A Challenging View: Antibiotics Play a Role in the Regulation of the Energetic Metabolism of the Producing Bacteria
Abstract
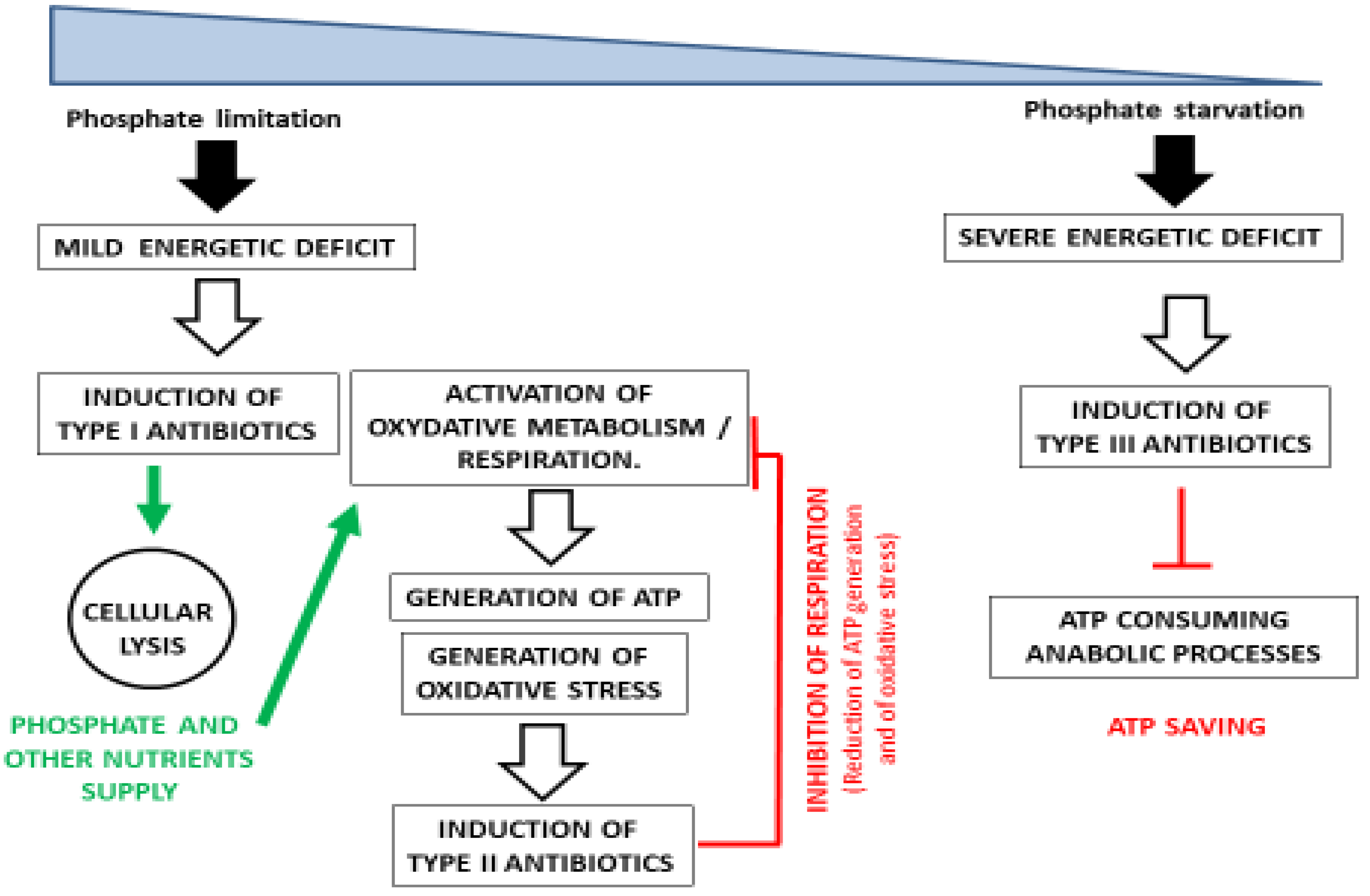
:1. Introduction
2. Discussion
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hopwood, D.A. Streptomyces in Nature and Medecine: The Antibiotic Makers; Oxford University Press: New York, NY, USA, 2007. [Google Scholar]
- Bakal, C.J.; Davies, J.E. No longer an exclusive club: Eukaryotic signalling domains in bacteria. Trends Cell Biol. 2000, 10, 32–38. [Google Scholar] [CrossRef]
- Li, J.; Kim, S.G.; Blenis, J. Rapamycin: One drug, many effects. Cell Metab. 2014, 19, 373–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Procopio, R.E.; Silva, I.R.; Martins, M.K.; Azevedo, J.L.; Araujo, J.M. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 2012, 16, 466–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, J.F. Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: An unfinished story. J. Bacteriol. 2004, 186, 5197–5201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esnault, C.; Dulermo, T.; Smirnov, A.; Askora, A.; David, M.; Deniset-Besseau, A.; Holland, I.B.; Virolle, M.J. Strong antibiotic production is correlated with highly active oxidative metabolism in Streptomyces coelicolor M145. Sci. Rep. 2017, 7, 200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos-Beneit, F. The Pho regulon: A huge regulatory network in bacteria. Front. Microbiol. 2015, 6, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allenby, N.E.; Laing, E.; Bucca, G.; Kierzek, A.M.; Smith, C.P. Diverse control of metabolism and other cellular processes in Streptomyces coelicolor by the PhoP transcription factor: Genome-wide identification of in vivo targets. Nucleic Acids Res. 2012, 40, 9543–9556. [Google Scholar] [CrossRef]
- Martin, J.F.; Rodriguez-Garcia, A.; Liras, P. The master regulator PhoP coordinates phosphate and nitrogen metabolism, respiration, cell differentiation and antibiotic biosynthesis: Comparison in Streptomyces coelicolor and Streptomyces avermitilis. J. Antibiot. 2017, 70, 534–541. [Google Scholar] [CrossRef] [Green Version]
- Polkade, A.V.; Mantri, S.S.; Patwekar, U.J.; Jangid, K. Quorum Sensing: An Under-Explored Phenomenon in the Phylum Actinobacteria. Front. Microbiol. 2016, 7, 131. [Google Scholar] [CrossRef] [Green Version]
- Latoscha, A.; Wormann, M.E.; Tschowri, N. Nucleotide second messengers in Streptomyces. Microbiology 2019, 165, 1153–1165. [Google Scholar] [CrossRef]
