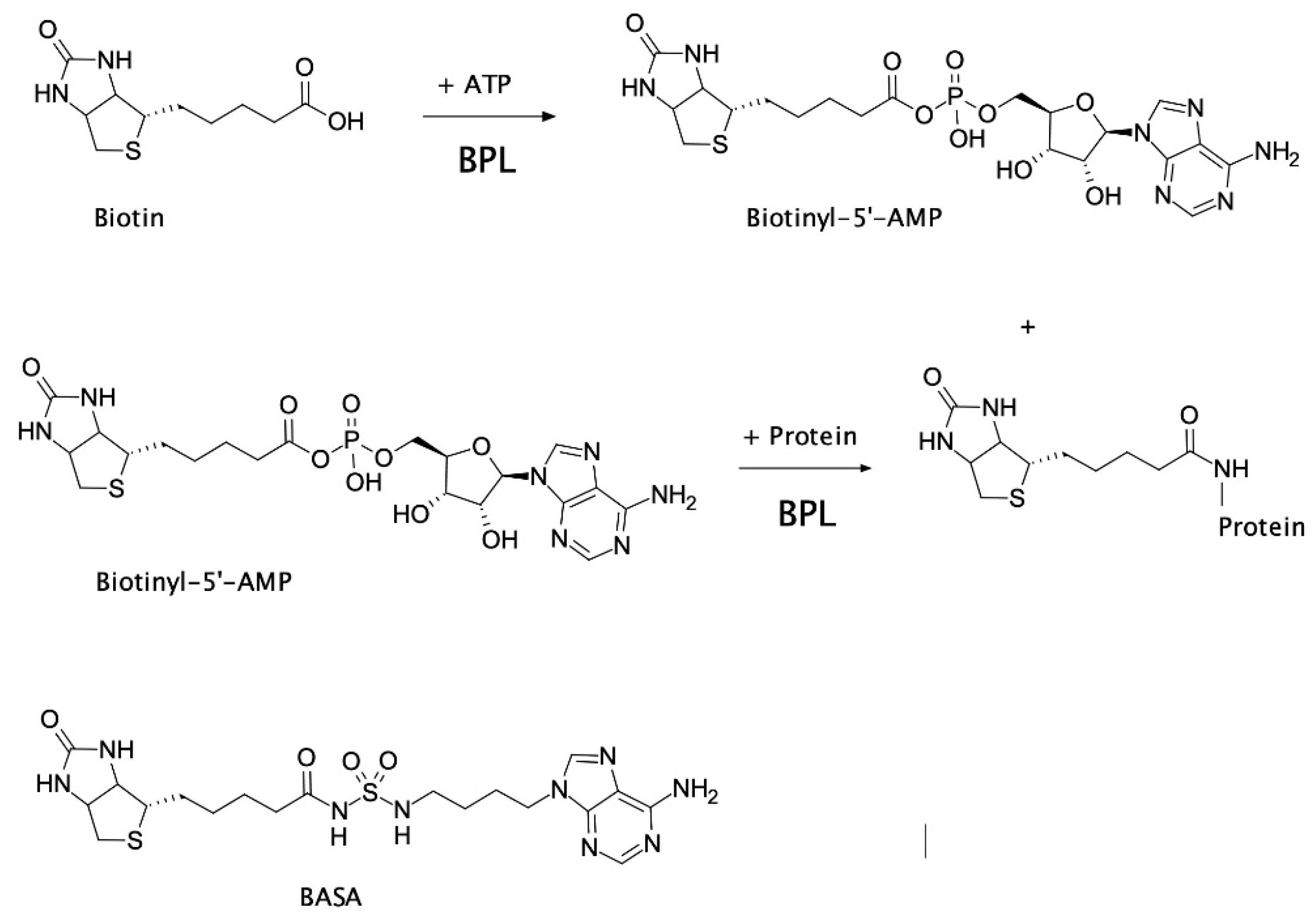
Abstract
Biotin protein ligase (BPL) inhibitors are a novel class of antibacterial that target clinically important methicillin-resistant Staphylococcus aureus (S. aureus). In S. aureus, BPL is a bifunctional protein responsible for enzymatic biotinylation of two biotin-dependent enzymes, as well as serving as a transcriptional repressor that controls biotin synthesis and import. In this report, we investigate the mechanisms of action and resistance for a potent anti-BPL, an antibacterial compound, biotinyl-acylsulfamide adenosine (BASA). We show that BASA acts by both inhibiting the enzymatic activity of BPL in vitro, as well as functioning as a transcription co-repressor. A low spontaneous resistance rate was measured for the compound (<10−9) and whole-genome sequencing of strains evolved during serial passaging in the presence of BASA identified two discrete resistance mechanisms. In the first, deletion of the biotin-dependent enzyme pyruvate carboxylase is proposed to prioritize the utilization of bioavailable biotin for the essential enzyme acetyl-CoA carboxylase. In the second, a D200E missense mutation in BPL reduced DNA binding in vitro and transcriptional repression in vivo. We propose that this second resistance mechanism promotes bioavailability of biotin by derepressing its synthesis and import, such that free biotin may outcompete the inhibitor for binding BPL. This study provides new insights into the molecular mechanisms governing antibacterial activity and resistance of BPL inhibitors in S. aureus.
1. Introduction
The need for new antimicrobial agents to combat the growing threat of drug resistance is currently not being met, with only five novel classes of antibiotics introduced since 2000 [1,2]. Strains that are now resistant to multiple classes of clinically approved antibiotics have been reported for Staphylococcus aureus, Enterococci species and Mycobacterium tuberculosis amongst others [3]. Without new antimicrobial agents, the inability to treat resistant microbial infections is predicted to result in 10 million deaths annually by 2050 [4]. Hence, there is a desperate need for new products with novel mechanisms of action that are not subject to existing resistance mechanisms. Precious few novel antimicrobial agents are currently in the drug development pipeline. One promising new drug target is biotin protein ligase (BPL) for which new potent, selective inhibitors are being developed (reviewed in [5,6]). In this report, we examine one such inhibitor that is active against drug-resistant S. aureus [7] and probe mechanisms of action and potential resistance.
In certain bacteria, such as clinically important S. aureus, BPL is a bifunctional protein with two important roles; (1) enzymatic biotinylation of biotin-dependent enzymes, and (2) biotin-dependent repression of certain target genes implicated in biotin homeostasis. This bifunctionality makes BPL the key master regulator of all biotin-mediated metabolic processes in S. aureus and a promising new antibiotic target [6]. S. aureus BPL (SaBPL) catalyzes the covalent attachment of a biotin prosthetic group onto two key biotin-dependent enzymes, namely acetyl-CoA carboxylase (ACC) and pyruvate carboxylase (PC). Protein biotinylation occurs through two partial reactions. Biotin is first ligated to ATP yielding the reaction intermediate biotinyl-5′-AMP (Figure 1). BPL then employs the labile biotinyl-5′-AMP to attach the biotinyl moiety onto a single target lysine residue present in the active site of ACC or PC [8]. The appended biotin is necessary for the biotin-dependent enzymes to fulfill their important metabolic roles. ACC catalyzes the first committed step in fatty acid synthesis, whilst PC replenishes the tricarboxylic acid cycle with oxaloacetate. ACC and BPL have been identified as essential gene products for growth in genetic knockout studies performed on S. aureus [9,10,11,12] and, consequently, have been the targets of drug discovery programs. The antibacterial efficacy of ACC inhibitors has been demonstrated in vivo (reviewed in [13,14]) and BPL inhibitors have shown efficacy in vitro against S. aureus and M. tuberculosis [15,16,17,18,19]. Whilst dispensable for growth in culture, PC has been shown to be an important virulence factor in bacteremia models in mice and nematodes [20,21]. As protein biotinylation is essential for the activity of ACC and PC, pharmacological inhibition of S. aureus BPL (SaBPL) is proposed to impact bacterial viability and pathogenesis due to the disruption of both enzymes (reviewed [6]).
SaBPL also regulates the transcription of three genetic elements, namely the biotin biosynthesis operon (bioO), a gene encoding the substrate-specific subunit of the biotin transport protein BioY, as well as a putative fatty acid ligase operon yhfT-yhfS [22,23]. Recent in vivo characterization in S. aureus has revealed a hierarchy in the control of these genes by SaBPL. The biotin biosynthesis operon was shown to be the most responsive to treatment with exogenous biotin, being repressed by 10 nM biotin 15 min post-treatment [23]. Transcripts for bioY and yhfT-yhfS persisted longer after biotin treatment than the biotin biosynthesis genes. It is proposed that this mechanism may assist with metabolic adaptation of the bacteria to their environment, as de novo biotin biosynthesis is an energetically expensive process that can be bypassed when exogenous biotin is readily available for import. This was supported experimentally where the growth rate of S. aureus was enhanced in media containing exogenous biotin, even though the bacteria were capable of biotin synthesis [23]. We propose that this ability is important during infection as S. aureus occupies a variety of niche microhabitats with varying biotin availability [23]. These findings also suggest that pharmacological induction of transcriptional repressor activity may lead to biotin starvation through the dual actions of reduced de novo biotin synthesis and reduced biotin import. Hence, small molecules that serve as co-repressors are beneficial in the design of antibacterials that target SaBPL.
