Staphylococcal-Produced Bacteriocins and Antimicrobial Peptides: Their Potential as Alternative Treatments for Staphylococcus aureus Infections
Abstract
:1. Introduction
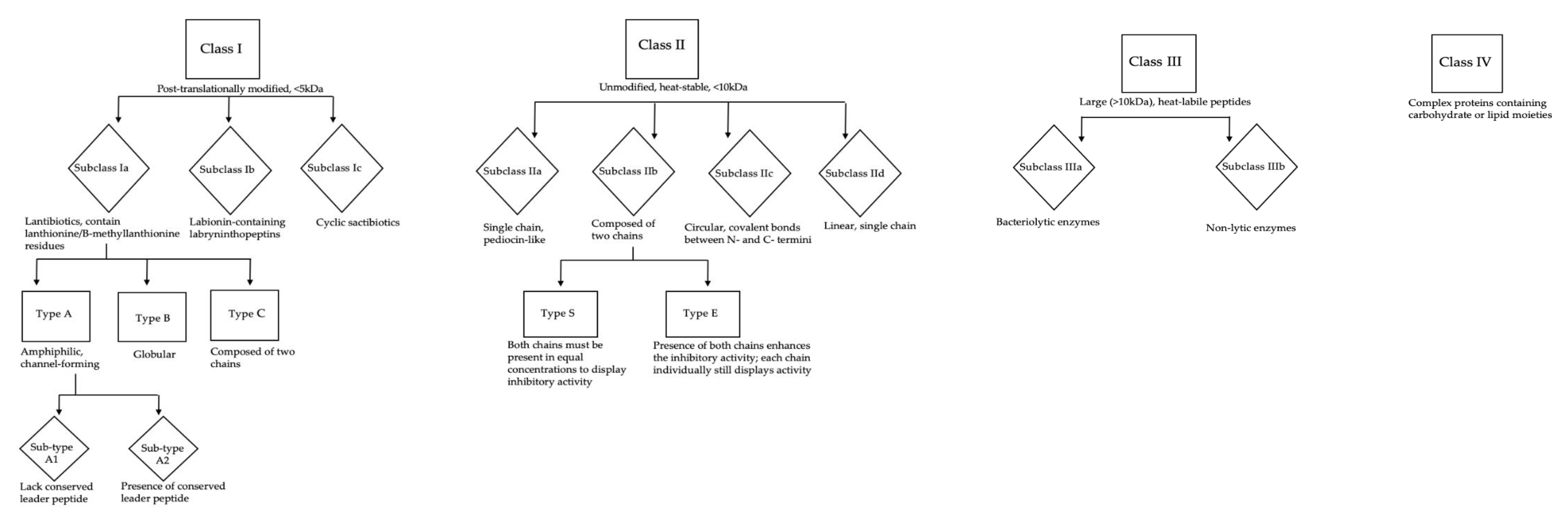
Bacteriocins as Novel Antimicrobials
2. Staphylococcins
2.1. Studies Showing in vitro Inhibitory Activity against Staphylococcus aureus
2.2. Models of in vivo Bacteriocin Therapy for Staphylococcus aureus Infection
3. Other Antimicrobial Substances with Anti-Staphylococcus aureus Activity
3.1. Bacteriocin-Like Inhibitory Substances
3.2. Inhibitory Staphylococcal Strains
3.3. Staphylococcal-Produced Antimicrobial Substances
4. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef]
- Ellington, J.K.; Harris, M.; Webb, L.; Smith, B.; Smith, T.; Tan, K.; Hudson, M. Intracellular Staphyloccus aureus. J. Bone Jt. Surg. Ser. B 2003, 85, 918–921. [Google Scholar] [CrossRef]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fowler, V.G., Jr.; Miro, J.M.; Hoen, B.; Cabell, C.H.; Abrutyn, E.; Rubinstein, E.; Corey, G.R.; Spelman, D.; Bradley, S.F.; Barsic, B.; et al. Staphylococcus aureus endocarditis: A consequence of medical progress. JAMA 2005, 293, 3012–3021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saeed, K.; Bal, A.M.; Gould, I.M.; David, M.Z.; Dryden, M.; Giannitsioti, E.; Hijazi, K.; Meisner, J.A.; Esposito, S.; Scaglione, F.; et al. An update on Staphylococcus aureus infective endocarditis from the International Society of Antimicrobial Chemotherapy (ISAC). Int. J. Antimicrob. Agents 2019, 53, 9–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pletz, M.W.; Burkhardt, O.; Welte, T. Nosocomial methicillin-resistant Staphylococcus aureus (MRSA) pneumonia: Linezolid or vancomycin?—Comparison of pharmacology and clinical efficacy. Eur. J. Med. Res. 2010, 15, 507–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lesher, B.; Gao, X.; Chen, Y.; Liu, Z. Methicillin-resistant Staphylococcus aureus nosocomial pneumonia: Role of linezolid in the People’s Republic of China. Clin. Outcomes Res. 2016, 8, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Kluytmans, J.; Van Belkum, A.; Verbrugh, H. Nasal carriage of Staphylococcus aureus: Epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 1997, 10, 505–520. [Google Scholar] [CrossRef]
- Von Eiff, C.; Becker, K.; Machka, K.; Stammer, H.; Peters, G. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 2001, 344, 11–16. [Google Scholar] [CrossRef]
- Weidenmaier, C.; Goerke, C.; Wolz, C. Staphylococcus aureus determinants for nasal colonization. Trends Microbiol. 2012, 20, 243–250. [Google Scholar] [CrossRef]
- Wertheim, H.F.L.; Melles, D.C.; Vos, M.C.; Van Leeuwen, W.; Van Belkum, A.; Verbrugh, H.A.; Nouwen, J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus aureus toxins. Curr. Opin. Microbiol. 2014, 17, 32–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powers, M.E.; Wardenburg, J.B. Igniting the Fire: Staphylococcus aureus Virulence Factors in the Pathogenesis of Sepsis. PLoS Pathog. 2014, 10, e1003871. [Google Scholar] [CrossRef] [PubMed]
- Archer, N.K.; Mazaitis, M.J.; William Costerton, J.; Leid, J.G.; Powers, M.E.; Shirtliff, M.E. Staphylococcus aureus biofilms: Properties, regulation and roles in human disease. Virulence 2011, 2, 445–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Paterson, G.K.; Harrison, E.M.; Holmes, M.A. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2014, 22, 42–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peacock, S.J.; Paterson, G.K. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef] [PubMed]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Thomas, D.G.; Wilson, J.M.; Day, M.J.; Russell, A.D. Mupirocin resistance in staphylococci: Development and transfer of isoleucyl-tRNA synthetase-mediated resistance In Vitro. J. Appl. Microbiol. 1999, 86, 715–722. [Google Scholar] [CrossRef]
- O’Sullivan, J.N.; Rea, M.C.; O’Connor, P.M.; Hill, C.; Ross, R.P. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol. 2019, 95. [Google Scholar] [CrossRef]
- Weese, J.S. Methicillin-resistant Staphylococcus aureus in animals. ILAR J. 2010, 51, 233–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fluit, A.C. Livestock-associated Staphylococcus aureus. Clin. Microbiol. Infect. 2012, 18, 735–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paterson, G.K.; Larsen, A.R.; Robb, A.; Edwards, G.E.; Pennycott, T.W.; Foster, G.; Mot, D.; Hermans, K.; Baert, K.; Peacock, S.J.; et al. The newly described mecA homologue, mecALGA251, is present in methicillin-resistant Staphylococcus aureus isolates from a diverse range of host species. J. Antimicrob. Chemother. 2012, 67, 2809–2813. [Google Scholar] [CrossRef] [PubMed]
