Scorpion-Venom-Derived Antimicrobial Peptide Css54 Exerts Potent Antimicrobial Activity by Disrupting Bacterial Membrane of Zoonotic Bacteria
Abstract
:1. Introduction
2. Results
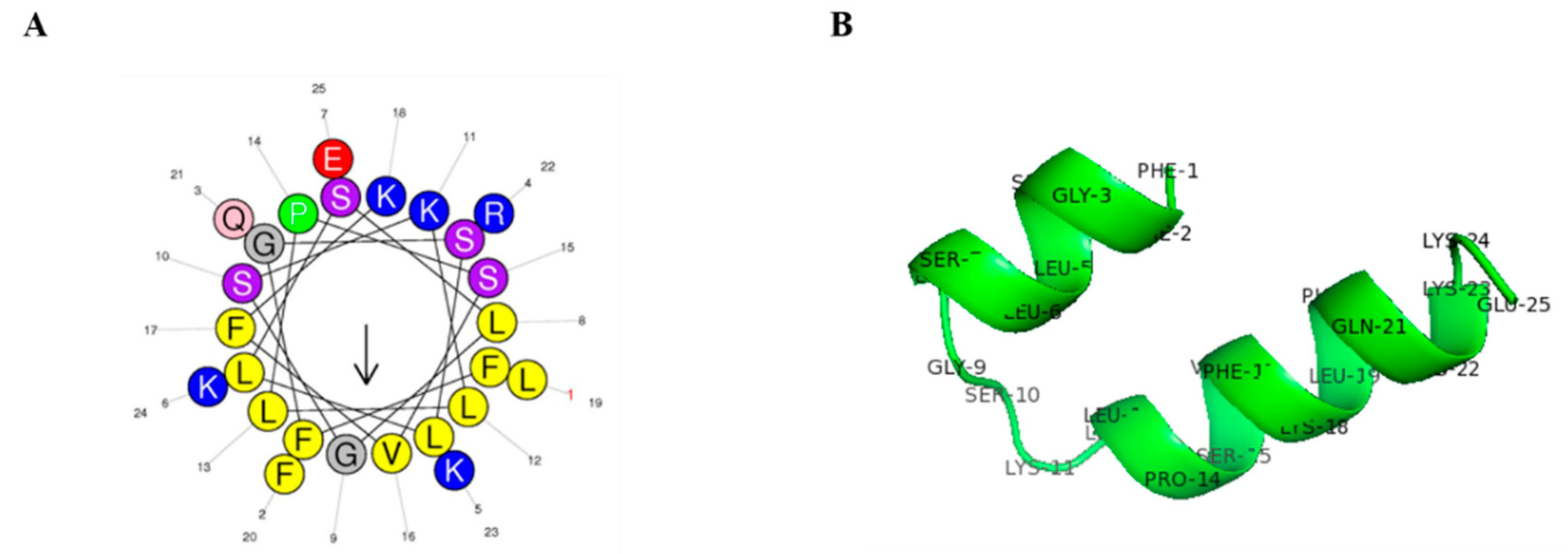
2.1. Peptide Structure and Characterization
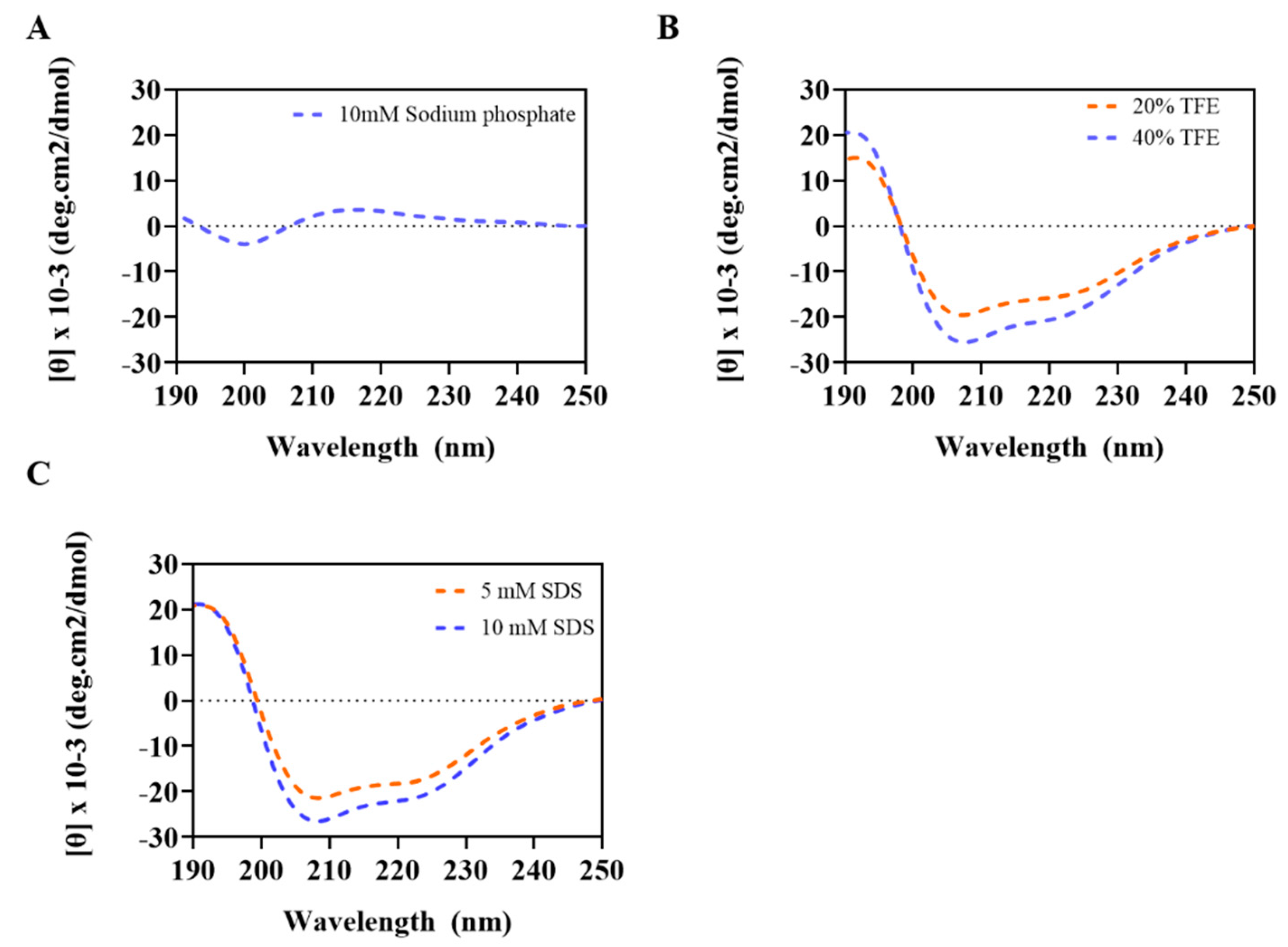
2.2. Circular Dichroism Measurements
2.3. Antimicrobial Activities against Zoonotic Pathogens
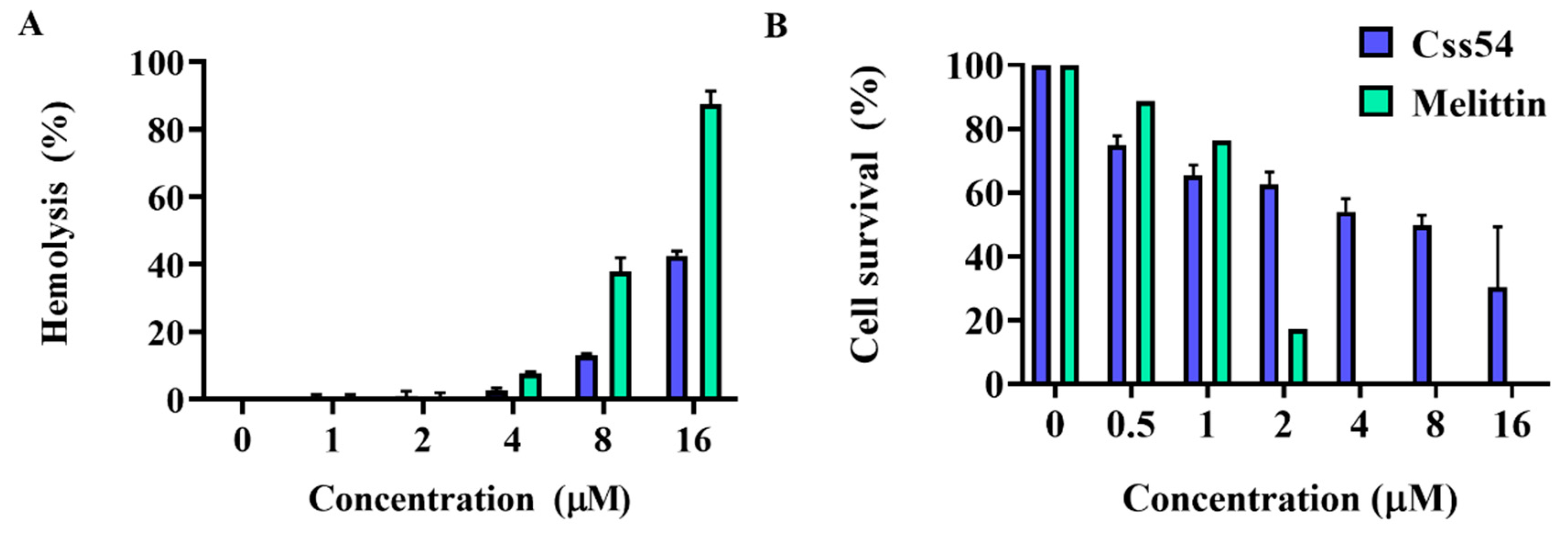
2.4. Cytotoxicity and Hemolytic Activities of Antimicrobial Peptides
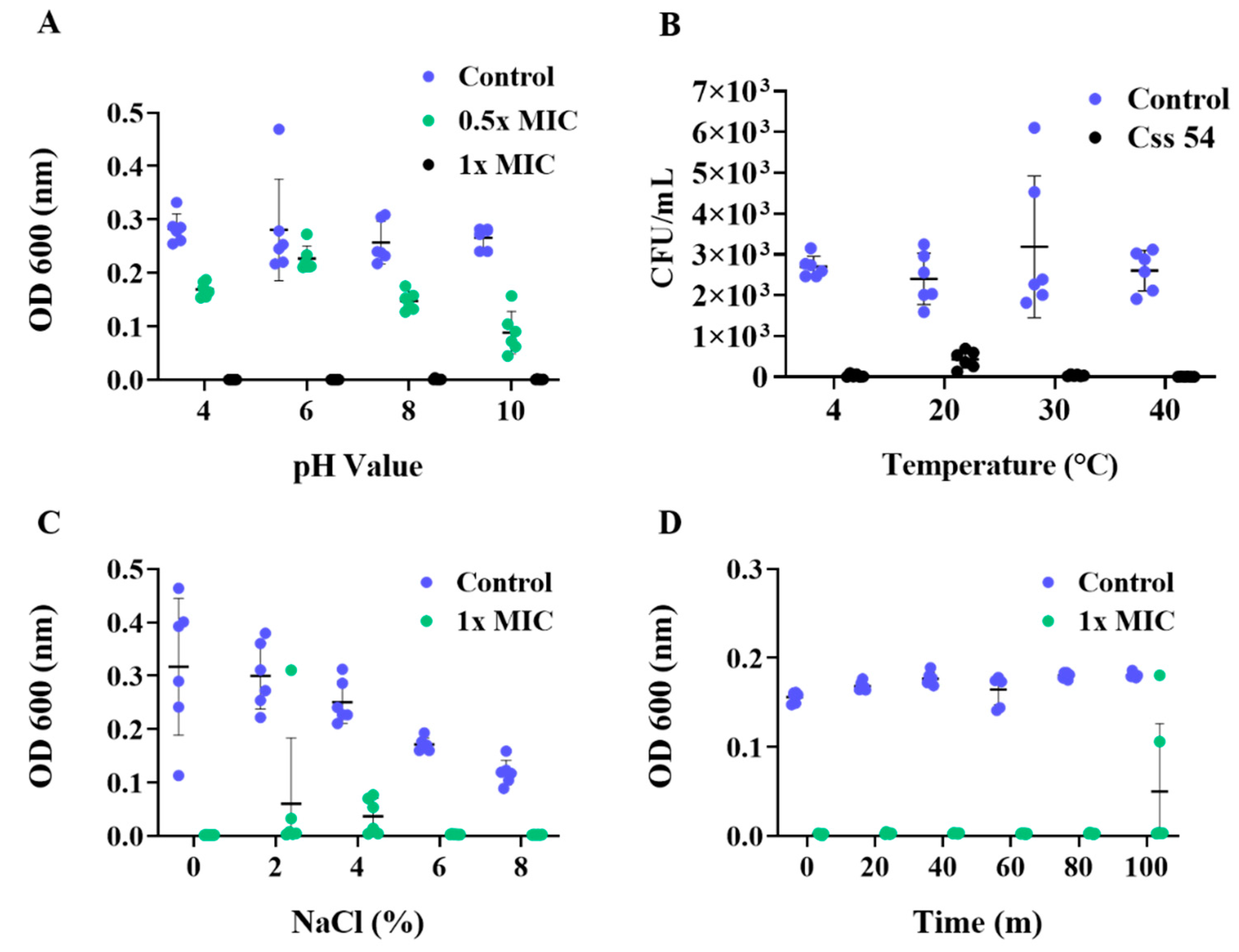
2.5. Antimicrobial Activities in Various Environments and Stability against Heat
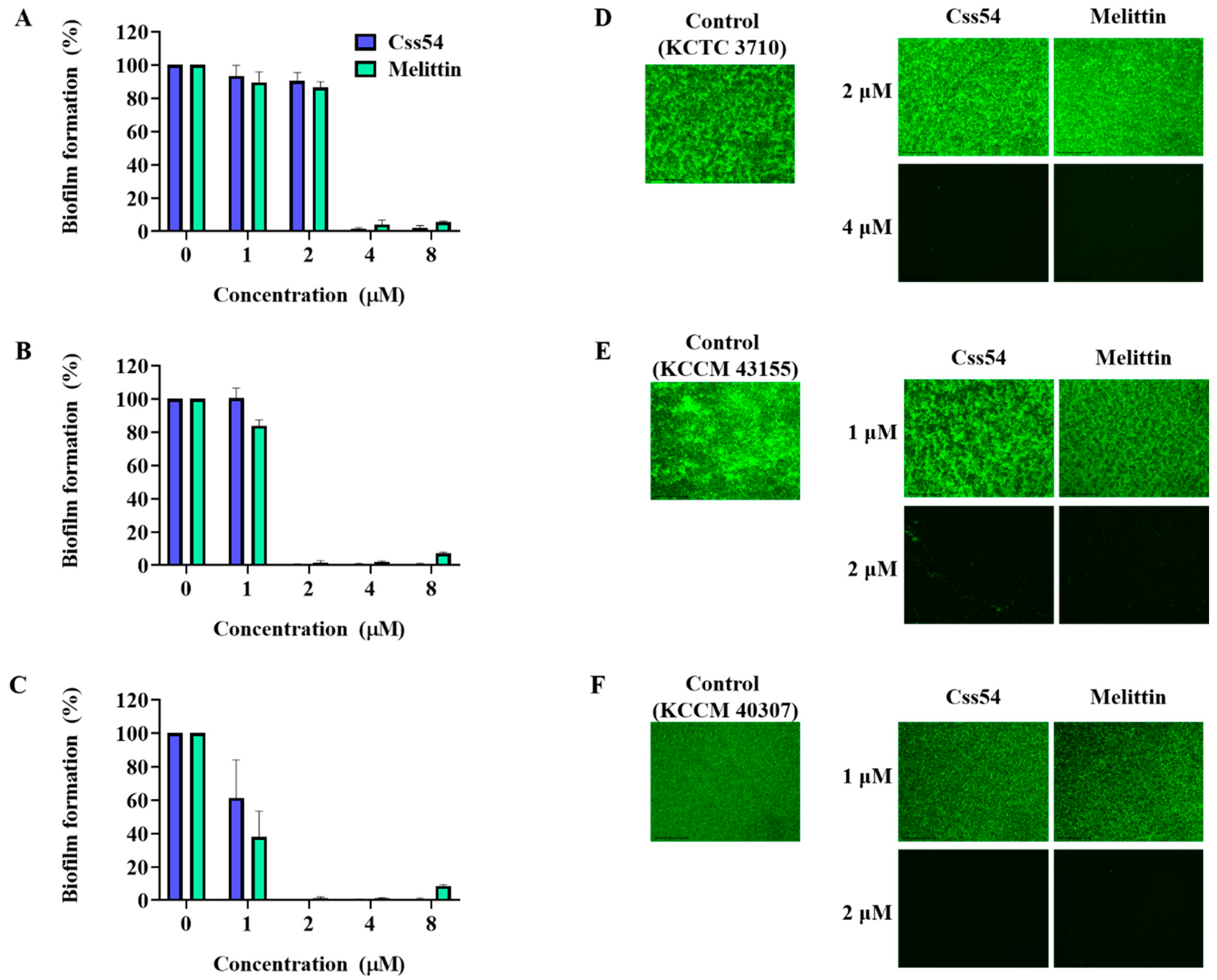
2.6. Biofilm Inhibition Assay
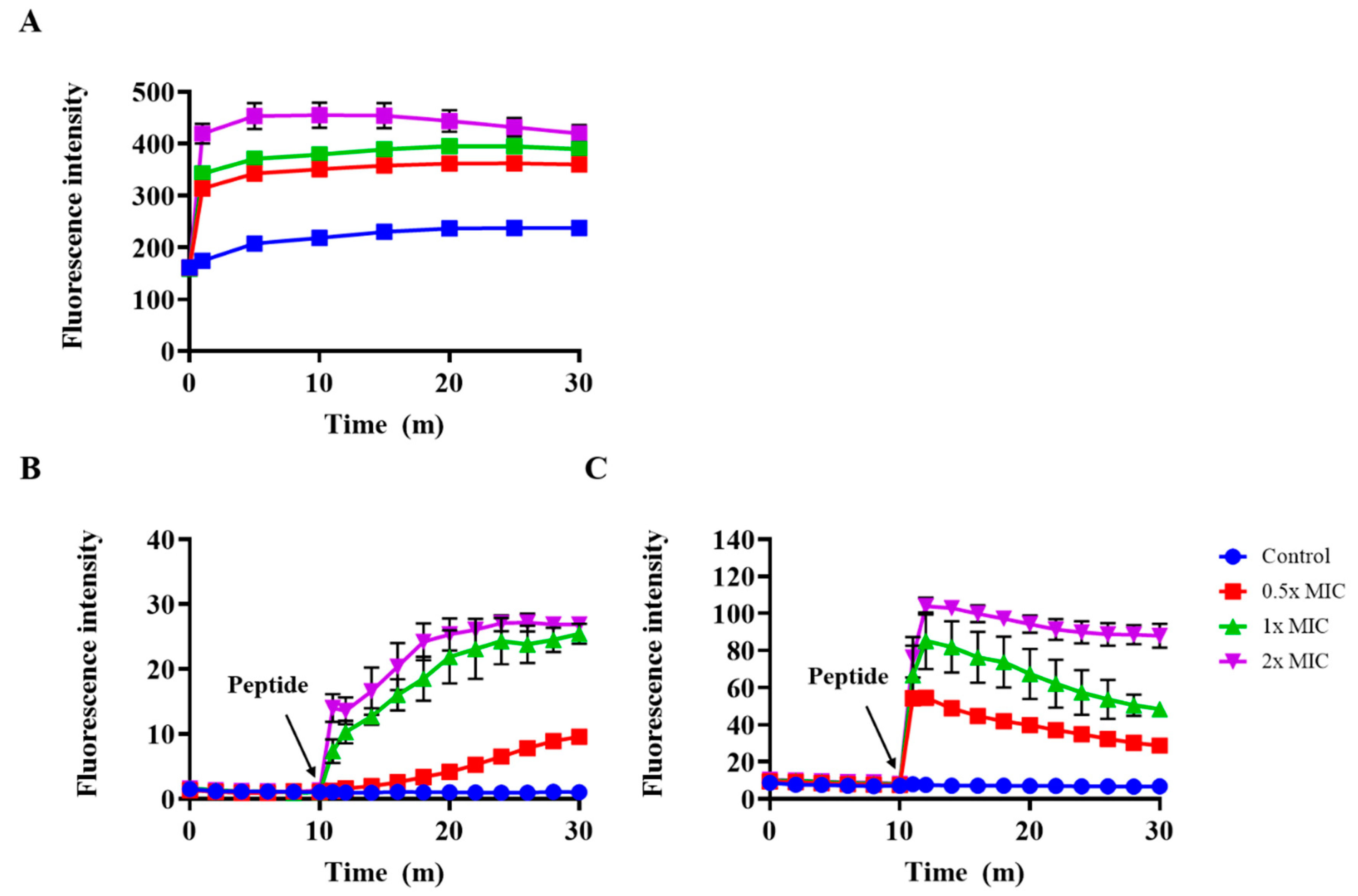
2.7. Outer Membrane Permeability and Membrane Depolarization Assay
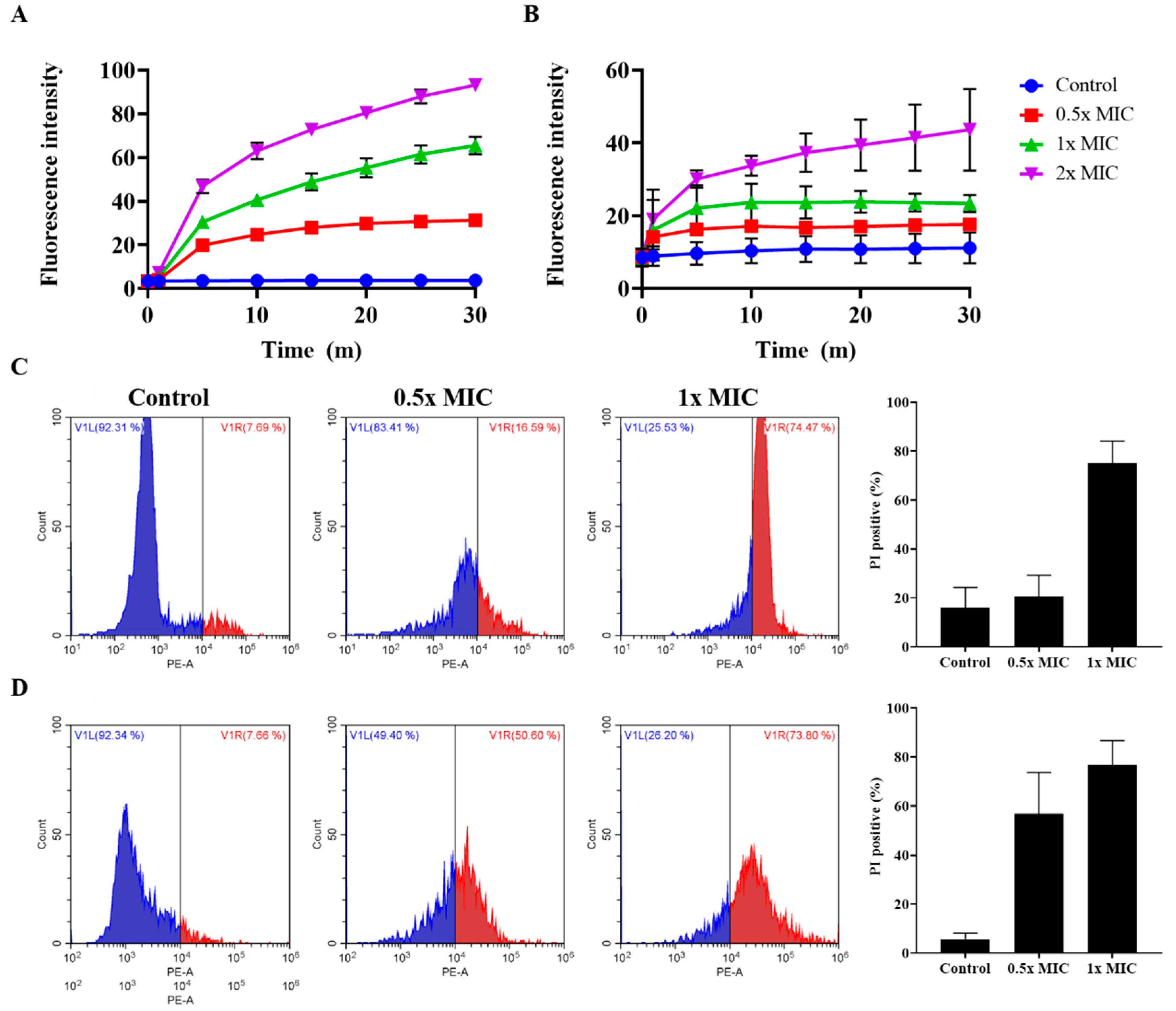
2.8. Effect of Peptides on Membrane Integrity
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Microorganisms
4.3. Peptide Synthesis and Sequence Analysis
4.4. Antimicrobial Activity Assay
4.5. Hemolytic Activity
4.6. Cytotoxicity Activity
4.7. Stability of Peptides
4.8. Biofilm Inhibition Assay
4.9. Circular Dichroism (CD) Spectroscopy
4.10. Outer Membrane Permeability Assay
4.11. Membrane Depolarization Assay
4.12. SYTOX Green Uptake Assay
4.13. Flow Cytometry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Veltri, D.; Kamath, U.; Shehu, A. Deep learning improves antimicrobial peptide recognition. Bioinformatics 2018, 34, 2740–2747. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Li, L. China’s national plan to combat antimicrobial resistance. Lancet Infect. Dis. 2016, 16, 1216–1218. [Google Scholar] [CrossRef]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J. Antibiotic resistance genes from livestock waste: Occurrence, dissemination, and