Genomic Analysis of CTX-M-Group-1-Producing Extraintestinal Pathogenic E. coli (ExPEc) from Patients with Urinary Tract Infections (UTI) from Colombia
Abstract
:1. Introduction
2. Results
2.1. Antibiotic Susceptibility
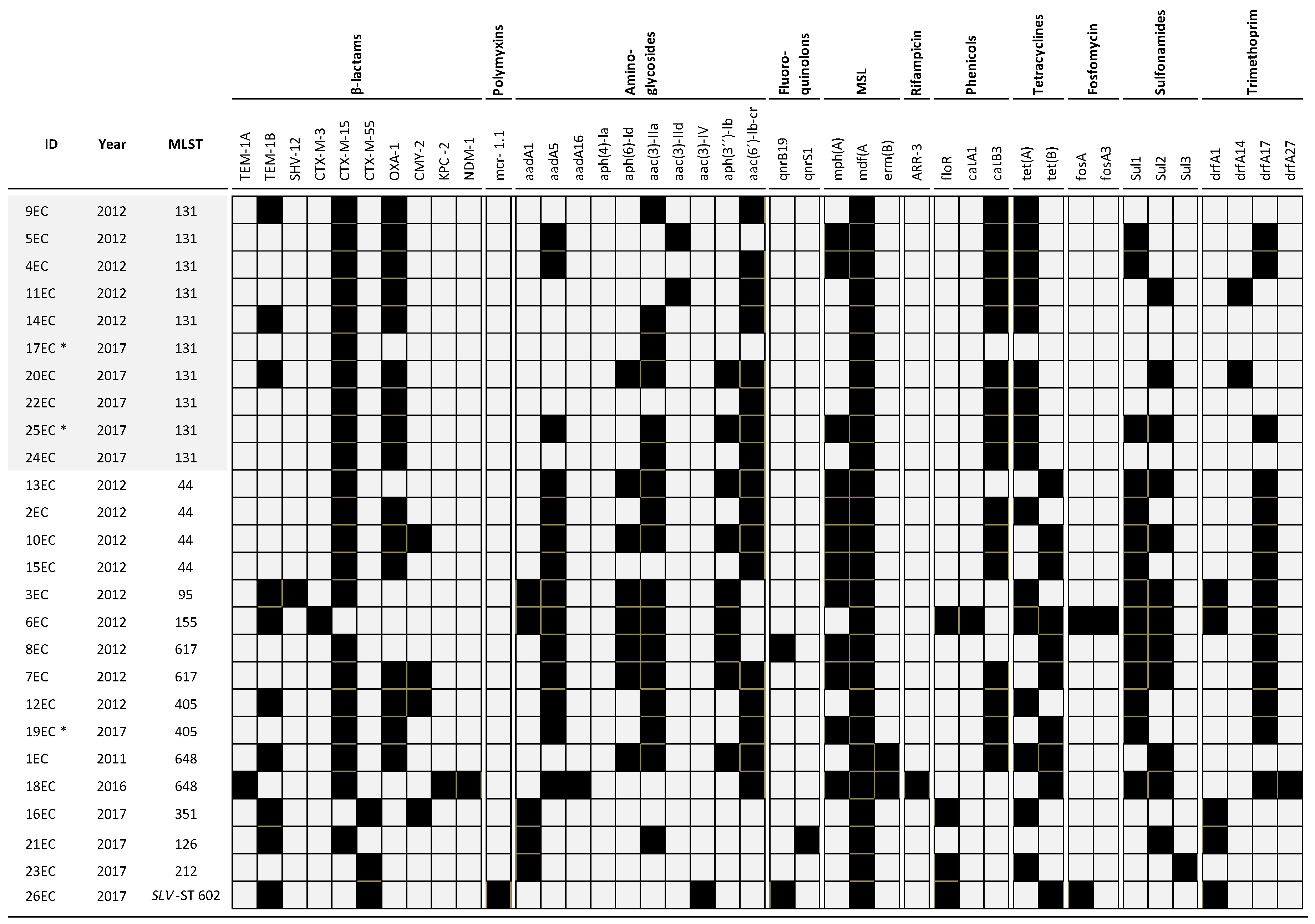
2.2. Molecular Characterization of β-Lactamase Genes
2.3. Multilocus Sequence Typing (MLST) and Phylogenetic Analysis
2.4. O25b-ST131 E. coli Clone Detection
2.5. Genes Related with Ciprofloxacin Resistance
2.6. Virulence Factor-Encoding Genes and Incompatibility Groups
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates
4.2. Susceptibility Testing
4.3. Whole-Genome Sequencing (WGS) and Analysis
4.4. Ethical Approval
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Marchaim, D.; Gottesman, T.; Schwartz, O.; Korem, M.; Maor, Y.; Rahav, G.; Karplus, R.; Lazarovitch, T.; Braun, E.; Sprecher, H.; et al. National Multicenter Study of Predictors and Outcomes of Bacteremia upon Hospital Admission Caused by Enterobacteriaceae Producing Extended-Spectrum β-Lactamases. Antimicrob. Agents Chemother. 2010, 54, 5099–5104. [Google Scholar] [CrossRef] [Green Version]
- Paterson, D.L.; Bonomo, R.A. Extended-Spectrum β-Lactamases: A Clinical Update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef] [Green Version]
- Russo, T.A.; Johnson, J.R. Medical and economic impact of extraintestinal infections due to Escherichia coli: Focus on an increasingly important endemic problem. Microbes Infect. 2003, 5, 449–456. [Google Scholar] [CrossRef]
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Iorga, B.I. Beta-lactamase database (BLDB)—Structure and function. J. Enzym. Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar] [CrossRef]
- WHO. Antimicrobial Resistance: Global Report on Surveillance. Available online: http://www.ncbi.nlm.nih.gov/pubmed/22247201%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2536104&tool=pmcentrez&rendertype=abstract (accessed on 10 February 2020).
- Stoesser, N.; Sheppard, A.E.; Pankhurst, L.; De Maio, N.; Moore, C.E.; Sebra, R.; Turner, P.; Anson, L.W.; Kasarskis, A.; Batty, E.M.; et al. Evolutionary History of the Global Emergence of the Escherichia coli Epidemic Clone ST131. mBio 2016, 7, e02162-15. [Google Scholar] [CrossRef] [Green Version]
- Price, L.B.; Johnson, J.R.; Aziz, M. The Epidemic of Extended-Spectrum-β-Lactamase-Producing Subclone, H 30-Rx. mBio 2013, 4, e00377-13. [Google Scholar] [CrossRef] [Green Version]
- Coque, T.M.; Baquero, F.; Cantón, R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Eurosurveillance 2008, 13, 1–11. [Google Scholar]