- Haag, A.F.; Fitzgerald, J.R.; Penadés, J.R. Staphylococcus aureus in Animals. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- McNamee, P.T.; McCullagh, J.J.; Thorp, B.H.; Ball, H.J.; Graham, D.; McCullough, S.J.; McConaghy, D.; Smyth, J.A. Study of leg weakness in two commercial broiler flocks. Vet. Rec. 1998, 143, 131–135. [Google Scholar] [CrossRef]
- McNamee, P.T.; Smyth, J.A. Bacterial chondronecrosis with osteomyelitis (‘femoral head necrosis’) of broiler chickens: A review. Avian Pathol. 2000, 29, 253–270. [Google Scholar] [CrossRef]
- Tenhagen, B.A.; Köster, G.; Wallmann, J.; Heuwieser, W. Prevalence of mastitis pathogens and their resistance against antimicrobial agents in dairy cows in Brandenburg, Germany. J. Dairy Sci. 2006, 89, 2542–2551. [Google Scholar] [CrossRef] [Green Version]
- Bradley, A.J.; Leach, K.A.; Breen, J.E.; Green, L.E.; Green, M.J. Survey of the incidence and aetiology of mastitis on diary farms in England and Wales. Vet. Rec. 2007, 160, 253–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barkema, H.W.; Schukken, Y.H.; Zadoks, R.N. Invited review: The role of cow, pathogen, and treatment regimen in the therapeutic success of bovine Staphylococcus aureus mastitis. J. Dairy Sci. 2006, 89, 1877–1895. [Google Scholar] [CrossRef] [Green Version]
- Kinross, P.T.N.; Petersen, A.; Skov, R.; Van Hauwermeiren, E.; Pantosti, A.; Laurent, F.; Voss, A.; Kluytmans, J.; Struelens, M.J.; Heuer, O.; et al. Livestock-associated meticillin-resistant Staphylococcus aureus (MRSA) among human MRSA isolates, European Union/European Economic Area countries, 2013. Euro Surveill. 2017, 22. [Google Scholar] [CrossRef] [Green Version]
- Erskine, R.J. Vaccination Strategies for Mastitis. Vet. Clin. N. Am. Food Anim. Pract. 2012, 28, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Leitner, G.; Lubashevsky, E.; Glickman, A.; Winkler, M.; Saran, A.; Trainin, Z. Development of a Staphylococcus aureus vaccine against mastitis in dairy cows: I. Challenge trials. Vet. Immunol. Immunopathol. 2003, 93, 31–38. [Google Scholar] [CrossRef]
- Keefe, G. Update on control of Staphylococcus aureus and Streptococcus agalactiae for management of mastitis. Vet. Clin. N. Am. Food Anim. Pract. 2012, 28, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Heringstad, B.; Klemetsdal, G.; Ruane, J. Selection for mastitis resistance in dairy cattle: A review with focus on the situation in the Nordic countries. Livest. Prod. Sci. 2000, 64, 95–106. [Google Scholar] [CrossRef]
- Rupp, R.; Boichard, D. Genetics of resistance to mastitis in dairy cattle. Vet. Res. 2003, 34, 671–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Götz, F.; Perconti, S.; Popella, P.; Werner, R.; Schlag, M. Epidermin and gallidermin: Staphylococcal lantibiotics. Int. J. Med. Microbiol. 2014, 304, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Round, J.L.; Mazmanian, S.K. The gut microbiome shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system HHS Public Access. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef] [Green Version]
- Cogen, A.L.; Nizet, V.; Gallo, R.L.; Richard Gallo, C.L. Skin microbiota: A source of disease or defence? NIH Public Access. Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef] [Green Version]
- Rosenthal, M.; Goldberg, D.; Aiello, A.; Larson, E.; Foxman, B. Skin microbiota: Microbial community structure and its potential association with health and disease. Infect. Genet. Evol. 2011, 11, 839–848. [Google Scholar] [CrossRef] [Green Version]
- Christensen, G.J.M.; Brüggemann, H. Bacterial skin commensals and their role as host guardians. Benef. Microbes 2014, 5, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Ross, R.P.; Hill, C.; Ross, R.P. Bacteriocins: Developing innate immunity for food. Food Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef]
- Jack, R.W.; Tagg, J.R.; Ray, B. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 1995, 59, 171–200. [Google Scholar] [CrossRef] [PubMed]
- Peschel, A.; Götz, F. Analysis of the Staphylococcus epidermidis genes epiF, -E, and -G involved in epidermin immunity. J. Bacteriol. 1996, 178, 531–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Havarstein, L.S.; Diep, D.B.; Nes, I.F. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 1995, 16, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Mak, P. Staphylococcal Bacteriocins. In Pet-to-Man Travelling Staphylococci: A World in Progress; Elsevier: Amsterdam, The Netherlands, 2018; pp. 161–171. ISBN 9780128135488. [Google Scholar]