treatment. Npj Clean Water 2020, 3, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Van Boeckel, T.P.; Glennon, E.E.; Chen, D.; Gilbert, M.; Robinson, T.P.; Grenfell, B.T.; Levin, S.A.; Bonhoeffer, S.; Laxminarayan, R. Reducing antimicrobial use in food animals. Science 2017, 357, 1350–1352. [Google Scholar] [CrossRef] [Green Version]
- Koch, B.J.; Hungate, B.A.; Price, L.B. Food-animal production and the spread of antibiotic resistance: The role of ecology. Front. Ecol. Environ. 2017, 15, 309–318. [Google Scholar] [CrossRef]
- Karesh, W.B.; Dobson, A.; Lloyd-Smith, J.O.; Lubroth, J.; Dixon, M.A.; Bennett, M.; Aldrich, S.; Harrington, T.; Formenty, P.; Loh, E.H. Ecology of zoonoses: Natural and unnatural histories. Lancet 2012, 380, 1936–1945. [Google Scholar] [CrossRef]
- Collaboration, A. Interagency Food Safety Analytics Collaboration (Ifsac) Strategic Plan Calendar Year 2017–2021. Available online: https://www.cdc.gov/foodsafety/pdfs/IFSAC-Strategic-Plan-2017-2021.pdf (accessed on 8 October 2020).
- Chlebicz, A.; Śliżewska, K. Campylobacteriosis, salmonellosis, yersiniosis, and listeriosis as zoonotic foodborne diseases: A review. Int. J. Environ. Res. Public Health 2018, 15, 863. [Google Scholar] [CrossRef] [Green Version]
- Farber, J.; Peterkin, P. Listeria monocytogenes, a food-borne pathogen. Microbiol. Mol. Biol. Rev. 1991, 55, 476–511. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Migura, L.; Hendriksen, R.S.; Fraile, L.; Aarestrup, F.M. Antimicrobial resistance of zoonotic and commensal bacteria in europe: The missing link between consumption and resistance in veterinary medicine. Vet. Microbiol. 2014, 170, 1–9. [Google Scholar] [CrossRef]