Structural biology has greatly assisted in understanding the bifunctional activity of SaBPL. The protein contains an N-terminal helix-turn-helix domain necessary for DNA binding (Figure S1; reviewed in [8,24]). When the cellular demand for biotin is low, and biotinyl-5′-AMP is not required for protein biotinylation, this reaction intermediate also serves as a co-repressor by inducing protein homodimerization, a pre-requisite for high-affinity DNA binding to the operator sequences and repression of the target operons and genes [25,26]. A series of well-characterized conformational changes occur in SaBPL to facilitate this protein dimerization [27]. Following the binding of biotin, residues 118–129 (known as the biotin-binding loop) undergo a disordered-to-ordered transition that creates a pocket that allows binding of ATP. An adenosine-binding loop, formed by residues 224–228, also undergoes structural changes to stabilize the binding of the adenylate moiety [27]. Further allosteric conformational changes occur to loops located in the dimerization interface upon ligand binding. These include the ordering of loops contained between amino acids 142 and 150 (extending an alpha-helix by three residues) and amino acids 198–201 [27]. Together these structural rearrangements facilitate the transition from monomer to homodimer. Arg122 and Phe123 in the biotin-binding loop stabilize the dimer by interacting directly with the side chain of Asp200 in the opposing subunit (Figure S1) [26]. Homodimerization appropriately aligns the two N-terminal DNA binding domains to facilitate their interaction with DNA (Figure S1) [27]. Protein dimerization has been reported to be induced by chemical analogs of biotin and biotinyl-5′-AMP [15,26,28,29].
Several groups have reported on the development of anti-BPL compounds with antibacterial activity [15,16,17,18,19,30,31,32,33,34]. Bisubstrate analogs that occupy both the biotin and ATP-binding pockets and mimic the mode of binding adopted by biotinyl-5′-AMP have been particularly promising. Non-hydrolyzable analogs of biotinyl-5′-AMP have been generated by replacing the labile phosphoanhydride linker connecting the biotin and AMP moieties with alternative bioisosteres. These new inhibitors exploited intermolecular interactions with the bacterial BPLs that are absent in the human enzyme, to provide necessary selectivity. Recently, the bisubstrate analog biotinyl-acylsulfamide adenosine (BASA, Figure 1) was reported with potent anti-S. aureus activity (minimal inhibitory concentration (MIC) RN4220 = 0.125 µg/mL, ATCC 49775 = 0.25 µg/mL) and necessary in vitro selectivity over the human equivalent (Ki SaBPL = 2.4 nM, Ki human BPL >10 μM; >4000-fold selectivity) [7]. A full description of the synthesis of BASA and its chemical characterization has already been described [7]. A crystal structure of SaBPL in complex with BASA revealed that the compound adopted the same mode of binding and induced the same conformational changes necessary for DNA binding and repressor activity as biotinyl-5′-AMP (Figure S1) [7]. In the current study, the cellular mechanisms that contribute to the antibacterial efficacy of BASA were explored. Two discrete resistance mechanisms were also discovered using advanced resistance studies and whole-genome sequencing analysis. This study provides new insights into the mechanism of action of anti-BPL compounds and possible resistance mechanisms to a novel class of antibiotics.
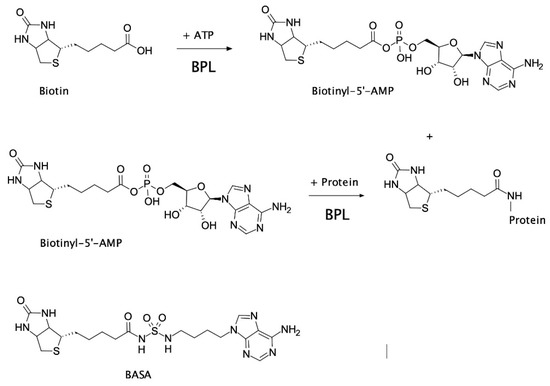
Figure 1.
Protein biotinylation catalyzed by biotin protein ligase (BPL). Biotin ligates with ATP to produce the reaction intermediate, biotinyl-5′-AMP. Biotinyl-5′-AMP is employed to covalently attach the biotinyl moiety onto a specific lysine residue present in the biotin-accepting protein substrate. The chemical structure of biotinyl-acylsulfamide adenosine (BASA), a chemical analog of the reaction intermediate, is shown.
2. Results
2.1. Biological Evaluation of BASA
The antibacterial spectrum, therapeutic window and mechanism of action of BASA were first established using a series of microbiological and cellular assays. Antimicrobial susceptibility assays performed against a panel of clinical isolates of S. aureus (n = 23) revealed that BASA was active against both methicillin-sensitive and resistant strains with minimal inhibitory concentration (MIC) measurements ranging from 0.125 to 0.5 µg/mL (Table 1). There was no change to the MIC when the antimicrobial susceptibility assay was performed in growth media supplemented with 10% fetal calf serum but it did increase eight-fold with 20% serum, suggesting slightly reduced efficacy in the presence of serum fatty acids (Table 1). This potent antibacterial activity was restricted to Staphylococci as other bacterial pathogens were insensitive to BASA including Gram-positive Streptococcus pneumoniae, and Gram-negative Escherichia coli and Enterobacter species. Mycobacterium tuberculosis was the only other bacterium susceptible to BASA (MIC = 55 µg/mL, 100 µM), albeit with weaker potency than S. aureus. Cytotoxicity testing against two human cell lines HepG2 (liver-derived) and HEK293 (kidney-derived) showed both were insensitive to BASA at concentrations up to 250 μg/mL (Table 1, Figure S2), providing a >100-fold therapeutic window for S. aureus. Finally, concentration and time-dependent kill kinetics experiments were performed by viable cell counts following 24 hours of treatment with BASA at one, two and four times the MIC. Growth ceased at all concentrations, however, killing was not observed consistent with BASA functioning as a bacteriostatic antibacterial (Figure S3). The bacteriostatic activity of BASA implies that host-induced killing and/or combination therapy may be required for in vivo efficacy. Consequently, synergy assays with other clinically used antibacterial agents were performed using a checkerboard titration assay [35]. Of the eight antibacterial agents tested, none were antagonistic with BASA, and synergy was detected with both methicillin and streptomycin as determined by a fractional inhibitory concentration (FIC) of <0.5 (Figure S4) [35,36]. Together these biological data establish BASA as a promising new antibacterial agent.

Table 1.
Antibacterial susceptibility and cytotoxicity of BASA.
2.2. Mechanism of Action of BASA
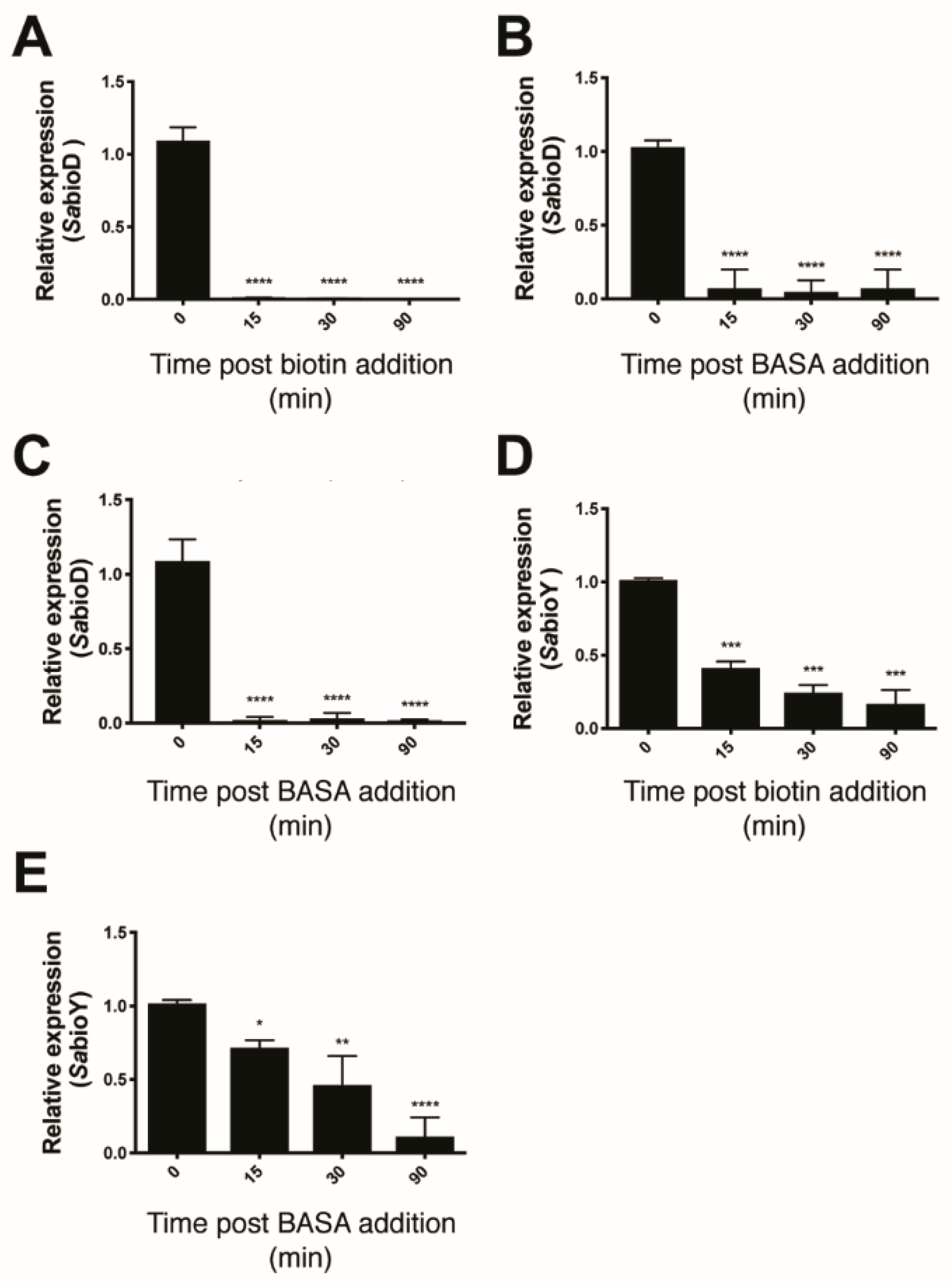
Three lines of evidence were consistent with BASA acting through direct binding to the BPL target. Firstly, the compound showed reduced antibacterial activity against a strain of S. aureus engineered to overexpress SaBPL [19]. The increased concentration of recombinant BPL in the bacteria elevated the MIC by eight-fold over an isotypic control strain (Figure S5A). This response was specific for BPL inhibition with a similar increase observed with another chemical analog of the reaction intermediate, biotinol-5′-AMP [32], but not the unrelated β-lactam amoxicillin (Figure S5B,C). In further support, increased concentrations of the BPL substrate biotin in the growth media also reduced the antibacterial activity of BASA. Super-physiological concentrations of 1 μM (i.e., >1000-fold serum concentrations) of exogenous biotin increased the MIC of BASA by 64-fold (Figure S6). Finally, we demonstrated that BASA functions as a co-repressor in vivo with a potency equivalent to that of biotin. Electrophoretic mobility shift assays (EMSA) were first performed and confirmed that BASA could promote SaBPL binding to DNA in vitro through the same mechanism utilized by the natural co-repressor biotinyl-5′-AMP (Figure S7). Here, the SaBPL-BASA and SaBPL-biotinyl-5′-AMP complexes were compared for binding to both the bioO and bioY promoter sequences. Prior to EMSA analysis, unliganded (apo) SaBPL was incubated with either BASA, or biotin and MgATP to allow