- Ruiz, S.J.; Montealegre, M.C.; Ruiz-Garbajosa, P.; Correa, A.; Briceño, D.F.; Martinez, E.; Rosso, F.; Muñoz, M.; Quinn, J.P.; Cantón, R.; et al. First Characterization of CTX-M-15-Producing Escherichia coli ST131 and ST405 Clones Causing Community-Onset Infections in South America. J. Clin. Microbiol. 2011, 49, 1993–1996. [Google Scholar] [CrossRef] [Green Version]
- Nicolas-Chanoine, M.H.; Bertrand, X.; Madec, J.Y. Escherichia coli ST131, an Intriguing Clonal Group. Clin. Microbiol. Rev. 2014, 27, 543–574. [Google Scholar] [CrossRef] [Green Version]
- Meletis, G.; Chatzidimitriou, D.; Malisiovas, N. Double- and multi-carbapenemase-producers: The excessively armored bacilli of the current decade. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1487–1493. [Google Scholar] [CrossRef]
- Miyakis, S.; Pefanis, A.; Tsakris, A. The Challenges of Antimicrobial Drug Resistance in Greece. Clin. Infect. Dis. 2011, 53, 177–184. [Google Scholar] [CrossRef]
- Titelman, E.; Iversen, A.; Kahlmeter, G.; Giske, C.G. Antimicrobial susceptibility to parenteral and oral agents in a largely polyclonal collection of CTX-M-14 and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae. APMIS 2011, 119, 853–863. [Google Scholar] [CrossRef]
- Kitchel, B.; Rasheed, J.K.; Endimiani, A.; Hujer, A.M.; Anderson, K.F.; Bonomo, R.A.; Patel, J.B. Genetic Factors Associated with Elevated Carbapenem Resistance in KPC-Producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2010, 54, 4201–4207. [Google Scholar] [CrossRef] [Green Version]
- Endimiani, A.; Perez, F.; Bajaksouzian, S.; Windau, A.R.; Good, C.E.; Choudhary, Y.; Hujer, A.M.; Bethel, C.R.; Bonomo, R.A.; Jacobs, M.R. Evaluation of updated interpretative criteria for categorizing Klebsiella pneumoniae with reduced carbapenem susceptibility. J. Clin. Microbiol. 2010, 48, 4417–4425. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside Modifying Enzymes. Drug Resist. Updates 2010, 16, 151–171. [Google Scholar] [CrossRef] [Green Version]
- Hojabri, Z.; Darabi, N.; Arab, M.; Saffari, F.; Pajand, O. Clonal diversity, virulence genes content and subclone status of Escherichia coli sequence type 131: Comparative analysis of E. coli ST131 and non-ST131 isolates from Iran. BMC Microbiol. 2019, 19, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Blanc, V.; Leflon-Guibout, V.; Blanco, J.; Haenni, M.; Madec, J.-Y.; Rafignon, G.; Bruno, P.; Mora, A.; Lopez, C.; Dahbi, G.; et al. Prevalence of day-care centre children (France) with faecal CTX-M-producing Escherichia coli comprising O25b:H4 and O16:H5 ST131 strains. J. Antimicrob. Chemother. 2014, 69, 1231–1237. [Google Scholar] [CrossRef]