- Rath, E.C.; Gill, H.; Bai, Y. Identification of potential antimicrobials against salmonella typhimurium and listeria monocytogenes using quantitative structure-activity relation modeling. PLoS ONE 2017, 12, e0189580. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Yang, F.; Li, F.; Li, Z.; Lang, Y.; Shen, B.; Wu, Y.; Li, W.; Harrison, P.L.; Strong, P.N. Therapeutic potential of a scorpion venom-derived antimicrobial peptide and its homologs against antibiotic-resistant gram-positive bacteria. Front. Microbiol. 2018, 9, 1159. [Google Scholar] [CrossRef]
- Attarde, S.; Pandit, S. Scorpion venom as therapeutic agent-current perspective. Int. J. Curr. Pharm. Res. 2016, 7, 59–72. [Google Scholar]
- Harrison, P.L.; Abdel-Rahman, M.A.; Miller, K.; Strong, P.N. Antimicrobial peptides from scorpion venoms. Toxicon 2014, 88, 115–137. [Google Scholar] [CrossRef]
- Tossi, A.; Sandri, L.; Giangaspero, A. Amphipathic, α-helical antimicrobial peptides. Pept. Sci. 2000, 55, 4–30. [Google Scholar] [CrossRef]
- Kuroda, K.; Caputo, G.A. Antimicrobial polymers as synthetic mimics of host-defense peptides. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2013, 5, 49–66. [Google Scholar] [CrossRef] [Green Version]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Kang, H.-K.; Kim, C.; Seo, C.H.; Park, Y. The therapeutic applications of antimicrobial peptides (amps): A patent review. J. Microbiol. 2017, 55, 1–12. [Google Scholar] [CrossRef]
- Kumar, S.; Varela, M.F. Molecular mechanisms of bacterial resistance to antimicrobial agents. Chemotherapy 2013, 14, 522–534. [Google Scholar]
- Garcia, F.; Villegas, E.; Espino-Solis, G.P.; Rodriguez, A.; Paniagua-Solis, J.F.; Sandoval-Lopez, G.; Possani, L.D.; Corzo, G. Antimicrobial peptides from arachnid venoms and their microbicidal activity in the presence of commercial antibiotics. J. Antibiot. 2013, 66, 3–10. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Wei, D.; Yan, P.; Zhu, X.; Shan, A.; Bi, Z. Characterization of cell selectivity, physiological stability and endotoxin neutralization capabilities of α-helix-based peptide amphiphiles. Biomaterials 2015, 52, 517–530. [Google Scholar] [CrossRef]