the formation of biotinyl-5′-AMP. Equipotent binding was observed for the two complexes to both the bioY and bioO sequences (Figure S7). Having established the DNA binding activity of the SaBPL-BASA complex in vitro, the co-repressor activity of BASA was then investigated using transcriptome studies on whole cells. S. aureus NCTC 8325 was grown in biotin depleted media until log phase and then treated with 10 nM of either biotin or BASA. Cells were also treated with 3.9 μM of BASA, a level four times the MIC. Cells were harvested at 0, 15, 30 and 90 min post-treatment and RNA extracted for qRT–PCR analysis to measure the levels of the bioD (the first gene in the biotin biosynthesis operon) and bioY (biotin transporter gene) transcripts (Figure 2). S. aureus treated with 10 nM biotin displayed a 111-fold decrease in bioD expression at 15 min post-treatment and maintained this level of tight repression throughout the 90-minute time course (Figure 2A). The addition of 10 nM of BASA resulted in a 14-fold repression of bioD expression 15 min post-addition (p ≤ 0.0001, Figure 2B). Similarly, this level of repression was maintained throughout the 90-minute time course. Stronger repression of bioD was achieved by increasing the dosage of BASA to 3.9 μM. A 52-fold decrease in bioD transcripts was measured at 15 min, and bioD expression was maintained over the time course with a 32-fold decrease measured at 30 min and 71-fold decrease at 90 min (p ≤ 0.0001, Figure 2C). The bioY transcript exhibited less dramatic repression than bioD for both supplemented biotin and BASA. With the addition of 10 nM biotin, bioY was only repressed two-fold at 15 min (p < 0.01), five-fold at 30 min (p < 0.01) and six-fold at 90 min (p < 0.01; Figure 2D). S. aureus treated with 10 nM of BASA exhibited a similar response, with bioY expression reduced by 1.5-fold at 15 min (p < 0.01), two-fold at 30 min (p < 0.01) and six-fold at 90 min (p ≤ 0.0001, Figure 2E). Together, these three lines of evidence demonstrated that BASA binds directly to the BPL target and functions as a co-repressor, promoting transcriptional repression comparable to that of the natural co-repressor biotinyl-5′-AMP.
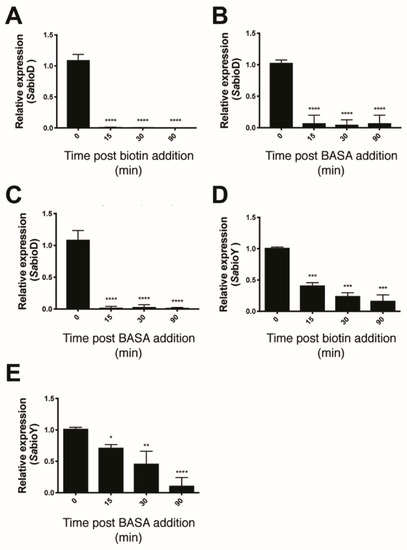
Figure 2.
BASA represses expression of bioD and bioY. Quantitative RT–PCR was employed to measure the repression of bioD following treatment with (A) 10 nM biotin, (B) 10 nM of BASA (0.01× minimal inhibitory concentration (MIC)) and (C) 3.9 μM (4× MIC) of BASA, and the repression of bioY in response to (D) 10 nM biotin and (E) 10 nM of BASA. Transcript levels were normalized against an internal control (16s rRNA) and calculated relative to time = 0. Error bars represent SEM from 3 independent biological replicates (n = 3), *p < 0.05, **p < 0.01, ***p > 0.001, ****p < 0.0001.
2.3. Advanced Resistance Studies Identify Two Resistance Mechanisms
The ability of S. aureus to develop resistance to BASA was investigated using both spontaneous resistance and serial passage approaches. The S. aureus NCTC 8325 strain was selected due to the availability of a sequenced reference genome for comparative sequence analysis. To measure the rate at which spontaneous resistance develops, 109 S. aureus cells were plated onto media containing four times the MIC of BASA. No resistant mutants grew after 24 hours which equates to a calculated resistance frequency of <10−9 at this inhibitor concentration. BASA resistant S. aureus strains were subsequently generated by the serial passage of seven populations of S. aureus NCTC 8325 in sub-inhibitory concentrations of BASA for 18 days at which time individual colonies were isolated. These were then passaged on non-selective media to verify that the inherited resistance was genetically stable. The sensitivity of the strains to BASA was then established using antimicrobial sensitivity assays (Table 2 and Figure S8A). The isotypic parent strain was highly sensitive to BASA, with an MIC of 0.0625 µg/mL. Interestingly, the most resistant strain, B7, still retained a low MIC of 4 μg/mL (64-fold greater than non-treated control) suggesting complete resistance to this BASA is difficult to readily evolve. The MIC values for B1–B6 were a modest 2–16-fold increase over the parent strain.

Table 2.
Strains used in this study and relevant genetic and phenotypic changes associated with BASA resistance. MICs were determined against BASA. Significance for doubling times was determined by ANOVA analysis using Dunnett’s multiple comparison test. (***p < 0.001). A full genotypic analysis of strains B1–B7 is shown in Supplementary Table S2.
To determine if there was a fitness cost associated with these mutations, in vitro growth curves for the seven strains were analyzed and doubling times calculated (Table 2, Figure S8B) [37]. All strains had growth kinetics comparable to the parental strain NCTC8325 (52 min), except B3, which exhibited significantly inhibited growth (doubling time, 193 min, p < 0.0001 B3 vs. NCTC8325) preventing it from reaching half the maximum optical density of the other strains. Collectively, these data suggest that in vitro resistance to the BPL inhibitor is possible without significant associated fitness costs. To test if the resistance mechanisms identified were specific for BASA, all seven strains were subjected to antibacterial susceptibility assays against a panel of diverse antibiotics that included two known BPL inhibitors, namely biotinol-5′-AMP [32] and BPL223 (compound 4c in [30]). The evolved resistance mechanisms appeared specific to the BPL inhibitors as there was no change in susceptibility to β-lactams amoxicillin and methicillin (Table S1). The only exception was observed for strain B3 which exhibited a modest two- to four-fold decrease in susceptibility to both β-lactams. Conversely, all strains exhibited decreased susceptibility to both BPL inhibitors. Whereas parent strain NCTC8325 exhibited an MIC of 1 μg/mL against biotinol-5′-AMP, all strains were resistant to this compound (MIC >64 μg/mL), except B5, which exhibited an MIC of 8 μg/mL. Similarly, NCTC8325 also exhibited an MIC of 8 μg/mL for BPL223, however B2, B4, B6 and B7 were not susceptible again (MIC >64 μg/mL). B1 (MIC = 64 μg/mL) and B3 (MIC = 16 μg/mL) exhibited a modest decrease in susceptibility, and B5 (MIC = 8 μg/mL) showed no change.
Whole-genome sequencing was subsequently employed to characterize mutations arising in the seven strains generated from advanced resistance studies, alongside a non-treated NCTC 8325 parent strain. DNA sequencing was performed with 150 bp paired-end reads on the Illumina Miseq platform, with each strain having 434,000–809,000 reads (between 22 and 40 times coverage of genome) sufficient for detection of single nucleotide variants [38]. The parent NCTC 8325 strain was compared to the reference sequence and differences observed between the parental strain and reference genome were accounted for in a new reference sequence. The resistant strains were then compared to this new parental reference genome. Between two and four mutations were identified in each strain (phenotypes summarized in Table 2, full characterization of genotypes in Table S2). Collectively, this encompassed 15 non-synonymous mutations in the coding regions of 11 independent genes. The majority of these were single nucleotide variants. However, two episodes of insertions that mediated frame shifts in coding regions were observed as were five deletion events, ranging from three base pairs (i.e., a single codon) to larger deletions (194 to 200 bp). Four strains also contained 94 to 139 bp deletions between open reading frames, SAOUHSC_01476 and SAOUHSC_01477, approximately 1 kb upstream of the putative operon containing BPL encoding gene birA (ORF SAOUHSC_01473) which is predicted to span SAOUHSC_01476 to SAOUHSC_01471 [39].