- Shin, J.M.; Gwak, J.W.; Kamarajan, P.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. Biomedical Applications of Nisin HHS Public Access. J. Appl. Microbiol. 2016, 120, 1449–1465. [Google Scholar] [CrossRef] [Green Version]
- Dobson, A.; Cotter, P.D.; Paul Ross, R.; Hill, C. Bacteriocin production: A probiotic trait? Appl. Environ. Microbiol. 2012, 78, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Krismer, B.; Weidenmaier, C.; Zipperer, A.; Peschel, A. The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat. Rev. Microbiol. 2017, 15, 675–687. [Google Scholar] [CrossRef]
- Schauber, J.; Gallo, R.L. Antimicrobial peptides and the skin immune defense system. J. Allergy Clin. Immunol. 2008, 122, 261–266. [Google Scholar] [CrossRef] [Green Version]
- Bastos, M.; Ceotto, H.; Coelho, M.; Nascimento, J. Staphylococcal Antimicrobial Peptides: Relevant Properties and Potential Biotechnological Applications. Curr. Pharm. Biotechnol. 2009, 10, 38–61. [Google Scholar] [CrossRef]
- Wladyka, B.; Piejko, M.; Bzowska, M.; Pieta, P.; Krzysik, M.; Mazurek, Ł.; Guevara-Lora, I.; Bukowski, M.; Sabat, A.J.; Friedrich, A.W.; et al. A peptide factor secreted by Staphylococcus pseudintermedius exhibits properties of both bacteriocins and virulence factors. Sci. Rep. 2015, 5, 14569. [Google Scholar] [CrossRef] [PubMed]
- Klaenhammer, T.R. Bacteriocins of lactic acid bacteria. Biochimie 1988, 70, 337–349. [Google Scholar] [CrossRef]
- Gálvez, A.; Abriouel, H.; López, R.L.; Omar, N. Ben Bacteriocin-based strategies for food biopreservation. Int. J. Food Microbiol. 2007, 120, 51–70. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.L. The business of developing antibacterials. Nat. Biotechnol. 2006, 24, 1521–1528. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins-a viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Kokai-Kun, J.F.; Walsh, S.M.; Chanturiya, T.; Mond, J.J. Lysostaphin cream eradicates Staphylococcus aureus nasal colonization in a cotton rat model. Antimicrob. Agents Chemother. 2003, 47, 1589–1597. [Google Scholar] [CrossRef] [Green Version]
- Bastos, M.D.; Coutinho, B.G.; Coelho, M.L. Lysostaphin: A staphylococcal bacteriolysin with potential clinical applications. Pharmaceuticals 2010, 3, 1139–1161. [Google Scholar] [CrossRef] [Green Version]
- Field, D.; Gaudin, N.; Lyons, F.; O’Connor, P.M.; Cotter, P.D.; Hill, C.; Ross, R.P. A bioengineered nisin derivative to control biofilms of Staphylococcus pseudintermedius. PLoS ONE 2015, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Cleveland, J.; Montville, T.J.; Nes, I.F.; Chikindas, M.L. Bacteriocins: Safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 2001, 71, 1–20. [Google Scholar] [CrossRef]
- Quinto, E.J.; Caro, I.; Villalobos-delgado, L.H.; Mateo, J.; De-mateo-silleras, B.; Redondo-del-r, P. Food Safety through Natural Antimicrobials. Antibiotics 2019, 81, 208. [Google Scholar] [CrossRef] [Green Version]
- Nakatsuj, T.; Kim, J.-N.; Lockhart, A.; Nakatsuji, T.; Hata, T.R.; Dorrestein, P.C.; David, G.; Gallo, R.L.; Shafiq, F.; Latif, H.; et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef] [Green Version]
- Otto, M. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev. Dermatol. 2010, 5, 183–195. [Google Scholar] [CrossRef] [Green Version]
- Sandiford, S.; Upton, M. Identification, characterization, and recombinant expression of epidermicin NI01, a novel unmodified bacteriocin produced by Staphylococcus epidermidis that displays potent activity against staphylococci. Antimicrob. Agents Chemother. 2012, 56, 1539–1547. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.; Kim, J.; Quan, L.H.; Heu, S.; Roh, E. Purification and characterization of pasteuricin produced by Staphylococcus pasteuri RSP-1 and active against multidrug-resistant Staphylococcus aureus. J. Food Prot. 2018, 81, 1768–1775. [Google Scholar] [CrossRef]
- Netz, D.J.; Pohl, R.; Beck-Sickinger, A.G.; Selmer, T.; Pierik, A.J.; Bastos, M.C.; Sahl, H.G. Biochemical characterisation and genetic analysis of aureocin A53, a new, atypical bacteriocin from Staphylococcus aureus. J. Mol. Biol. 2002, 319, 745–756. [Google Scholar] [CrossRef]
- Navaratna, M.A.; Sahl, H.G.; Tagg, J.R. Two-component anti-Staphylococcus aureus lantibiotic activity produced by Staphylococcus aureus C55. Appl. Environ. Microbiol. 1998, 64, 4803–4808. [Google Scholar] [CrossRef] [Green Version]