- Chung, E.M.; Dean, S.N.; Propst, C.N.; Bishop, B.M.; van Hoek, M.L. Komodo dragon-inspired synthetic peptide drgn-1 promotes wound-healing of a mixed-biofilm infected wound. npj Biofilms Microbiomes 2017, 3, 1–13. [Google Scholar] [CrossRef]
- Colagiorgi, A.; Bruini, I.; Di Ciccio, P.A.; Zanardi, E.; Ghidini, S.; Ianieri, A. Listeria monocytogenes biofilms in the wonderland of food industry. Pathogens 2017, 6, 41. [Google Scholar] [CrossRef] [Green Version]
- Dosler, S.; Karaaslan, E. Inhibition and destruction of pseudomonas aeruginosa biofilms by antibiotics and antimicrobial peptides. Peptides 2014, 62, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Picoli, T.; Peter, C.M.; Zani, J.L.; Waller, S.B.; Lopes, M.G.; Boesche, K.N.; de Oliveira Hübner, S.; Fischer, G. Melittin and its potential in the destruction and inhibition of the biofilm formation by staphylococcus aureus, escherichia coli and pseudomonas aeruginosa isolated from bovine milk. Microb. Pathog. 2017, 112, 57–62. [Google Scholar] [CrossRef]
- Sun, S.; Zhao, G.; Huang, Y.; Cai, M.; Yan, Q.; Wang, H.; Chen, Y. Enantiomeric effect of d-amino acid substitution on the mechanism of action of α-helical membrane-active peptides. Int. J. Mol. Sci. 2018, 19, 67. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Dong, W.; Sun, L.; Ma, L.; Shang, D. Insights into the membrane interaction mechanism and antibacterial properties of chensinin-1b. Biomaterials 2015, 37, 299–311. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [Green Version]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [Green Version]