The only gene with non-synonymous mutations in five independent strains was pyc which encodes the biotin-dependent enzyme PC. Strains B1, B2, B4, B6 and B7 all contained mutations that led to premature truncation of the PC protein. These strains also exhibited the greatest (4–64 fold) increases in MIC (Table 2). Streptavidin blot analysis performed on whole cell lysates to detect biotinylated protein confirmed the loss of biotinylated PC (Figure S9). In addition to the deletion of pyc, the most resistant strain B7 also contained the missense mutation D200E in the birA gene that encodes BPL. This was accompanied by a nonsense mutation in the fmtA gene, involved in removing D-alanine from teichoic acids in the cell wall [40], and an intergenic deletion between SAOUHSC_01476 and SAOUHSC_01477 (Table S2). Other mutations present in strains containing the pyc deletion included the following: Strain B1 contained a frame-shift mutation, leading to premature truncation of the potassium transport protein trkA and a missense mutation, E20K, in a putative sensor histidine kinase. Strains B2, B4 and B6 contained deletions in the same intergenic region as B7, and strain B6 contained two additional missense mutations, P626L in the RNA polymerase β-subunit and R296H in a glycosyltransferase-A-like gene. It is worth noting that isolates from B6 and B7 were suspected to be derived from the same intermediate strain, possibly due to cross-contamination, as they contained identical mutations in two separate locations in the genome. Strains B3 and B5 that exhibited the lowest (two- to four-fold) reduced susceptibility to BASA were devoid of mutations in the pyc gene. Strain B3 contained two mutations; a missense mutation in the ci-di-AMP phosphodiesterase gdpP [41] that converted histidine 442 in the presumptive catalytic triad in the DHH domain to proline and a 3bp deletion which led to the loss of glutamine at position 213 in the disulfide stress effector yjbH [42]. Strain B5 contained a 111 bp deletion in a gene predicted as a greA transcription factor elongation homolog and a missense mutation P15L in a putative bacterial ferredoxin, SAOUHSC_01504. Interestingly, none of the strains harbored mutations in biotin-dependent ACC. Together these data suggest S. aureus can evolve at least two possible resistance mechanisms in vitro to overcome BASA: (1) deletion of pyc and (2) mutation of the BPL target.
2.4. Resistance Mechanism 1: Deletion of pyc
Of all the mutations identified in the advanced resistance study, four were predicted to cause a loss of function due to protein truncation (pyc, trkA, fmtA, greA), whilst two encoded for missense mutations (gdpP, yjbH). To determine if the loss of function in any of these gene products was sufficient for reduced susceptibility, strains containing transposon disrupted target genes were obtained from the Nebraska transposon mutant library [43]. As all mutants in the library were generated in S. aureus JE2 (CA-MRSA, USA300) this served as the isotypical control strain. Of the 10 discrete genes harboring mutations identified in B1–B7, five transposons disrupted strains were available for further analysis, namely pyc, trkA, fmtA, gtfA and yjbH. All five strains were initially tested for susceptibility to BASA by monitoring growth for 20 hours in the presence of varying concentrations of the compound. Parental strain JE2 exhibited an MIC of 0.5 μg/mL. Only transposon disruption of Δpyc and ΔyjbH lead to an increased MIC. Both strains exhibited an MIC of 4 µg/mL, the same susceptibility as that observed in the initial advanced resistance studies with isolate B7 (Table 2). To test if the deletions leading to reduced susceptibility also resulted in changed growth kinetics (as had been observed in strain B3 containing the mutation in yjbH), the growth kinetics were explored further for the Δpyc and ΔyjbH strains by monitoring growth over two days in the presence of 0, 0.5, 2 and 4 μg/mL BASA (Figure S10). The strains showed no difference in growth in the untreated control, suggesting that there was no growth defect associated with the mutations (Figure S10A). Consistent with the MIC determination, the parental strain had no growth at concentrations higher than 0.5 μg/mL. Whilst growth of the Δpyc and ΔyjbH strains was still partially inhibited by BASA, even at low concentrations, both strains showed significantly increased growth over the wildtype with 50% and 25% maximal growth relative to untreated control after two days (Figure S10B). The Δpyc strain also grew at 2 μg/mL (Figure S10C) and ΔyjbH at both 2 and 4 μg/mL BASA (Figure S10D). The growth kinetics of the strains under antibiotic pressure were also markedly different. The Δpyc strain exhibited a pronounced lag phase before entering a relatively rapid log phase growth. In contrast, the ΔyjbH strain showed slower steady growth throughout the time course. The fact the Δpyc strain was able to grow at four times the MIC suggested this mutation alone was sufficient for the four-fold increase in resistance observed in the NCTC-8325-derived strains containing this mutation.
2.5. Resistance Mechanism 2: Missense Mutation in the BPL Target
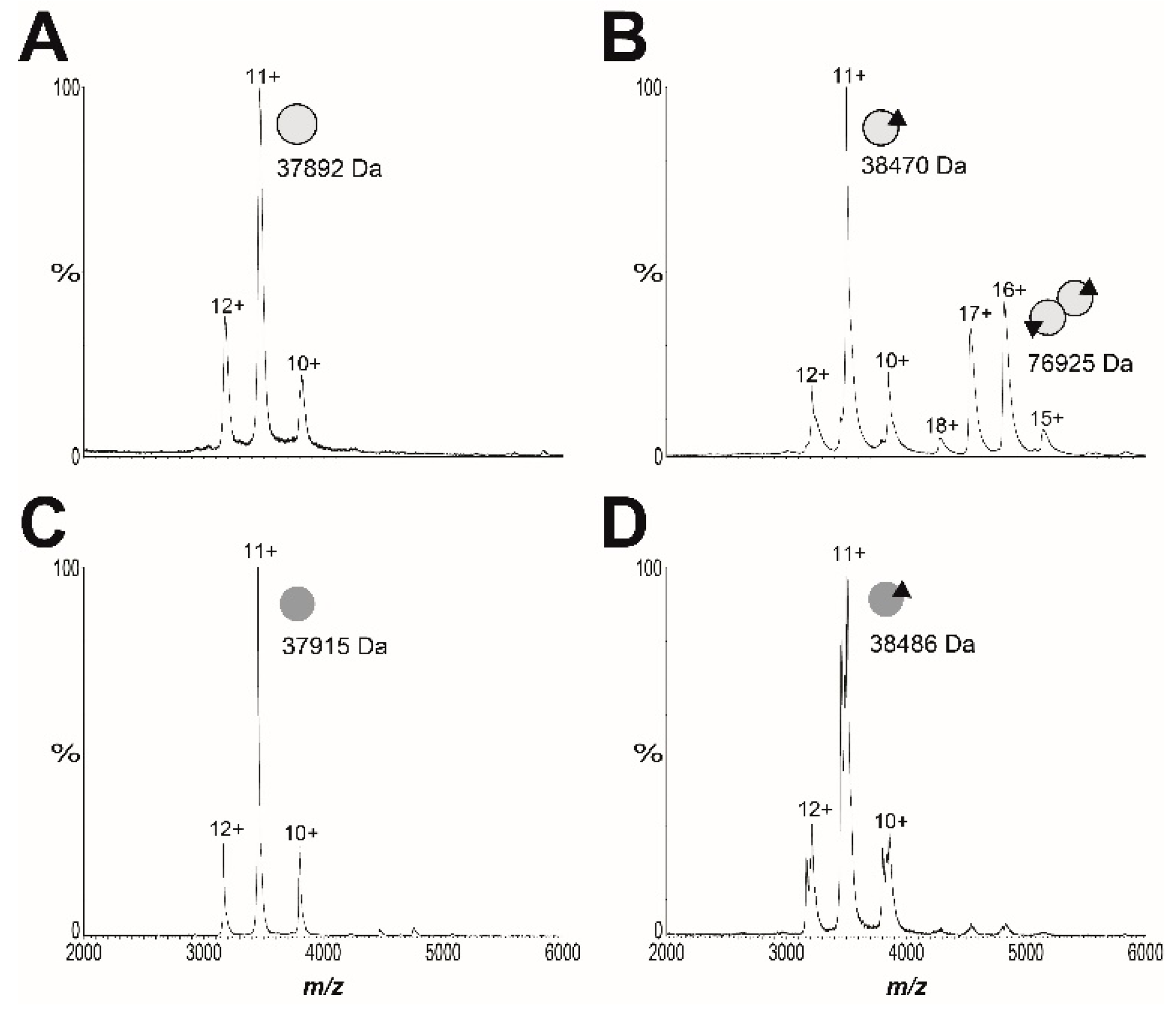
The advanced resistance studies also identified the missense mutation SaBPL-D200E in clone B7. Analysis of the available structural data revealed that this mutation resides in the dimerization interface between the two BPL subunits (Figure S1D). Based on this observation, we subsequently explored the proposal that SaBPL-D200E disrupted dimerization activity. Site-directed mutagenesis was employed to re-engineer the D200E substitution into the protein sequence from S. aureus Mu50 for which structural data has been published [27,44]. Recombinant SaBPL and SaBPL-D200E were purified in their apo state for further characterization. Denatured mass spectrometry (MS) confirmed the desired mutation was present in SaBPL-D200E (measured mass: 37,904 Da; predicted mass: 37,904 Da) and native nano-electrospray ionization–mass spectrometry (nESI–MS) together with a previously described streptavidin blot method both confirmed that the proteins were devoid of any co-purified biotinyl-5′-AMP (Figure 3 and Figure S11). The catalytic properties of wildtype and SaBPL-D200E were then measured using an in vitro protein biotinylation assay. The biochemical data revealed that enzyme activity was not significantly affected by the D200E mutation, with the affinity for biotin increased modestly by two-fold (wildtype SaBPL Km 1.8 ± 0.3 μM, SaBPL-D200E Km 3.8 ± 0.4 μM (p < 0.05)). Likewise, the D200E mutation had a minimal effect upon the inhibitory activity of BASA in the enzyme assay (Ki wildtype SaBPL 4.8 ± 2.1 nM; Ki SaBPL-D200E = 10.9 ± 3.5 nM; p = 0.18). Native nESI–MS was then utilized to investigate the oligomeric state of SaBPL-D200E (Figure 3, Figure S12 and Table S3). This technique has previously been employed to study co-repressor induced dimerization of wildtype SaBPL [25]. Whilst SaBPL underwent a well-characterized monomer-to-dimer transition induced by biotinyl-5′-AMP (Figure 3A,B), no dimerization was observed for SaBPL-D200E in both its apo and holo forms across a range of concentrations (Figure 3C,D). This point mutation, thus, prevents protein dimerization. The native nESI–MS provided sufficient mass resolution to confirm the presence of bound biotinyl-5′-AMP, indicating that lack of protein dimerization is not due to the inability to catalyze the formation of co-repressor.