- Sivapragasam, S.; Grove, A. The Link between Purine Metabolism and Production of Antibiotics in Streptomyces. Antibiotics 2019, 8, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Lih, C.J.; Pan, K.H.; Cohen, S.N. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 2001, 15, 3183–3192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakey, J.H.; Lea, E.J.; Rudd, B.A.; Wright, H.M.; Hopwood, D.A. A new channel-forming antibiotic from Streptomyces coelicolor A3 (2) which requires calcium for its activity. J. Gen. Microbiol. 1983, 129, 3565–3573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsao, S.W.; Rudd, B.A.; He, X.G.; Chang, C.J.; Floss, H.G. Identification of a red pigment from Streptomyces coelicolor A3 (2) as a mixture of prodigiosin derivatives. J. Antibiot. 1985, 38, 128–131. [Google Scholar] [CrossRef] [Green Version]
- Malpartida, F.; Hopwood, D.A. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host. Nature 1984, 309, 462–464. [Google Scholar] [CrossRef]
- Novotna, J.; Vohradsky, J.; Berndt, P.; Gramajo, H.; Langen, H.; Li, X.M.; Minas, W.; Orsaria, L.; Roeder, D.; Thompson, C.J. Proteomic studies of diauxic lag in the differentiating prokaryote Streptomyces coelicolor reveal a regulatory network of stress-induced proteins and central metabolic enzymes. Mol. Microbiol. 2003, 48, 1289–1303. [Google Scholar] [CrossRef]
- Takano, E.; Gramajo, H.C.; Strauch, E.; Andres, N.; White, J.; Bibb, M.J. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3 (2). Mol. Microbiol. 1992, 6, 2797–2804. [Google Scholar] [CrossRef] [Green Version]
- Gramajo, H.C.; Takano, E.; Bibb, M.J. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3 (2) is transcriptionally regulated. Mol. Microbiol. 1993, 7, 837–845. [Google Scholar] [CrossRef] [Green Version]
- Hurdle, J.G.; O’Neill, A.J.; Chopra, I.; Lee, R.E. Targeting bacterial membrane function: An underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 2011, 9, 62–75. [Google Scholar] [CrossRef] [Green Version]
- Dias, C.; Rauter, A.P. Membrane-targeting antibiotics: Recent developments outside the peptide space. Future Med. Chem. 2019, 11, 211–228. [Google Scholar] [CrossRef] [PubMed]
- Manteca, A.; Mader, U.; Connolly, B.A.; Sanchez, J. A proteomic analysis of Streptomyces coelicolor programmed cell death. Proteomics 2006, 6, 6008–6022. [Google Scholar] [CrossRef] [PubMed]
- Filippova, S.N.; Vinogradova, K.A. Programmed cell death as one of the stages of streptomycete differentiation. Microbiology 2017, 86, 439–454. [Google Scholar] [CrossRef]
- Chater, K.F. Recent advances in understanding Streptomyces. F1000Research 2016, 5, 2795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tenconi, E.; Traxler, M.F.; Hoebreck, C.; van Wezel, G.P.; Rigali, S. Production of Prodiginines Is Part of a Programmed Cell Death Process in Streptomyces coelicolor. Front. Microbiol. 2018, 9, 1742. [Google Scholar] [CrossRef] [PubMed]