- Le, C.-F.; Fang, C.-M.; Sekaran, S.D. Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [Green Version]
- Pfalzgraff, A.; Brandenburg, K.; Weindl, G. Antimicrobial peptides and their therapeutic potential for bacterial skin infections and wounds. Front. Pharmacol. 2018, 9, 281. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Feix, J.B. Peptide–membrane interactions and mechanisms of membrane destruction by amphipathic α-helical antimicrobial peptides. Biochim. Biophys. Acta (BBA) Biomembr. 2006, 1758, 1245–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leveritt, J.M., III; Pino-Angeles, A.; Lazaridis, T. The structure of a melittin-stabilized pore. Biophys. J. 2015, 108, 2424–2426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dempsey, C.E. The actions of melittin on membranes. Biochim. Biophys. Acta (BBA) Rev. Biomembr. 1990, 1031, 143–161. [Google Scholar] [CrossRef]
- Oruko, R.O.; Odiyo, J.O.; Edokpayi, J.N. The role of leather microbes in human health. In Role of Microbes in Human Health and Diseases; IntechOpen: Rijeka, Croatia, 2019. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Segura, M. Streptococcus suis research: Progress and Challenges. Pathogenes 2020, 9, 707. [Google Scholar] [CrossRef]
- Abu-Zinadah, O.; Rahmy, T.; Alahmari, A.; Abdu, F. Effect of melittin on mice stomach. Saudi J. Biol. Sci. 2014, 21, 99–108. [Google Scholar] [CrossRef] [Green Version]
- Rahmy, T.; Alahmari, A.; Abdu, F.; Abu-Zinadah, O. Protective effect of melittin against gastric inflammation in mice. Life Sci. J. 2013, 10, 1369–1384. [Google Scholar]
- Duffy, C.; Sorolla, A.; Wang, E.; Golden, E.; Woodward, E.; Davern, K.; Ho, D.; Johnstone, E.; Pfleger, K.; Redfern, A. Honeybee venom and melittin suppress growth factor receptor activation in her2-enriched and triple-negative breast cancer. NPJ Precis. Oncol. 2020, 4, 1–16. [Google Scholar] [CrossRef]