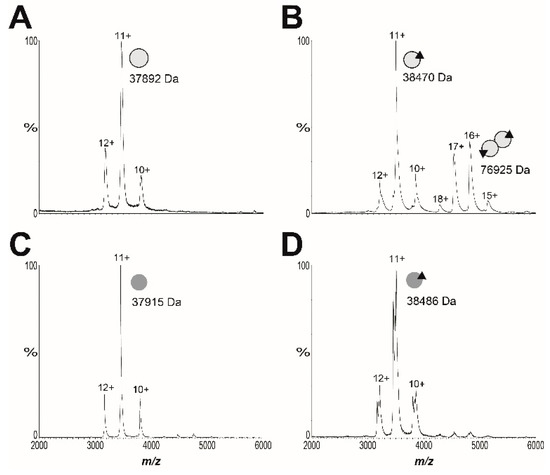
Figure 3.
Native native nano-electrospray ionization–mass spectrometry (nESI–MS) reveals S. aureus BPL (SaBPL) D200E is monomeric in the presence and absence of ligands. The oligomeric states of 10 μM of (A) wildtype apo SaBPL and (B) holo SaBPL were compared with 90 μM of (C) apo SaBPL-D200E and (D) holo SaBPLD200E. Monomeric protein is diagrammatically represented by a single grey sphere, homodimer by conjoined spheres and the presence of biotinyl-5′-AMP by a black triangle. Spectra showing SaBPL D200E at varying protein concentrations between 1.4 and 90 μM are presented in Figure S12 and masses extracted from the spectra are in Table S3.
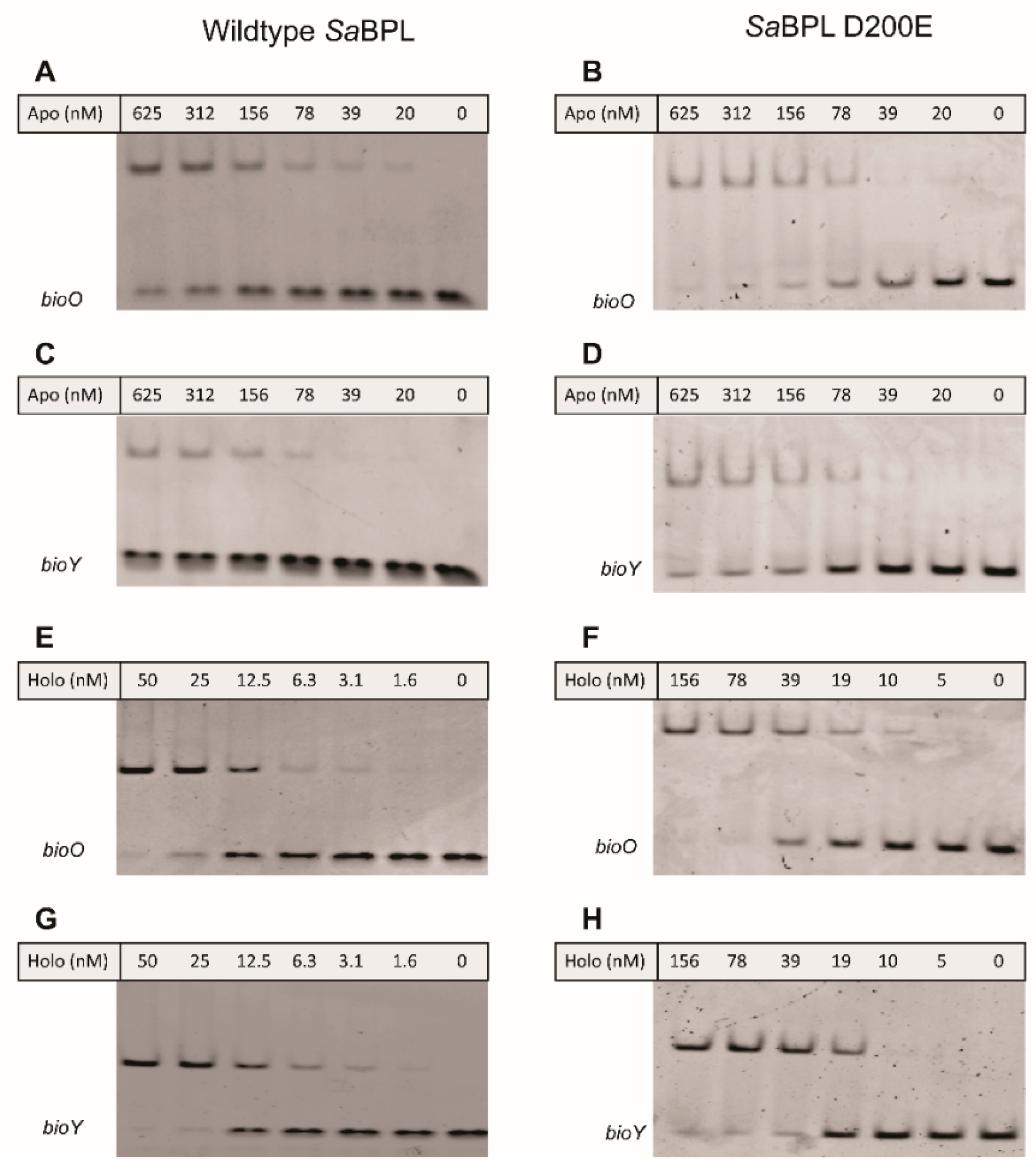
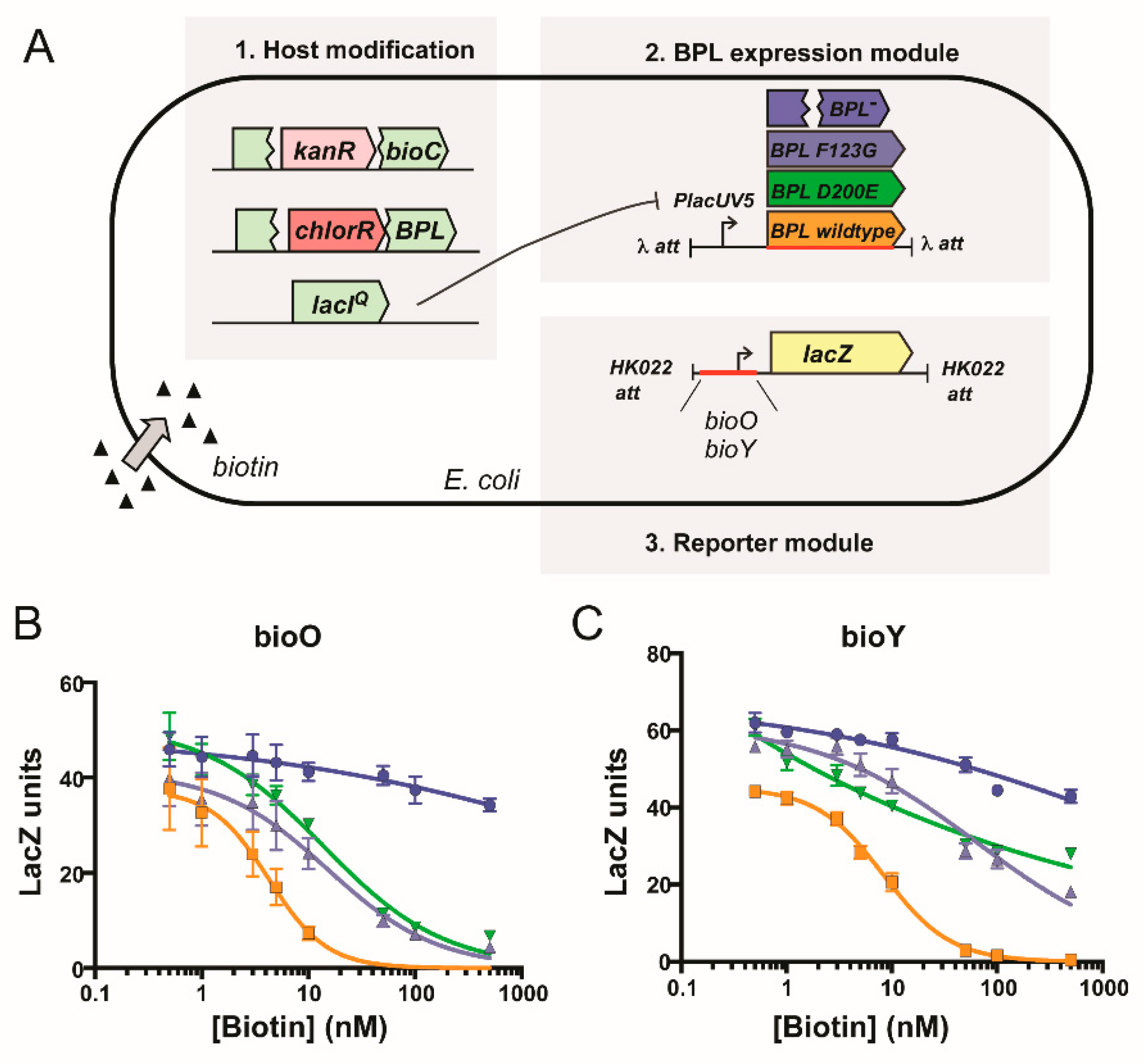
The effect of impaired homodimerization ability on DNA binding and transcriptional repression was then assessed using a combination of in vitro EMSA analysis and a previously established in vivo transcriptional reporter system in E. coli [23]. Both wildtype and SaBPL-D200E demonstrated weak DNA binding activity in their apo states in the EMSA, with unbound bioO and bioY probes observed even at the highest protein concentration tested (i.e., 625 nM; Figure 4A–D). EMSA analysis was then repeated in the presence of biotin and MgATP to measure DNA binding in response to co-repressor biotinyl-5′-AMP. Unlike the apo protein, 50 nM of wildtype SaBPL was sufficient to bind all of the oligonucleotide probes consistent with the homodimer having a higher affinity for the DNA (Figure 4E,G) [25,26]. Conversely, the D200E mutation had a significant impact on DNA binding. Approximately three-fold higher concentrations of holo SaBPL-D200E were required for the same response with bioO (156 nM; Figure 4F) and the mutant failed to achieve complete binding to the bioY oligonucleotide even at the highest protein concentration tested (156 nM; Figure 4G,H). These data reveal that the D200E mutation impaired SaBPL dimerization and reduced binding to DNA.