- Banerjee, R.; Robicsek, A.; Kuskowski, M.A.; Porter, S.; Johnston, B.D.; Sokurenko, E.; Tchesnokova, V.; Price, L.B.; Johnson, J.R. Molecular Epidemiology of Escherichia coli Sequence Type 131 and Its H30 and H30-Rx Subclones among Extended-Spectrum-β-Lactamase-Positive and -Negative E. coli Clinical Isolates from the Chicago Region, 2007 to 2010. Antimicrob. Agents Chemother. 2013, 57, 6385–6388. [Google Scholar] [CrossRef] [Green Version]
- Pitout, J.D.D.; DeVinney, R. Escherichia coli ST131: A multidrug-resistant clone primed for global domination. F1000Research 2017, 6, 195. [Google Scholar] [CrossRef] [Green Version]
- Johnson, T.J.; Wannemuehler, Y.M.; Nolan, L.K. Evolution of the iss Gene in Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2360–2369. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.R.; Porter, S.B.; Zhanel, G.; Kuskowski, M.A.; Denamur, E. Virulence of Escherichia coli Clinical Isolates in a Murine Sepsis Model in Relation to Sequence Type ST131 Status, Fluoroquinolone Resistance, and Virulence Genotype. Infect. Immun. 2012, 80, 1554–1562. [Google Scholar] [CrossRef] [Green Version]
- Carattoli, A. Resistance Plasmid Families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 2227–2238. [Google Scholar] [CrossRef] [Green Version]
- Blanco, M.; Alonso, M.P.; Nicolas-Chanoine, M.-H.; Dahbi, G.; Mora, A.; Blanco, J.E.; López, C.; Cortés, P.; Llagostera, M.; Leflon-Guibout, V.; et al. Molecular epidemiology of Escherichia coli producing extended-spectrum β-lactamases in Lugo (Spain): Dissemination of clone O25b:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2009, 63, 1135–1141. [Google Scholar] [CrossRef] [Green Version]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and Simple Determination of the Escherichia coli Phylogenetic Group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [Green Version]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef] [Green Version]
| ID | City | Collection Date | Source | Ward | MLST | MIC (ug/mL) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CRO | CTX | CAZ | FEP | TZP | ETP | CIP | FOS | ||||||
| 1EC | Bogotá | 2011 | Urine | ER | 648 | >4 | >4 | 16 | 32 | 16/4 | ≤0.25 | >4 | ≤32 |
| 2EC | Cali | 2012 | Urine | ER | 44 | >4 | >4 | 32 | 16 | 16/4 | ≤0.25 | >4 | ≤32 |
| 3EC | Cali | 2012 | Urine | ER | 95 | >4 | >4 | 16 | 8 | 8/4 | ≤0.25 | >4 | ≤32 |
| 4EC | Cali | 2012 | Urine | ER | 131 | >4 | >4 | 16 | 8 | 4/4 | ≤0.25 | 4 | ≤32 |
| 5EC | Cali | 2012 | Urine | ER | 131 | >4 | >4 | 8 | 16 | ≤2/4 | ≤0.25 | >4 | ≤32 |
| 6EC | Cali | 2012 | Urine | ER | 155 | >4 | >4 | 1 | 4 | ≤2/4 | ≤0.25 | >4 | >128 |