- Russo, P.; Hadjilouka, A.; Beneduce, L.; Capozzi, V.; Paramithiotis, S.; Drosinos, E.H.; Spano, G. Effect of different conditions on listeria monocytogenes biofilm formation and removal. Czech J. Food Sci. 2018, 36, 208–214. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Lawrence, M.L.; Ainsworth, A.J.; Austin, F.W. Comparative assessment of acid, alkali and salt tolerance in listeria monocytogenes virulent and avirulent strains. FEMS Microbiol. Lett. 2005, 243, 373–378. [Google Scholar] [CrossRef] [Green Version]
- Dong, N.; Chou, S.; Li, J.; Xue, C.; Li, X.; Cheng, B.; Shan, A.; Xu, L. Short symmetric-end antimicrobial peptides centered on β-turn amino acids unit improve selectivity and stability. Front. Microbiol. 2018, 9, 2832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, X.; Liu, H.; Zheng, Y.; Guo, D.; Shi, C.; Xu, Y.; Xia, X. Inhibitory effect of tq on listeria monocytogenes atcc 19115 biofilm formation and virulence attributes critical for human infection. Front. Cell. Infect. Microbiol. 2019, 9, 304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mojsoska, B.; Jenssen, H. Peptides and peptidomimetics for antimicrobial drug design. Pharmaceuticals 2015, 8, 366–415. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Rajasekaran, G.; Shin, S.Y. Ll-37-derived short antimicrobial peptide kr-12-a5 and its d-amino acid substituted analogs with cell selectivity, anti-biofilm activity, synergistic effect with conventional antibiotics, and anti-inflammatory activity. Eur. J. Med. Chem. 2017, 136, 428–441. [Google Scholar] [CrossRef] [PubMed]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. Pep-fold3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef] [Green Version]