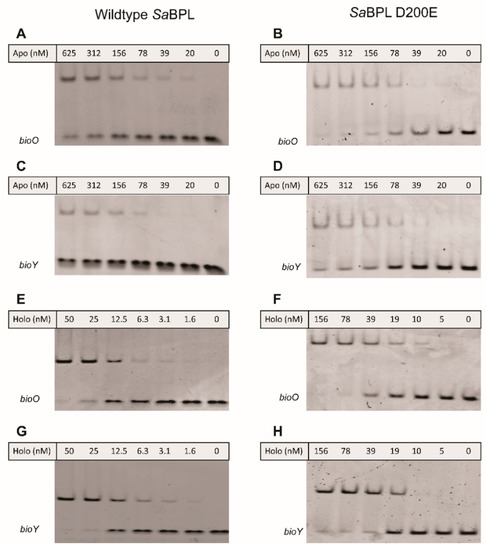
Figure 4.
SaBPL D200E has reduced DNA binding activity compared to wildtype SaBPL. Electrophoretic mobility shift assays (EMSA) analysis was performed to measure the binding of (A) apo SaBPL wildtype binding to bioO, (B) apo SaBPL D200E binding to bioO, (C) apo SaBPL wildtype binding to bioY and (D) apo SaBPL D200E binding to bioY, (E) holo SaBPL wildtype binding to bioO, (F) holo SaBPL D200E binding to bioO, (G) holo SaBPL wildtype binding to bioY and (H) holo SaBPL D200E binding to bioY.
Finally, the biological consequence of the D200E mutation upon repressor activity was determined using an in vivo reporter assay. This in vivo reporter assay was previously developed in a biotin auxotrophic E. coli strain that had the DNA binding activity of its endogenous BPL removed [45]. The assay was adapted such that the transcriptional repressor activities of both wildtype SaBPL and SaBPL-D200E could be investigated against the S. aureus bioO and bioY promoters (Figure 5A). Expression of a β-galactosidase reporter gene was measured in the presence of varying biotin concentrations, and a repression constant determined (KR biotin i.e., the biotin concentration required for a 50% reduction of β-galactosidase activity). A previously characterized SaBPL-F123G mutant that similarly disrupts dimerization activity was also included for comparison [25,26]. Modest three-fold increases (p < 0.05) in biotin concentration were required for repression of the bioO promoter by either of the mutant proteins (D200E KR biotin = 13.9 ± 3.4 nM; F123G KR biotin = 15.3 ± 3.5 nM) compared to wildtype SaBPL (KR biotin = 4.3 ± 1.9 nM; Figure 5B). Greater differences were observed for the bioY promoter, where wildtype BPL (KR biotin = 8.2 ± 0.7 nM) repressed at vastly lower biotin concentrations than either mutant (KR biotin >500 nM), neither of which reached basal levels of expression even at 500 nM biotin (Figure 5C). These data were consistent with the reduced affinity of SaBPL-D200E for the bioY oligonucleotide measured in the EMSA analysis. Together these data revealed that the dimerization-impaired mutant, SaBPL D200E, reduced affinity for DNA binding in vitro and transcriptional repressor activity in vivo. The data also suggest impaired repression of the genes required for biotin synthesis as a resistance mechanism to combat BASA in S. aureus.
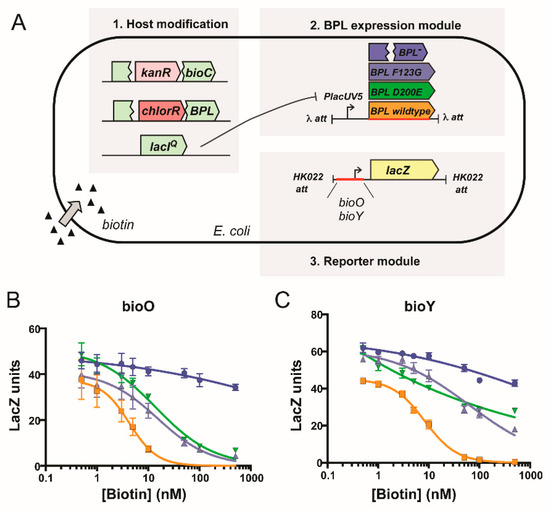
Figure 5.
Biotin-regulated repressor activity of SaBPL and mutants. (A) In vivo expression assays were performed in an E. coli reporter strain containing chromosomally integrated repressor and promoter constructs [23]. LacZ units were calculated by subtracting the value generated by the control strain containing no integrated promoter construct (LacZ unit ≤10). Expression of the β-galactosidse reporter gene, under the control of promoters (B) bioO or (C) bioY, was measured in media containing varying concentrations of biotin. Transcription factors SaBPL (orange), SaBPL D200E (green) and SaBPL F123G (black) were analyzed alongside a control strain that harbored no repressor protein (blue). Error bars represent SEM from independent biological replicates (n = 6). The amount of biotin to reach half-maximum repression (KR biotin) was calculated from the curves using GraphPad Prism. The KR biotin for wildtype SaBPL repressing bioO and bioY was 4.3 ± 1.9 nM and 8.2 ± 0.7 nM, respectively. The KR biotin for SaBPL-D200E repressing bioO and bioY was 13.9 ± 3.4 nM and >500 nM, respectively. The KR biotin for SaBPL-F123G repressing bioO and bioY was 15.3 ± 3.5 nM and >500 nM.
3. Discussion
BPL has been proposed as a drug target for the development of novel anti-infective therapeutics against antibiotic-resistant bacteria. In this report, we first investigated the mechanism of action of the potent BPL inhibitor BASA as an exemplar for this new class of antibacterial. Multiple lines of evidence were consistent with BASA acting directly through the BPL target. It has previously been proposed that anti-BPL compounds exert their antibacterial activity through inhibition of the enzymatic function of BPL and consequent reduced biotinylation and activity of the two biotin-dependent enzymes present in S. aureus, namely PC and ACC [7]. The data presented here provide evidence that anti-BPL compounds can also act as co-repressors to repress biotin synthesis and transport. In support, transcriptomic studies revealed that BASA induced repression of both bioY (biotin transport) and bioD (biotin synthesis) in S. aureus with similar potency and kinetics as the natural ligand biotin. These data are consistent with the hypothesis that the pharmacological inhibition of BPL induces biotin starvation within the bacteria by compromising both the supply and utilization of biotin. The combined effect of these mechanisms contributes to the antibacterial efficacy of BASA. Biotin deprivation is detrimental to the bacteria as neither PC nor ACC can be post-translationally activated by BPL without bioavailable biotin. In response, the bacteria attempt to increase biotin supply through either de novo synthesis (using bioO) or import of exogenous sources (using bioY). The importance of the biotin starvation state in the activity of anti-BPL compounds has not been previously reported.