- Beites, T.; Oliveira, P.; Rioseras, B.; Pires, S.D.; Oliveira, R.; Tamagnini, P.; Moradas-Ferreira, P.; Manteca, A.; Mendes, M.V. Streptomyces natalensis programmed cell death and morphological differentiation are dependent on oxidative stress. Sci. Rep. 2015, 5, 12887. [Google Scholar] [CrossRef] [Green Version]
- Beites, T.; Pires, S.D.; Santos, C.L.; Osorio, H.; Moradas-Ferreira, P.; Mendes, M.V. Crosstalk between ROS homeostasis and secondary metabolism in S. natalensis ATCC 27448: Modulation of pimaricin production by intracellular ROS. PLoS ONE 2011, 6, e27472. [Google Scholar]
- Miranda, R.U.; Gomez-Quiroz, L.E.; Mendoza, M.; Perez-Sanchez, A.; Fierro, F.; Barrios-Gonzalez, J. Reactive oxygen species regulate lovastatin biosynthesis in Aspergillus terreus during submerged and solid-state fermentations. Fungal Biol. 2014, 118, 979–989. [Google Scholar] [CrossRef]
- Bibian, M.E.; Perez-Sanchez, A.; Mejia, A.; Barrios-Gonzalez, J. Penicillin and cephalosporin biosyntheses are also regulated by reactive oxygen species. Appl. Microbiol. Biotechnol. 2020, 104, 1773–1783. [Google Scholar] [CrossRef]
- Lu, J.M.; Rosokha, S.V.; Neretin, I.S.; Kochi, J.K. Quinones as electron acceptors X-ray structures, spectral (EPR, UV-vis) characteristics and electron-transfer reactivities of their reduced anion radicals as separated vs. contact ion pairs. J. Am. Chem. Soc. 2006, 128, 16708–16719. [Google Scholar] [CrossRef]
- Prajapati, D.; Kumari, N.; Dave, K.; Chatupale, V.; Pohnerkar, J. Chromomycin, an antibiotic produced by Streptomyces flaviscleroticus might play a role in the resistance to oxidative stress and is essential for viability in stationary phase. Environ. Microbiol. 2018, 21, 814–826. [Google Scholar] [CrossRef] [PubMed]
- Balemans, W.; Vranckx, L.; Lounis, N.; Pop, O.; Guillemont, J.; Vergauwen, K.; Mol, S.; Gilissen, R.; Motte, M.; Lancois, D.; et al. Novel antibiotics targeting respiratory ATP synthesis in Gram-positive pathogenic bacteria. Antimicrob. Agents Chemother. 2012, 56, 4131–4139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paudel, A.; Hamamoto, H.; Panthee, S.; Sekimizu, K. Menaquinone as a potential target of antibacterial agents. Drug Discov. Ther. 2016, 10, 123–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sellamuthu, S.; Singh, M.; Kumar, A.; Singh, S.K. Type-II NADH Dehydrogenase (NDH-2): A promising therapeutic target for antitubercular and antibacterial drug discovery. Expert Opin. Ther. Targets 2017, 21, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Symersky, J.; Osowski, D.; Walters, D.E.; Mueller, D.M. Oligomycin frames a common drug-binding site in the ATP synthase. Proc. Natl. Acad. Sci. USA 2012, 109, 13961–13965. [Google Scholar] [CrossRef] [Green Version]