| 7EC | Cali | 2012 | Urine | ER | 617 | >4 | >4 | 64 | 32 | 8/4 | ≤0.25 | >4 | ≤32 |
| 8EC | Cali | 2012 | Urine | ER | 617 | >4 | >4 | 8 | 16 | 8/4 | ≤0.25 | >4 | ≤32 |
| 9EC | Cali | 2012 | Urine | ER | 131 | >4 | >4 | 16 | 16 | 16/4 | ≤0.25 | >4 | 128 |
| 10EC | Cali | 2012 | Urine | ER | 44 | >4 | >4 | 64 | 32 | 16/4 | ≤0.25 | >4 | ≤32 |
| 11EC | Cali | 2012 | Urine | ER | 131 | >4 | >4 | 4 | 4 | 4/4 | ≤0.25 | >4 | ≤32 |
| 12EC | Cali | 2012 | Urine | ER | 405 | >4 | >4 | 32 | 16 | 8/4 | ≤0.25 | >4 | 64 |
| 13EC | Cali | 2012 | Urine | ER | 44 | >4 | >4 | 16 | 16 | 8/4 | ≤0.25 | >4 | ≤32 |
| 14EC | Bogotá | 2012 | Urine | ER | 131 | >4 | >4 | 16 | 32 | 8/4 | ≤0.25 | >4 | ≤32 |
| 15EC | Bogotá | 2012 | Urine | ER | 44 | >4 | >4 | 16 | 8 | 8/4 | ≤0.25 | >4 | ≤32 |
| 16EC | Ibague | 2017 | Urine | ER | 351 | >4 | >4 | >32 | 64 | 32/4 | ≤0.25 | 0.5 | ≤32 |
| 17EC * | Cucuta | 2017 | Blood | ER | 131 | >4 | >4 | 16 | 16 | ≤2/4 | ≤0.25 | >4 | ≤32 |
| 18EC | Cali | 2016 | Urine | HOSP | 648 | >4 | >4 | >32 | 32 | >128/4 | >32 | >4 | ≤32 |
| 19EC * | Barranquilla | 2017 | Blood | HOSP | 405 | >4 | >4 | 16 | 32 | 8/4 | ≤0.25 | >4 | ≤32 |
| 20EC | Pereira | 2017 | Urine | HOSP | 131 | >4 | >4 | 32 | 32 | 8/4 | ≤0.25 | >4 | ≤32 |
| 21EC | Pasto | 2017 | Urine | ER | 126 | >4 | >4 | 16 | 8 | ≤2/4 | ≤0.25 | 4 | ≤32 |
| 22EC | Ibague | 2017 | Urine | ER | 131 | >4 | >4 | 32 | 32 | 32/4 | ≤0.25 | >4 | ≤32 |
| 23EC | Bucaramanga | 2017 | Urine | ER | 212 | >4 | >4 | >32 | >64 | ≤2/4 | 8 | ≤0.25 | 64 |
| 24EC | Cucuta | 2017 | Urine | ICU | 131 | >4 | >4 | >32 | 32 | >128/4 | ≤0.25 | >4 | ≤32 |
| 25EC* | Pasto | 2017 | Blood | HOSP | 131 | >4 | >4 | 16 | 8 | 4/4 | ≤0.25 | >4 | ≤32 |
| 26EC | Ibague | 2017 | Urine | HOSP | SLV-ST 602 | >4 | 4 | 8 | 8 | ≤2/4 | ≤0.25 | ≤0.25 | 64 |
| ID | MLST | CTX-M | Phylogroup | Serogroup | fimH | Subclone | Allele gyrA/parC | CIP |
|---|---|---|---|---|---|---|---|---|
| 4EC | 131 | CTX-M-15 | B2 | O25b:H4 | 30 | H30-Rx | gyrA1AB/parC1aAB | 4 |
| 5EC | 131 | CTX-M-15 | B2 | O25b:H4 | 30 | H30-Rx | gyrA1AB/parC1aAB | >4 |
| 9EC | 131 | CTX-M-15 | B2 | O25b:H4 | 35 | No-H30 | gyrA1AB/parC1aAB | >4 |
| 11EC | 131 | CTX-M-15 | B2 | O25b:H4 | 30 | H30-Rx | gyrA1AB/parC1aAB | >4 |
| 14EC | 131 | CTX-M-15 | B2 | O25b:H4 | 35 | No-H30 | gyrA1AB/parC1aAB | >4 |
| 17EC * | 131 | CTX-M-15 | B2 | O25b:H4 | 30 | H30-Rx | gyrA1AB/parC1aAB | >4 |
| 20EC | 131 | CTX-M-15 | B2 | O25b:H4 | 30 | H30-Rx | gyrA1AB/parC1aAB | >4 |
| 22EC | 131 | CTX-M-15 | B2 | O25b:H4 | 30 | H30-Rx | gyrA1AB/parC1aAB | >4 |