Having established a greater understanding of the mechanism of action of BASA, we next addressed possible mechanisms of resistance. The incidence rate for the generation of spontaneous resistance was low, with no resistant colonies growing after 24 hours, resulting in a resistance rate of less than 1 × 10−9. Similar low spontaneous resistance rates have been reported for another anti-BPL compound active against M. tuberculosis (less than 1.4 × 10−10) [33]. This highlights BPL as an important antibacterial target with low potential for resistance to develop. Our advanced resistance studies involving serial passaging of S. aureus in sub-inhibitory concentrations of BASA for 18 days and subsequent characterization of the resistant strains resulted in the identification of two discrete resistance mechanisms; (1) deletion of PC and (2) a missense mutation in the BPL target. Loss of PC was the most common resistance mechanism observed in laboratory testing. A premature truncation of PC was present in five of the seven strains evolved to be resistant to BASA. As the biotin-accepting domain of PC that is essential for catalysis is contained within the C-terminal region of the enzyme, these truncated proteins were unable to be biotinylated and, hence, inactive. The transposon disrupted Δpyc strain in the genetic background of MRSA strain USA300 JE2 confirmed that PC deletion conferred resistance independently of the other mutations and was a viable resistance mechanism in another genetic background, exhibiting a four-fold decrease in MIC over the isotypic wildtype parent strain. Notable was the observation that no mutations were observed in ACC. The current report suggests that bioavailable biotin is selectively utilized such that ACC is preferentially biotinylated over PC. This was consistent with previous genetic knockout studies which have demonstrated ACC, but not PC, is essential for growth in laboratory cultures [9,10]. Removal of PC from the cell, therefore, overcomes biotin starvation by removing the only other competing protein that requires biotin. However, this is unlikely to be a mechanism readily available to S. aureus during infection in vivo, as loss of PC and other tricarboxylic acid cycle genes have been shown to reduce virulence in murine and nematode models of infection [20,21].
The advanced resistance studies also identified a strain with a missense mutation in BPL. Biochemical characterization of this mutation revealed disrupted homodimerization, consequently leading to weakened DNA binding and reduced repressor activities. This residue, D200, is located on a loop within the dimer interface that becomes ordered upon ligand binding. The side chain of Asp200 forms hydrogen bonding interactions with Arg122 and hydrophobic interactions with Phe123 on the opposing subunit. By inhibiting dimer formation, D200E disrupted the DNA binding activity of SaBPL and repression of SaBPL target genes. The mechanism of resistance findings supports the proposal that the co-repressor function of BASA contributes to antibacterial efficacy. We propose that disruption of the repressor activity provides an opportunity for cells to resist the action of BASA by increasing the availability of intracellular biotin. The D200E mutant with reduced repression of bioO and bioY will result in increased de novo biotin synthesis and biotin transport, respectively. Increased accumulation of intracellular biotin could compete against BASA for binding within the active site of SaBPL. In support, we observed that increasing exogenous biotin concentrations decreased the antibacterial activity of BASA. It has also been shown that S. aureus can stimulate import and sequester biotin inside the cell in response to increased exogenous biotin concentrations [25]. Similar resistance mechanisms through the deregulation of target genes have also been observed for other antibiotic targets in the literature. The antibiotic pyrithiamine binds a riboswitch protein and causes the repression of thiamine biosynthesis genes, leading to thiamine starvation. However, resistance to pyrithiamine is generated through mutations in the binding site of the riboswitch protein such that the antibiotic can no longer bind and cause gene repression, leading to a de-repression of thiamine synthesis [46,47].
The mechanism of entry of anti-BPL inhibitors into S. aureus has not been established. One possibility is that their common biotinyl moiety facilitates import into the bacteria through BioY, the biotin-specific subunit of the multivitamin transport system. Hence, targeting BPL with an inhibitor that also acts as a co-repressor would reduce BioY expression and limit entry of the chemotherapeutic into the bacteria which would be an undesirable situation for an antibacterial. However, several lines of unpublished data suggest that the BASA enters the bacteria through an alternative mechanism, likely via membrane-associated aquaporins. No change in the antibacterial potency of BASA was observed when tested against a bioY knockout S. aureus strain (data not shown). Similarly, increasing the concentration of exogenous biotin in the growth media did not impede the internalization of a fluorescent BPL inhibitor containing the biotinyl group [30]. Together these observations suggest that BASA is imported into bacteria independently of BioY. Hence, the altered expression of BioY noted here is unlikely to impact the accumulation of BASA inside the bacteria and affect its efficacy.
The oligomeric state of the SaBPL D200E variant reported here supports previous in vitro studies on both S. aureus and E. coli BPLs, where modification of residues in the dimerization interface affect repressor activity. The mutations, F123G in SaBPL [25,26] and D197Y in E. coli (equivalent of D200 in S. aureus) [48,49] demonstrate impaired homodimerization and loss of repressor activity [25,48,49]. However, it should be noted that the mutants used in those studies were engineered using site-directed mutagenesis based on knowledge of the crystal structure. The current report provides the first example where this phenotype has been evolved in vivo. The interaction between Phe123 and Asp200 appears particularly important in SaBPL dimerization. Asp200 from one subunit forms hydrophobic interactions with Phe123 on the opposing subunit to facilitate dimerization. Mutation of these two amino acids result in enzymes with similar affinity for biotin, and only two-fold weaker than the wildtype enzyme [26]. Therefore, the ligand binding and catalytic properties of the mutant were not impaired. Conversely, homodimerization was significantly disrupted for the mutants, thereby perturbing DNA binding and repressor activities. It is proposed that these mutant strains require a much higher concentration of biotin before SaBPL can be induced to be a fully active transcriptional repressor. Additionally, crystal structures of SaBPL in complex with BASA reveal that the inhibitor becomes buried deep in the homodimer and is shielded away from the solvent. Consequently, it is difficult for the inhibitor to readily diffuse out from the binding site when SaBPL is in its dimeric state. This results in a stable inhibitor-protein complex that ensures the compound experiences high residency time with the BPL target, thus contributing to the overall efficacy of the antibacterial.
4. Materials and Methods
4.1. General Bacterial Culture and Molecular Biology Reagents
All chemicals and reagents were purchased as analytical grade or higher. All molecular biology enzymes (restriction enzymes and DNA polymerase) and buffers were acquired from New England Biolabs. Oligonucleotides were purchased from Geneworks Ptd Ltd. Details of oligonucleotides used for cloning (Table S4), RT–PCR (Table S5) and EMSA analysis (Table S6), bacterial strains (Tables S7 and S8) and plasmids (Table S9) used in the study are supplied in the supplementary information. Unless otherwise stated, all bacterial strains were purchased from the American Tissue Culture Collection. The following strains were provided by the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for distribution by BEI Resources (BEIResources.org), NIAID, NIH: Staphylococcus aureus subsp. aureus, Strain USA300 (JE2), Transposon Mutants NE754, NE896, NE1022, NE788 and NE792. Cation-adjusted Mueller Hinton II (Becton, Dickinson and Company, MD, USA) was used to propagate S. aureus. Antibacterial agents were obtained from Sigma-Aldrich.
4.2. Quantification of Gene Expression Using qRT–PCR
The qRT–PCR protocols used to measure the expression of bioD and bioY were completed as outlined previously [23]. However, the sub-cultured S. aureus NCTC 8325 cultures were treated with either 10 nM biotin or BASA at 10 nM or 3.9 μM (i.e., 4× MIC) once reaching mid-log phase, prior to cell lysis, RNA extraction and qRT–PCR analysis.
4.3. Electrophoretic Mobility Shift Assay (EMSA)
The EMSA protocol was adapted from methods previously published [23,25,50]. For analysis of BASA, 100 μM of the compound was added to the EMSA binding buffer. As BASA was dissolved in 100% DMSO, the final DMSO concentration in the binding buffer was adjusted to 2.5% (v/v). Control reactions were prepared by adding 100 μM biotin and 2.5% (v/v) DMSO to the binding reaction.
4.4. Spontaneous Resistance Rate
A culture containing 109 CFU of S. aureus NCTC 8325 was plated onto cation-adjusted Mueller Hinton broth (CAMHB) Agar containing 4× MIC of BASA. Nine technical replicate cultures were plated from the same starting colony to control for priming mutations in the initial colony. Plates were assessed for growth after 24 h at 37 °C. As the determination of true resistance rates using fluctuation analysis requires some colonies to grow [51,52,53], a resistance frequency was instead calculated.
4.5. Generation of Resistant Mutants by Serial Passaging
Mutants were selected by serial passage in CAMHB in 96-well plates as previously described [54]. Briefly, a 2-fold dilution series of BASA was plated across 7 wells. Initial concentrations ranged from 1 μg/mL to 0.016 μg/mL, with these concentrations increasing (up to 64 μg/mL) as the MIC increased. Approximately 104 CFU of S. aureus NCTC 8325 at mid-log phase were added to each well and allowed to grow at 37 °C for 20 h. The OD at 620 nm was measured and the highest concentration of BASA that permitted growth (OD620 > 0.1 used as the threshold) was diluted 1000-fold to inoculate a new concentration series of BASA. This process was repeated for 18 days, and on the final day, single isolates were recovered by streaking out the strains from the highest concentrations of BASA that still allowed growth. Genomic DNA was isolated with the Wizard Genomic DNA purification kit (Promega, Wisconsin, USA) according to the manufacturer’s guidelines. Bacteria were treated with 10 μg of lysostaphin (Sigma-Aldrich Inc, MO, USA) and 1 mg/mL lysozyme prior to lysis. These selected strains were also propagated for 6 days on CAMHB agar in the absence of antibiotic selection, before re-testing these strains for antibacterial susceptibility to assess the stability of the generated resistance.