- Hong, S.; Pedersen, P.L. ATP synthase and the actions of inhibitors utilized to study its roles in human health, disease, and other scientific areas. Microbiol. Mol. Biol. Rev. 2008, 72, 590–641. [Google Scholar] [CrossRef] [Green Version]
- Seipke, R.F.; Hutchings, M.I. The regulation and biosynthesis of antimycins. Beilstein J. Org. Chem. 2013, 9, 2556–2563. [Google Scholar] [CrossRef] [Green Version]
- Coccetti, P.; Nicastro, R.; Tripodi, F. Conventional and emerging roles of the energy sensor Snf1/AMPK in Saccharomyces cerevisiae. Microb. Cell 2018, 5, 482–494. [Google Scholar] [CrossRef] [Green Version]
- Godinez, O.; Dyson, P.; del Sol, R.; Barrios-Gonzalez, J.; Millan-Pacheco, C.; Mejia, A. Targeting the Osmotic Stress Response for Strain Improvement of an Industrial Producer of Secondary Metabolites. J. Microbiol. Biotechnol. 2015, 25, 1787–1795. [Google Scholar] [CrossRef]
- Hopwood, D.A. How do antibiotic-producing bacteria ensure their self-resistance before antibiotic biosynthesis incapacitates them? Mol. Microbiol. 2007, 63, 937–940. [Google Scholar] [CrossRef]
- Wencewicz, T.A. Crossroads of Antibiotic Resistance and Biosynthesis. J. Mol. Biol. 2019, 431, 3370–3399. [Google Scholar] [CrossRef] [PubMed]
- Abrudan, M.I.; Smakman, F.; Grimbergen, A.J.; Westhoff, S.; Miller, E.L.; van Wezel, G.P.; Rozen, D.E. Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc. Natl. Acad. Sci. USA 2015, 112, 11054–11059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumbhar, C.; Mudliar, P.; Bhatia, L.; Kshirsagar, A.; Watve, M. Widespread predatory abilities in the genus Streptomyces. Arch. Microbiol. 2014, 196, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Stubbendieck, R.M.; Straight, P.D. Multifaceted Interfaces of Bacterial Competition. J. Bacteriol. 2016, 198, 2145–2155. [Google Scholar] [CrossRef] [Green Version]
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Virolle, M.-J. A Challenging View: Antibiotics Play a Role in the Regulation of the Energetic Metabolism of the Producing Bacteria. Antibiotics 2020, 9, 83. https://doi.org/10.3390/antibiotics9020083
Virolle M-J. A Challenging View: Antibiotics Play a Role in the Regulation of the Energetic Metabolism of the Producing Bacteria. Antibiotics. 2020; 9(2):83. https://doi.org/10.3390/antibiotics9020083
Chicago/Turabian StyleVirolle, Marie-Joelle. 2020. "A Challenging View: Antibiotics Play a Role in the Regulation of the Energetic Metabolism of the Producing Bacteria" Antibiotics 9, no. 2: 83. https://doi.org/10.3390/antibiotics9020083