- Augustin, J.; Rosenstein, R.; Wieland, B.; Schneider, U.; Schnell, N.; Engelke, G.; Entian, K.-D.; Götz, F. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur. J. Biochem. 1992, 204, 1149–1154. [Google Scholar] [CrossRef]
- Van De Kamp, M.; Van Den Hooven, H.W.; Konings, R.N.H.; Bierbaum, G.; Sahl, H.-G.; Kuipers, O.P.; Siezen, R.J.; De Vos, W.M.; Hilbers, C.W.; Van De Ven, F.J.M. Elucidation of the Primary Structure of the Lantibiotic Epilancin K7 from Staphylococcus epidermidis K7: Cloning and Characterisation of the Epilancin-K7–Encoding Gene and NMR Analysis of Mature Epilancin K7. Eur. J. Biochem. 1995, 230, 587–600. [Google Scholar] [CrossRef]
- Heidrich, C.; Pag, U.; Josten, M.; Metzger, J.; Jack, R.W.; Bierbaum, G.; Jung, G.; Sahl, H.G. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl. Environ. Microbiol. 1998, 64, 3140–3146. [Google Scholar] [CrossRef] [Green Version]
- McAuliffe, O.; Ross, R.P.; Hill, C. Lantibiotics: Structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 2001, 25, 285–308. [Google Scholar] [CrossRef] [Green Version]
- Ennahar, S.; Sashihara, T.; Sonomoto, K.; Ishizaki, A. Class IIa bacteriocins: Biosynthesis, structure and activity. FEMS Microbiol. Rev. 2000, 24, 85–106. [Google Scholar] [CrossRef] [PubMed]
- Kaletta, C.; Entian, K.D.; Kellner, R.; Jung, G.; Reis, M.; Sahl, H.G. Pep5, a new lantibiotic: Structural gene isolation and prepeptide sequence. Arch. Microbiol. 1989, 152, 16–19. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, J.S.; Ceotto, H.; Nascimento, S.B.; Giambiagi-DeMarval, M.; Santos, K.R.N.; Bastos, M.C.F. Bacteriocins as alternative agents for control of multiresistant staphylococcal strains. Lett. Appl. Microbiol. 2006, 42, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Saising, J.; Dube, L.; Ziebandt, A.-K.; Voravuthikunchai, S.P.; Nega, M.; Götz, F. Activity of gallidermin on Staphylococcus aureus and Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 2012, 56, 5804–5810. [Google Scholar] [CrossRef] [Green Version]
- Bierbaum, G.; Sahl, H.-G. Lantibiotics: Mode of Action, Biosynthesis and Bioengineering. Curr. Pharm. Biotechnol. 2009, 10, 2–18. [Google Scholar] [CrossRef]
- de Freire Bastos, M.D.C.; Varella Coelho, M.L.; da Silva Santos, O.C. Resistance to bacteriocins produced by gram-positive bacteria. Microbiology 2015, 161, 683–700. [Google Scholar] [CrossRef] [Green Version]
- Bonelli, R.R.; Schneider, T.; Sahl, H.G.; Wiedemann, I. Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode-of-action studies. Antimicrob. Agents Chemother. 2006, 50, 1449–1457. [Google Scholar] [CrossRef] [Green Version]
- Brötz, H.; Josten, M.; Wiedemann, I.; Schneider, U.; Götz, F.; Bierbaum, G.; Sahl, H.G. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 1998, 30, 317–327. [Google Scholar] [CrossRef] [Green Version]
- Piper, C.; Draper, L.A.; Cotter, P.D.; Ross, R.P.; Hill, C. A comparison of the activities of lacticin 3147 and nisin against drug-resistant Staphylococcus aureus and Enterococcus species. J. Antimicrob. Chemother. 2009, 64, 546–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, E.; Radler, F. Caseicin, a bacteriocin from Lactobacillus casei. Folia Microbiol. 1993, 38, 441–446. [Google Scholar] [CrossRef]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of lactic acid bacteria: Extending the family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Souza Duarte, A.F.; Ceotto, H.; Coelho, M.L.V.; de Paiva Brito, M.A.V.; de Freire Bastos, M.C. Identification of new staphylococcins with potential application as food biopreservatives. Food Control 2013, 32, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Varella Coelho, M.L.; Santos Nascimento, J.D.; Fagundes, P.C.; Madureira, D.J.; Oliveira, S.S.; Vasconcelos de Paiva Brito, M.A.; Freire Bastos, M.C. Activity of staphylococcal bacteriocins against Staphylococcus aureus and Streptococcus agalactiae involved in bovine mastitis. Res. Microbiol. 2007, 158, 625–630. [Google Scholar] [CrossRef]
- Joerger, M.C.; Klaenhammer, T.R. Characterization and purification of helveticin J and evidence for a chromosomally determined bacteriocin produced by Lactobacillus helveticus 481. J. Bacteriol. 1986, 167, 439–446. [Google Scholar] [CrossRef] [Green Version]
- Schindler, C.A.; Schuhardt, V.T. Lysostaphin: A new bacteriolytic agent for the staphylococcus. Proc. Natl. Acad. Sci. USA 1964, 51, 414–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Climo, M.W.; Patron, R.L.; Goldstein, B.P.; Archer, G.L. Lysostaphin treatment of experimental methicillin-resistant Staphylococcus aureus aortic valve endocarditis. Antimicrob. Agents Chemother. 1998, 42, 1355–1360. [Google Scholar] [CrossRef] [Green Version]
