and exploiting the mechanistic basis for selectivity of polyketide inhibitors of F0F1-ATPase. Proc. Natl. Acad. Sci. USA 2000, 97, 14766–14771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldberg, J.; Currais, A.; Prior, M.; Fischer, W.; Chiruta, C.; Ratliff, E.; Daugherty, D.; Dargusch, R.; Finley, L.; Esparza-Moltó, P.B.; et al. The mitochondrial ATP synthase is a shared drug target for aging and dementia. Aging Cell. 2018, 17, e12715. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Alavian, K.N.; Beutner, G.; Lazrove, E.; Sacchetti, S.; Park, H.A.; Licznerski, P.; Li, H.; Nabili, P.; Hockensmith, K.; Graham, M.; et al. An uncoupling channel within the c-subunit ring of the F1F0 ATP synthase is the mitochondrial permeability transition pore. Proc. Natl. Acad. Sci. USA 2014, 111, 10580–10585. [Google Scholar] [CrossRef] [Green Version]
- Bernardi, P.; Rosola, A.; Forte, M.; Lippe, G. The mitochondrial permeability transition pore: Channel formation by F-ATP synthase, integration in signal transduction, and role in pathophysiology. Physiol. Rev. 2015, 95, 1111–1115. [Google Scholar] [CrossRef]
- Martínez-Reyes, I.; Cuezva, J.M. The H+-ATP synthase: A gate to ROS-mediated cell death or cell survival. Biochim. Biophys. Acta 2014, 1837, 1099–1112. [Google Scholar] [CrossRef] [Green Version]
- Sebaugh, J.L. Guidelines for accurate EC50/IC50 estimation. Pharm. Stat. 2011, 10, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Bettelheim, F.A.; Brown, W.H.; Campbell, M.K.; Farrell, S.H.O.; Torres, O. Introduction to Organic and Biochemistry, 8th ed.; Cengage learning: Boston, MA, USA, 2011. [Google Scholar]
- Chou, T.-C. Theoretical Basis, Experimental Design, and Computerized Simulation of Synergism and Antagonism in Drug Combination Studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nehme, H.; Ayde, H.; El Obeid, D.; Sabatier, J.M.; Fajloun, Z. Potential Inhibitory Effect of Apis mellifera’s Venom and of Its Two Main Components—Melittin and PLA2—on Escherichia coli F1F0-ATPase. Antibiotics 2020, 9, 824. https://doi.org/10.3390/antibiotics9110824
Nehme H, Ayde H, El Obeid D, Sabatier JM, Fajloun Z. Potential Inhibitory Effect of Apis mellifera’s Venom and of Its Two Main Components—Melittin and PLA2—on Escherichia coli F1F0-ATPase. Antibiotics. 2020; 9(11):824. https://doi.org/10.3390/antibiotics9110824
Chicago/Turabian StyleNehme, Hala, Helena Ayde, Dany El Obeid, Jean Marc Sabatier, and Ziad Fajloun. 2020. "Potential Inhibitory Effect of Apis mellifera’s Venom and of Its Two Main Components—Melittin and PLA2—on Escherichia coli F1F0-ATPase" Antibiotics 9, no. 11: 824. https://doi.org/10.3390/antibiotics9110824
APA StyleNehme, H., Ayde, H., El Obeid, D., Sabatier, J. M., & Fajloun, Z. (2020). Potential Inhibitory Effect of Apis mellifera’s Venom and of Its Two Main Components—Melittin and PLA2—on Escherichia coli F1F0-ATPase. Antibiotics, 9(11), 824. https://doi.org/10.3390/antibiotics9110824