- Yuan, S.; Chan, H.S.; Hu, Z. Using pymol as a platform for computational drug design. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2017, 7, e1298. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (mic) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-K.; Park, S.-C.; Hahm, K.-S.; Park, Y. Antimicrobial hpa3nt3 peptide analogs: Placement of aromatic rings and positive charges are key determinants for cell selectivity and mechanism of action. Biochim. Biophys. Acta (BBA) Biomembr. 2013, 1828, 443–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.K.; Kang, N.H.; Ko, S.J.; Park, J.; Park, E.; Shin, D.W.; Kim, S.H.; Lee, S.A.; Lee, J.I.; Lee, S.H. Antibacterial and antibiofilm activity and mode of action of magainin 2 against drug-resistant acinetobacter baumannii. Int. J. Mol. Sci. 2018, 19, 3041. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-Y.; Park, S.-C.; Yoon, M.-Y.; Hahm, K.-S.; Park, Y. C-terminal amidation of pmap-23: Translocation to the inner membrane of gram-negative bacteria. Amino Acids 2011, 40, 183–195. [Google Scholar] [CrossRef]
- Lee, J.-K.; Luchian, T.; Park, Y. New antimicrobial peptide kills drug-resistant pathogens without detectable resistance. Oncotarget 2018, 9, 15616–15634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, J.Y.; Kim, M.K.; Mereuta, L.; Seo, C.H.; Luchian, T.; Park, Y. Mechanism of action of antimicrobial peptide p5 truncations against pseudomonas aeruginosa and staphylococcus aureus. AMB Express 2019, 9, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Name | Sequence | Molecular Mass (Da) | Net Charge | H a |
|---|---|---|---|---|
| Css54 | FFGSLLSLGSKLLPSVFKLFQRKKE-NH2 | 2869.5 | 5 | 0.451 |
| Strains | MICs (μM) | |||||
|---|---|---|---|---|---|---|
| Css 54 | Melittin | Cefotaxime | Vancomycin | Colistin | Ampicillin | |
| Gram-positive (+) | ||||||
| Listeria monocytogenes | 2 | 2 | 64 | 0.5 | 64 | 1 |