- Kiri, N.; Archer, G.; Climo, M.W. Combinations of lysostaphin with beta-lactams are synergistic against oxacillin-resistant Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2002, 46, 2017–2020. [Google Scholar] [CrossRef] [Green Version]
- Von Eiff, C.; Kokai-Kun, J.F.; Becker, K.; Peters, G. In Vitro Activity of Recombinant Lysostaphin against Staphylococcus aureus Isolates from Anterior Nares and Blood. Antimicrob. Agents Chemother. 2003, 47, 3613–3615. [Google Scholar] [CrossRef] [Green Version]
- Potter, A.; Ceotto, H.; Coelho, M.L.V.; Guimarães, A.J.; Bastos, M.D.C.F. The gene cluster of aureocyclicin 4185: The first cyclic bacteriocin of Staphylococcus aureus. Microbiology 2014, 160, 917–928. [Google Scholar] [CrossRef] [Green Version]
- Wladyka, B.; Wielebska, K.; Wloka, M.; Bochenska, O.; Dubin, G.; Dubin, A.; Mak, P. Isolation, biochemical characterization, and cloning of a bacteriocin from the poultry-associated Staphylococcus aureus strain CH-91. Appl. Microbiol. Biotechnol. 2013, 97, 7229–7239. [Google Scholar] [CrossRef] [Green Version]
- Fontana, M.B.; de Bastos, M.C.; Brandelli, A. Bacteriocins Pep5 and epidermin inhibit Staphylococcus epidermidis adhesion to catheters. Curr. Microbiol. 2006, 52, 350–353. [Google Scholar] [CrossRef] [PubMed]
- Ekkelenkamp, M.B.; Hanssen, M.; Hsu, S.T.D.; De Jong, A.; Milatovic, D.; Verhoef, J.; Van Nuland, N.A.J. Isolation and structural characterization of epilancin 15X, a novel lantibiotic from a clinical strain of Staphylococcus epidermidis. FEBS Lett. 2005, 579, 1917–1922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kellner, R.; Jung, G.; Hörner, T.; Zähner, H.; Schnell, N.; Entian, K.-D.; Götz, F. Gallidermin: A new lanthionine-containing polypeptide antibiotic. Eur. J. Biochem. 2005, 177, 53–59. [Google Scholar] [CrossRef]
- Bengtsson, T.; Lönn, J.; Khalaf, H.; Palm, E. The lantibiotic gallidermin acts bactericidal against Staphylococcus epidermidis and Staphylococcus aureus and antagonizes the bacteria-induced proinflammatory responses in dermal fibroblasts. Microbiologyopen 2018, 7, e00606. [Google Scholar] [CrossRef] [Green Version]
- Sung, C.; Kim, B.G.; Kim, S.; Joo, H.S.; Kim, P.I. Probiotic potential of Staphylococcus hominis MBBL 2-9 as anti-Staphylococcus aureus agent isolated from the vaginal microbiota of a healthy woman. J. Appl. Microbiol. 2010, 108, 908–916. [Google Scholar] [CrossRef]
- Kim, P.I.; Sohng, J.K.; Sung, C.; Joo, H.S.; Kim, E.M.; Yamaguchi, T.; Park, D.; Kim, B.G. Characterization and structure identification of an antimicrobial peptide, hominicin, produced by Staphylococcus hominis MBBL 2-9. Biochem. Biophys. Res. Commun. 2010, 399, 133–138. [Google Scholar] [CrossRef]
- Fagundes, P.C.; Ceotto, H.; Potter, A.; Vasconcelos Paiva Brito, M.A.; Brede, D.A.; Nes, I.F.; de Freire Bastos, M.C. Hyicin 3682, a bioactive peptide produced by Staphylococcus hyicus 3682 with potential applications for food preservation. Res. Microbiol. 2011, 162, 1052–1059. [Google Scholar] [CrossRef]
- Fagundes, P.C.; Nascimento de Sousa Santos, I.; Silva Mrancisco, M.; Mattos Albano, R.; de Freire Bastos, M.D. Genetic and biochemical characterization of hyicin 3682, the first bacteriocin reported for Staphylococcus hyicus. Microbiol. Res. 2017, 198, 36–46. [Google Scholar] [CrossRef]
- Islam, M.R.; Nishie, M.; Nagao, J.I.; Zendo, T.; Keller, S.; Nakayama, J.; Kohda, D.; Sahl, H.G.; Sonomoto, K. Ring A of nukacin ISK-1: A lipid II-binding motif for type-A(II) lantibiotic. J. Am. Chem. Soc. 2012, 134, 3687–3690. [Google Scholar] [CrossRef]
- Minamikawa, M.; Kawai, Y.; Inoue, N.; Yamazaki, K. Purification and characterization of warnericin RB4, anti-Alicyclobacillus bacteriocin, produced by Staphylococcus warneri RB4. Curr. Microbiol. 2005, 51, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.F.S.; Ceotto-Vigoder, H.; Barrias, E.S.; Souto-Padrón, T.C.B.S.; Nes, I.F.; Bastos, M.D.C.F. Hyicin 4244, the first sactibiotic described in staphylococci, exhibits an anti-staphylococcal biofilm activity. Int. J. Antimicrob. Agents 2018, 51, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Netz, D.J.; Sahl, H.G.; Marcolino, R.; dos Santos Nascimento, J.; de Oliveira, S.S.; Soares, M.B.; do Carmo de Freire Bastos, M. Molecular characterisation of aureocin A70, a multi-peptide bacteriocin isolated from Staphylococcus aureus. J. Mol. Biol. 2001, 311, 939–949. [Google Scholar] [CrossRef]