| 24EC | 131 | CTX-M-15 | B2 | O25b:H4 | 30 | H30-Rx | gyrA1AB/parC1aAB | >4 |
| 25EC * | 131 | CTX-M-15 | B2 | O25b:H4 | 30 | H30-Rx | gyrA1AB/parC1aAB | >4 |
| 16EC | 351 | CTX-M-55 | B1 | O18a:H7 | 31 | - | 0.5 | |
| 2EC | 44 | CTX-M-15 | A | O89:H4 | 54 | gyrA1AB | >4 | |
| 10EC | 44 | CTX-M-15 | A | O89:H4 | 54 | gyrA1AB | >4 | |
| 13EC | 44 | CTX-M-15 | A | O89:H4 | 54 | gyrA1AB | >4 | |
| 15EC | 44 | CTX-M-15 | A | O89:H4 | 54 | gyrA1AB | >4 | |
| 3EC | 95 | CTX-M-15 | B2 | O50:H4 | 27 | gyrA1AB | >4 | |
| 21EC a | 126 | CTX-M-15 | B2 | H5 | 26 | - | 4 | |
| 6EC | 155 | CTX-M-3 | B1 | O162:H19 | 32 | gyrA1AB | >4 | |
| 23EC | 212 | CTX-M-55 | B2 | O18ac:H49 | 38 | - | ≤0.25 | |
| 12EC | 405 | CTX-M-15 | D | O102:H6 | 27 | gyrA1AB | >4 | |
| 19EC * | 405 | CTX-M-15 | D | O102:H6 | 27 | gyrA1AB | >4 | |
| 7EC | 617 | CTX-M-15 | A | O89:H10 | - | gyrA1AB | >4 | |
| 8EC | 617 | CTX-M-15 | A | O89:H10 | - | gyrA1AB | >4 | |
| 1EC | 648 | CTX-M-15 | F | O1:H6 | 27 | gyrA1AB | >4 | |
| 18EC | 648 | CTX-M-15 | F | H6 | 27 | gyrA1AB | >4 | |
| 26EC | SLV-ST 602 | CTX-M-15 | B1 | H21 | 86 | - | ≤0.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De La Cadena, E.; Mojica, M.F.; Castillo, N.; Correa, A.; Appel, T.M.; García-Betancur, J.C.; Pallares, C.J.; Villegas, M.V. Genomic Analysis of CTX-M-Group-1-Producing Extraintestinal Pathogenic E. coli (ExPEc) from Patients with Urinary Tract Infections (UTI) from Colombia. Antibiotics 2020, 9, 899. https://doi.org/10.3390/antibiotics9120899
De La Cadena E, Mojica MF, Castillo N, Correa A, Appel TM, García-Betancur JC, Pallares CJ, Villegas MV. Genomic Analysis of CTX-M-Group-1-Producing Extraintestinal Pathogenic E. coli (ExPEc) from Patients with Urinary Tract Infections (UTI) from Colombia. Antibiotics. 2020; 9(12):899. https://doi.org/10.3390/antibiotics9120899
Chicago/Turabian StyleDe La Cadena, Elsa, María Fernanda Mojica, Nathaly Castillo, Adriana Correa, Tobias Manuel Appel, Juan Carlos García-Betancur, Christian José Pallares, and María Virginia Villegas. 2020. "Genomic Analysis of CTX-M-Group-1-Producing Extraintestinal Pathogenic E. coli (ExPEc) from Patients with Urinary Tract Infections (UTI) from Colombia" Antibiotics 9, no. 12: 899. https://doi.org/10.3390/antibiotics9120899
APA StyleDe La Cadena, E., Mojica, M. F., Castillo, N., Correa, A., Appel, T. M., García-Betancur, J. C., Pallares, C. J., & Villegas, M. V. (2020). Genomic Analysis of CTX-M-Group-1-Producing Extraintestinal Pathogenic E. coli (ExPEc) from Patients with Urinary Tract Infections (UTI) from Colombia. Antibiotics, 9(12), 899. https://doi.org/10.3390/antibiotics9120899