| (KCTC 3710) | ||||||
| Listeria monocytogenes | 2 | 1 | 4 | 0.5 | 32 | 0.5 |
| (KCCM 40307) | ||||||
| Listeria monocytogenes | 2 | 1 | 4 | 0.5 | 16 | 1 |
| (KCCM 43155) | ||||||
| Streptococcus suis | 2 | 2 | <0.13 | <0.13 | >64 | 0.5 |
| (KCTC 3557) | ||||||
| Gram-negative (−) | ||||||
| Campylobacter jejuni | 2 | 2 | 32 | 0.5 | >64 | >64 |
| (KCTC 5327) | ||||||
| Salmonella typhimurium | 4 | 2 | 1 | 64 | 1 | 2 |
| (ATCC 14028) | ||||||
| Salmonella typhimurium | 4 | 2, | 0.5 | 64 | 2 | >64 |
| (CCARM 8009) | ||||||
| Salmonella typhimurium | 4 | 2 | 0.5 | 64 | 4 | >64 |
| (CCARM 8013) | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.; Oh, J.H.; Kang, H.K.; Choi, M.-C.; Seo, C.H.; Park, Y. Scorpion-Venom-Derived Antimicrobial Peptide Css54 Exerts Potent Antimicrobial Activity by Disrupting Bacterial Membrane of Zoonotic Bacteria. Antibiotics 2020, 9, 831. https://doi.org/10.3390/antibiotics9110831
Park J, Oh JH, Kang HK, Choi M-C, Seo CH, Park Y. Scorpion-Venom-Derived Antimicrobial Peptide Css54 Exerts Potent Antimicrobial Activity by Disrupting Bacterial Membrane of Zoonotic Bacteria. Antibiotics. 2020; 9(11):831. https://doi.org/10.3390/antibiotics9110831
Chicago/Turabian StylePark, Jonggwan, Jun Hee Oh, Hee Kyoung Kang, Moon-Chang Choi, Chang Ho Seo, and Yoonkyung Park. 2020. "Scorpion-Venom-Derived Antimicrobial Peptide Css54 Exerts Potent Antimicrobial Activity by Disrupting Bacterial Membrane of Zoonotic Bacteria" Antibiotics 9, no. 11: 831. https://doi.org/10.3390/antibiotics9110831
APA StylePark, J., Oh, J. H., Kang, H. K., Choi, M.-C., Seo, C. H., & Park, Y. (2020). Scorpion-Venom-Derived Antimicrobial Peptide Css54 Exerts Potent Antimicrobial Activity by Disrupting Bacterial Membrane of Zoonotic Bacteria. Antibiotics, 9(11), 831. https://doi.org/10.3390/antibiotics9110831