- Lynch, D.; O’Connor, P.M.; Cotter, P.D.; Hill, C.; Field, D.; Begley, M. Identification and characterisation of capidermicin, a novel bacteriocin produced by Staphylococcus capitis. PLoS ONE 2019, 14, e0223541. [Google Scholar] [CrossRef]
- Gibreel, T.M.; Upton, M. Synthetic epidermicin NI01 can protect Galleria mellonella larvae from infection with Staphylococcus aureus. J. Antimicrob. Chemother. 2013, 68, 2269–2273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halliwell, S.; Warn, P.; Sattar, A.; Derrick, J.P.; Upton, M. A single dose of epidermicin NI01 is sufficient to eradicate MRSA from the nares of cotton rats. J. Antimicrob. Chemother. 2017, 72, 778–781. [Google Scholar] [CrossRef] [Green Version]
- Sugai, M.; Fujiwara, T.; Akiyama, T.; Ohara, M.; Komatsuzawa, H.; Inoue, S.; Suginaka, H. Purification and molecular characterization of glycylglycine endopeptidase produced by Staphylococcus capitis EPK1. J. Bacteriol. 1997, 179, 1193–1202. [Google Scholar] [CrossRef] [Green Version]
- Patron, R.L.; Climo, M.W.; Goldstein, B.P.; Archer, G.L. Lysostaphin treatment of experimental aortic valve endocarditis caused by a Staphylococcus aureus isolate with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 1999, 43, 1754–1755. [Google Scholar] [CrossRef] [Green Version]
- Quickel, K.E.; Selden, R.; Caldwell, J.R.; Nora, N.F.; Schaffner, W. Efficacy and safety of topical lysostaphin treatment of persistent nasal carriage of Staphylococcus aureus. Appl. Microbiol. 1971, 22, 446–450. [Google Scholar] [CrossRef] [Green Version]
- Shah, A.; Mond, J.; Walsh, S. Lysostaphin-coated catheters eradicate Staphylococccus aureus challenge and block surface colonization. Antimicrob. Agents Chemother. 2004, 48, 2704–2707. [Google Scholar] [CrossRef] [Green Version]
- Dixon, R.E.; Goodman, J.S.; Koenig, M.G. Lysostaphin: An enzymatic approach to staphylococcal disease. Yale J. Biol. Med. 1968, 41, 62–68. [Google Scholar]
- Harrison, E.F.; Zygmunt, W.A. Lysostaphin in experimental renal infections. J. Bacteriol. 1967, 93, 520–524. [Google Scholar] [CrossRef] [Green Version]
- Zygmunt, W.A.; Harrison, E.F.; Browder, H.P.; Tavormina, P.A. Comparative inhibition of methicillin-resistant strains of Staphylococcus aureus by lysostaphin and other antibiotics. Appl. Microbiol. 1968, 16, 1174–1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.A.; Kusuma, C.; Mond, J.J.; Kokai-Kun, J.F. Lysostaphin disrupts Staphylococcus aureus and Staphylococcus epidermidis biofilms on artificial surfaces. Antimicrob. Agents Chemother. 2003, 47, 3407–3414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huber, M.M.; Huber, T.W. Susceptibility of methicillin-resistant Staphylococcus aureus to lysostaphin. J. Clin. Microbiol. 1989, 27, 1122–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bramley, A.J.; Foster, R. Effects of lysostaphin on Staphylococcus aureus infections of the mouse mammary gland. Res. Vet. Sci. 1990, 49, 120–121. [Google Scholar] [CrossRef]
- Schuhardt, V.T.; Schindler, C.A. Lysostaphin therapy in mice infected with Staphylococcus aureus. J. Bacteriol. 1964, 88, 815–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Placencia, F.X.; Kong, L.; Weisman, L.E. Treatment of methicillin-resistant Staphylococcus aureus in neonatal mice: Lysostaphin versus vancomycin. Pediatr. Res. 2009, 65, 420–424. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for In Vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Manosroi, A.; Khanrin, P.; Lohcharoenkal, W.; Werner, R.G.; Götz, F.; Manosroi, W.; Manosroi, J. Transdermal absorption enhancement through rat skin of gallidermin loaded in niosomes. Int. J. Pharm. 2010, 392, 304–310. [Google Scholar] [CrossRef]
- O’Sullivan, J.; O’Connor, P.; Rea, M.; O’Sullivan, O.; Walsh, C.; Healy, B.; Mathur, H.; Field, D.; Hill, C.; Paul, R. Nisin J, a novel natural nisin variant, is produced by Staphylococcus capitis sourced from the human skin microbiota. J. Bacteriol. 2019, 202. [Google Scholar] [CrossRef] [Green Version]
- Hurst, A. Nisin. Adv. Appl. Microbiol. 1981, 27, 85–123. [Google Scholar]
- Gross, E.; Morell, J.L. The Structure of Nisin. J. Am. Chem. Soc. 1971, 93, 4634–4635. [Google Scholar] [CrossRef] [PubMed]
- Zendo, T.; Fukao, M.; Ueda, K.; Higuchi, T.; Nakayama, J.; Sonomoto, K. Identification of the lantibiotic nisin q, a new natural nisin variant produced by Lactococcus lactis 61-14 isolated from a river in Japan. Biosci. Biotechnol. Biochem. 2003, 67, 1616–1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wirawan, R.E.; Klesse, N.A.; Jack, R.W.; Tagg, J.R. Molecular and genetic characterization of a novel nisin variant produced by Streptococcus uberis. Appl. Environ. Microbiol. 2006, 72, 1148–1156. [Google Scholar] [CrossRef] [Green Version]
- O’Connor, P.M.; O’Shea, E.F.; Guinane, C.M.; O’Sullivan, O.; Cotter, P.D.; Ross, R.P.; Hill, C. Nisin H is a new nisin variant produced by the gut-derived strain streptococcus hyointestinalis DPC6484. Appl. Environ. Microbiol. 2015, 81, 3953–3960. [Google Scholar] [CrossRef] [Green Version]
- De Kwaadsteniet, M.; Ten Doeschate, K.; Dicks, L.M.T. Characterization of the structural gene encoding nisin F, a new lantibiotic produced by a Lactococcus lactis subsp. lactis isolate from freshwater catfish (Clarias gariepinus). Appl. Environ. Microbiol. 2008, 74, 547–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannoehr, J.; Guardabassi, L. Staphylococcus pseudintermedius in the dog: Taxonomy, diagnostics, ecology, epidemiology and pathogenicity. Vet. Dermatol. 2012, 23, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Kadlec, K.; Schwarz, S.; Perreten, V.; Andersson, U.G.; Finn, M.; Greko, C.; Moodley, A.; Kania, S.A.; Frank, L.A.; Bemis, D.A.; et al. Molecular analysis of methicillinresistant Staphylococcus pseudintermedius of feline origin from different European countries and North America. J. Antimicrob. Chemother. 2010, 65, 1826–1828. [Google Scholar] [CrossRef]
- Frank, L.A.; Loeffler, A. Meticillin-resistant Staphylococcus pseudintermedius: Clinical challenge and treatment options. Vet. Dermatol. 2012, 23, 283–291. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, A.J.; Harrison, E.M.; Stanczak-Mrozek, K.; Leggett, B.; Waller, A.; Holmes, M.A.; Lloyd, D.H.; Lindsay, J.A.; Loeffler, A. Genomic insights into the rapid emergence and evolution of MDR in Staphylococcus pseudintermedius. J. Antimicrob. Chemother. 2014, 70, 997–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barré-Sinoussi, F.; Montagutelli, X. Animal models are essential to biological research: Issues and perspectives. Future Sci. OA 2015, 1. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, J.A. Innate immunity of insects. Curr. Opin. Immunol. 1995, 7, 4–10. [Google Scholar] [CrossRef]
- Kokai-Kun, J.F. The cotton rat as a model for Staphylococcus aureus nasal colonization in humans: Cotton Rat S. aureus Nasal Colonization Model. Methods Mol. Biol. 2007, 431, 241–254. [Google Scholar]
- Kumar, R.; Jangir, P.K.; Das, J.; Taneja, B.; Sharma, R. Genome Analysis of Staphylococcus capitis TE8 Reveals Repertoire of Antimicrobial Peptides and Adaptation Strategies for Growth on Human Skin. Sci. Rep. 2017, 7, 10447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khusro, A.; Aarti, C.; Salem, A.Z.M.; Buendía Rodríguez, G.; Rivas-Cáceres, R.R. Antagonistic trait of Staphylococcus succinus strain AAS2 against uropathogens and assessment of its in vitro probiotic characteristics. Microb. Pathog. 2018, 118, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Rahmdel, S.; Shekarforoush, S.S.; Hosseinzadeh, S.; Torriani, S.; Gatto, V. Antimicrobial spectrum activity of bacteriocinogenic Staphylococcus strains isolated from goat and sheep milk. J. Dairy Sci. 2019, 102, 2928–2940. [Google Scholar] [CrossRef] [Green Version]
- Janek, D.; Zipperer, A.; Kulik, A.; Krismer, B.; Peschel, A. High Frequency and Diversity of Antimicrobial Activities Produced by Nasal Staphylococcus Strains against Bacterial Competitors. PLoS Pathog. 2016, 12, e1005812. [Google Scholar] [CrossRef] [Green Version]
- Carson, D.A.; Barkema, H.W.; Naushad, S.; De Buck, J. Bacteriocins of non-aureus staphylococci isolated from bovine milk. Appl. Environ. Microbiol. 2017, 83, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- de Jong, A.; van Hijum, S.A.F.T.; Bijlsma, J.J.E.; Kok, J.; Kuipers, O.P. BAGEL: A web-based bacteriocin genome mining tool. Nucleic Acids Res. 2006, 34, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Iwase, T.; Uehara, Y.; Shinji, H.; Tajima, A.; Seo, H.; Takada, K.; Agata, T.; Mizunoe, Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 2010, 465, 346–349. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Krishnan, V.; Macon, K.; Manne, K.; Narayana, S.V.L.; Schneewind, O. Secreted proteases control autolysin-mediated biofilm growth of Staphylococcus aureus. J. Biol. Chem. 2013, 288, 29440–29452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mann, E.E.; Rice, K.C.; Boles, B.R.; Endres, J.L.; Ranjit, D.; Chandramohan, L.; Tsang, L.H.; Smeltzer, M.S.; Horswill, A.R.; Bayles, K.W. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE 2009, 4, e5822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huseby, M.J.; Kruse, A.C.; Digre, J.; Kohler, P.L.; Vocke, J.A.; Mann, E.E.; Bayles, K.W.; Bohach, G.A.; Schlievert, P.M.; Ohlendorf, D.H.; et al. Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proc. Natl. Acad. Sci. USA 2010, 107, 14407–14412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugimoto, S.; Iwamoto, T.; Takada, K.; Okuda, K.I.; Tajima, A.; Iwase, T.; Mizunoe, Y. Staphylococcus epidermidis Esp degrades specific proteins associatedwith Staphylococcus aureus biofilm formation and host-pathogen interaction. J. Bacteriol. 2013, 195, 1645–1655. [Google Scholar] [CrossRef] [Green Version]
- Zipperer, A.; Konnerth, M.C.; Laux, C.; Berscheid, A.; Janek, D.; Weidenmaier, C.; Burian, M.; Schilling, N.A.; Slavetinsky, C.; Marschal, M.; et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016, 535, 511–516. [Google Scholar] [CrossRef]
| Class | Subclass | Subtype | Bacteriocin | Producing Strain | Inhibits S. aureus: Strain (MIC) | in vivo Model | References |
|---|---|---|---|---|---|---|---|
| I | Ia | A1 | BacCh91 * | S. aureus CH9/DSM26258 | in vitro: ATCC25923, Newman, M-122, RN4220 (4.0–6.0 μM) | [92] | |
| Epicidin 280 * | S. epidermidis BN280 | [70] | |||||
| Epidermin * | S. epidermidis Tü3298 | in vitro | [74,93] | ||||
| Epilancin 15X * | S. epidermidis 15X154 | [94] | |||||
| Epilancin K7 * | S. epidermidis K7 | [69] | |||||
| Gallidermin * | S. gallinarum F16/P57 Tü3298 | in vitro: ATCC29213, CCUG35601 (1.25–8.0 μg/mL) | [75,95,96] | ||||
| Hominicin | S. hominis MBBL2–9 | in vitro: ATCC25923, ATCC11435, CCAR M3501 (0.06–3.82 μg/mL) | [97,98] | ||||
| Hyicin 3682 | S. hyicus 3682 | in vitro | [99,100] | ||||
| Nisin J | S. capitis APC2923 | in vitro | [20] | ||||
| Pep5 | S.epidermidis 5 | in vitro | [73,74,93] | ||||
| A2 | Nukacin ISK-1 ** | S. warneri Nukadoko/S. simulans 3299 | [101] | ||||
| Warnericin RB4 | S. warneri RB4 | [102] | |||||
| Ic | Hyicin 4244 | S. hyicus 4244 | in vitro | [83,103] | |||
| II | IIb | S | Aureocin A70 | S. aureus A70 | [104] | ||
| S | C55 * | S. aureus C55 | [67] | ||||
| IId | Aureocin A53 * | S. aureus A53 | [66,74] | ||||
| BacSp222 ** | S.pseudintermedius | in vitro: DSM26258, MRSA USA300, KB/8568, ATCC25923 (0.89–1.30 μM) | [52] | ||||
| II | IId | Capidermicin | S. capitis CIT060 | in vitro: NCDO1499, DPC5297, Newman, RF122 (3.1–10 μg/mL) | [105] | ||
| Epidermicin NI01 * | S. epidermidis 224 | in vitro: 1195, MRSA s37, MRSA s41, MRSA s71 (1.0–2.0 μg/mL) | greater wax moth, cotton rat | [64,106,107] | |||
| III | IIIa | Endopeptidase ALE-1 † | S. capitis EPk1 | [108] | |||
| Lysostaphin ** | S. simulans biovar Staphylolyticus ATCC1362 | in vitro (0.002–100 μg/mL) | rat, mouse, cotton rat, rabbit, human | [87,88,90,109,110,111,112,113,114,115,116,117,118,119] | |||
| IV | Aureocyclicin 4185 | S. aureus 4185 | [91] |
| Class I | Class II | |||
|---|---|---|---|---|
| Lantibiotics | Sactibiotics | Lasso Peptides | ||
| Number of Bacteriocin Clusters Identified | 29 | 3 | 4 | 69 |
| Number of Isolates that the Clusters are Present in | 29 | 3 | 2 | 68 |
| The Species that the Clusters are Present in | S. capitis | S. capitis | S. fleurettii | S. equorum |
| S. chromogenes | S. sciuri | S. gallinarum | ||
| S. cohnii | S. haemolyticus | |||
| S. epidermidis | S. hyicus | |||
| S. equorum | S. saprophyticus | |||
| S. gallinarum | S. sciuri | |||
| S. sciuri | S. simulans | |||
| S. simulans | S. succinus | |||
| S. succinus | S. warneri | |||
| S. vitulinus | S. xylosus | |||
| Number of Isolates Showing Inhibitory Activity in vitro | 15 | 2 | 1 | 9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Newstead, L.L.; Varjonen, K.; Nuttall, T.; Paterson, G.K. Staphylococcal-Produced Bacteriocins and Antimicrobial Peptides: Their Potential as Alternative Treatments for Staphylococcus aureus Infections. Antibiotics 2020, 9, 40. https://doi.org/10.3390/antibiotics9020040
Newstead LL, Varjonen K, Nuttall T, Paterson GK. Staphylococcal-Produced Bacteriocins and Antimicrobial Peptides: Their Potential as Alternative Treatments for Staphylococcus aureus Infections. Antibiotics. 2020; 9(2):40. https://doi.org/10.3390/antibiotics9020040
Chicago/Turabian StyleNewstead, Logan L., Katarina Varjonen, Tim Nuttall, and Gavin K. Paterson. 2020. "Staphylococcal-Produced Bacteriocins and Antimicrobial Peptides: Their Potential as Alternative Treatments for Staphylococcus aureus Infections" Antibiotics 9, no. 2: 40. https://doi.org/10.3390/antibiotics9020040