4.6. Genomic DNA Purification and Whole-Genome Sequencing
Genomic DNA was isolated with the Wizard Genomic DNA purification kit (Promega, Wisconsin, USA) according to the manufacturers’ guidelines. Bacteria were treated with 10 μg of lysostaphin (Sigma-Aldrich Inc, MO, USA) and 1 mg/mL lysozyme prior to lysis. DNA quantities ranged from 15–60 ng/μL. Genomic DNA from parental strain NCTC 8325 and isolates B1–B7 was submitted to ACRF (Cancer Genomics Facility, Adelaide, South Australia) for library preparation and sequencing. Whole-genome sequencing was performed with 150 bp paired-end reads on Illumina Miseq. The number of sequences for each strain ranged from 434,000–809,000 (between 22 and 40× coverage of genome), 32% GC content. Sequence reads are stored on the Biosample with accession numbers B1: SAMN14388873, B2: SAMN14388874, B3: SAMN14388875, B4: SAMN14388876, B5: SAMN14388877, B6: SAMN14388878, B7: SAMN14388879, S. aureus subsp. aureus NCTC 8325 SAMN14388880.
4.7. Bioinformatic Analysis
The Fastq reads were analyzed using the Breseq analysis pipeline software [55]. The parental NCTC 8325 strain that had not undergone antibacterial selection was mapped to the reference genome (NC_007795.1) and the differences were applied to generate a new reference sequence using gdtools. The de-multiplexed reads from isolates B1–B7 were then mapped to this new reference and mutations identified. More than 98% of reads from each isolate were mapped to the reference genome. Manual inspection of the resulting HTML files was used to determine true positives. Amongst those manually excluded from further analysis was a missense mutation in rsbU which is not transcribed in NCTC8325 [56], mislabeling of P626L (as P579L) due to a non-standard start codon and removal of mutations identified in all strains (including the parental NCTC 8325) that did not reach the threshold required for inclusion into the new reference genome.
4.8. Antimicrobial Susceptibility Testing/Cross Resistance
Antimicrobial susceptibility assays were performed using a broth microdilution method as recommended by CLSI (Clinical and Laboratory Standards Institute, Document M07-A8, 2009, Wayne, Pa.). Antibacterial compounds were diluted two-fold in cation-adjusted Mueller Hinton Broth (CAMHB) to a final concentration range of 64–0.06 μg/mL in 3.2% DMSO. To prepare the inoculum, an overnight culture of S. aureus was subcultured 1:1000 into fresh media and grown to mid-log phase. Then, 96-well microtiter plates containing diluted compounds were inoculated with 5 × 104 CFU and incubated at 37 °C for 20–22 h with shaking. Growth of the culture was quantified after shaking for 15 seconds to disrupt any sedimentation by measuring the absorbance at 620 nm or 630 nm using either a Thermo Multiskan Ascent plate reader or Biotek EL808 plate reader, respectively. Assays were performed in triplicate.
4.9. Measurement of the Kinetics of Bacterial Growth
The kinetics of bacterial growth were measured in Mueller Hinton broth devoid of antibiotics. Growth curves were obtained by subculturing an overnight culture 1:1000 and incubating cells at 37 °C to an approximate OD600 of 1 before a further 1:1000 dilution to yield an approximate initial inoculum of 105 cells in 100 μL. Cultures were grown for 24 h in a 96 well plate at 37 °C. Growth of the cultures was monitored at OD630 every 30 min for 24 h with shaking using an EL808™ Absorbance Microplate Reader (BioTek Instruments Inc, Winooski, VT, USA). Growth curves were analyzed by the method described in [37]. In brief, the log of the ODs from OD 0.1–0.3 was plotted against time and log(2)/gradient was determined giving the doubling time. Cultures were plated in technical duplicate and three independent biological replicates were collected.
4.10. Protein Methods
Expression and purification of non-liganded (i.e., apo) SaBPL were performed following previously described protocols [25,44,57]. Purification of apo protein was performed by incubating bacterial lysate with streptavidin agarose at 30 °C for 1 h prior to incubating with purified biotin domain-GST fusion protein from S. aureus [58] (SaPC90) at 30 °C for 1 h, as described previously [25,26]. Protein concentration was determined by Bradford assay. Purified apo protein was confirmed by native nano-electrospray ionization–mass spectrometry (nESI–MS) and by Western blot analysis as described previously [25,26]. An enzyme activity assay to measure the incorporation of biotin onto protein by BPL was performed as previously reported [30].
4.11. Mass Spectrometry
Native nESI–MS of apo SaBPL was carried out on a Waters Synapt G1 HDMS system completed according to [25]. MS parameters were as previously reported to maintain non-covalent interactions and included; capillary voltage, 1.5–1.7 kV; cone voltage, 40–80 V; trap collision energy, 20–50 V; transfer collision energy, 15–20 V; source temperature, 50 °C; extraction cone, 2.0–5.0 V; trap gas flow, 5–8 mL/min; backing pressure, 3.95 mbar. Data analysis was performed in MassLynx V4.1 using manual peak finding.
4.12. Chromosomal Integration and β-Galactosidase Reporter Assay
SaBPL-D200E was cloned into the established integration vector (methods provided in supplementary materials) and incorporated chromosomally into the E. coli reporter strains prepared previously [23,25], using established methods [45]. β-galactosidase assays were performed as previously reported [23,25,45].
5. Conclusions
In this report, we demonstrate that BASA exerts its antibacterial effect through two mechanisms inhibiting the catalytic biotinylation activity of BPL and serving as a co-repressor. BASA treatment results in a biotin starved state within the bacteria. S. aureus strains resistant to BASA were characterized and two discrete resistance mechanisms were discovered. However, the low spontaneous resistance rate would suggest that resistance may be difficult to evolve in vivo and confirms BPL as a bona fide antibacterial drug target. Further chemical development of BPL inhibitors will focus on maintaining both mechanisms of action. New analogous with improved pharmacology are under development to realize valid pre-clinical antibiotic candidates.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6382/9/4/165/s1, Figure S1: SaBPL structures; Figure S2: Cytotoxicity assays; Figure S3: Concentration kill kinetics; Figure S4: Synergism of BASA with various antibiotics; Figure S5: Mechanism of action studies; Figure S6: BASA activity with exogenous biotin; Figure S7: Electromobility shift assays; Figure S8: Growth curves of BASA resistant strains; Figure S9: Blots demonstrating loss of PC; Figure S10: Growth curves of transposon mutants with BASA; Figure S11: Blots demonstrating preparation of apo SaBPL; Figure S12: Native MS spectra; Table S1: MIC values of BASA resistant strains; Table S2: Genotypes of BASA resistant strains; Table S3: Molecular masses measured by native MS; Table S4: Oligonucleotides used for cloning; Table S5: Oligonucleotides used for qPCR; Table S6: Oligonucleotides used for EMSA analysis; Table S7: Bacterial strains used in this study; Table S8: Bacterial strains used in reporter assays; Table S9: Plasmids used in this study.
Author Contributions
Conceptualization, B.A.E., K.E.S., A.D.A., G.W.B. and S.W.P.; data curation, A.J.H., J.S., L.M.S., A.S.P., K.J.L., B.B.-R. and W.T.; formal analysis, B.A.E., K.E.S., T.L.P. and S.W.P; funding acquisition, A.D.A., G.W.B. and S.W.P.; investigation, A.J.H., J.S., L.M.S., A.S.P. and Z.F.; methodology, A.J.H., K.E.S. and T.L.P.; writing—original draft, A.J.H., J.S. and L.M.S.; writing—review and editing, B.A.E., K.E.S., A.D.A., G.W.B. and S.W.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Health and Medical Research Council (NHMRC; APP1068885) and the Australian Research Council (CE140100 003; DP160101450). S.W.P. is supported by the NHMRC (GN1147538). The authors are grateful to the Wallace and Carthew families for their financial support of this work.
Acknowledgments
The authors would like to thank J. Turnidge and J. Bell (University of Adelaide) for their assistance with antibacterial susceptibility assays.
Conflicts of Interest
The authors declare no competing financial interest.
References
- Cooper, M.A.; Shlaes, D. Fix the antibiotics pipeline. Nature. 2011, 472, 32. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.S.; Blaskovich, M.A.T.; Cooper, M.A. Antibiotics in the clinical pipeline at the end of 2015. J. Antibiot. 2016, 70, 3–24. [Google Scholar] [CrossRef] [PubMed]
- Rushton, J.; Stärk, K.D.C.; Ferreira, J.P. Antimicrobial Resistance; Organisation for Economic Co-Operation and Development (OECD): Paris, France, 2014. [Google Scholar]
- World Health Organisation. Global Action Plan on Antimicrobial Resistance; WHO: Geneva, Switzerland, 2015. [Google Scholar]
- Feng, J.; Paparella, A.; Booker, G.W.; Polyak, S.W.; Abell, A.D. Biotin Protein Ligase Is a Target for New Antibacterials. Antibiotics. 2016, 5, 26. [Google Scholar] [CrossRef] [PubMed]
- Paparella, A.; Da Costa, T.P.S.; Yap, M.; Tieu, W.; Wilce, M.; Booker, G.; Abell, A.; Polyak, S.W. Structure Guided Design of Biotin Protein Ligase Inhibitors for Antibiotic Discovery. Curr. Top. Med. Chem. 2013, 14, 4–20. [Google Scholar] [CrossRef]
- Lee, K.J.; Tieu, W.; Blanco-Rodriguez, B.; Paparella, A.S.; Yu, J.; Hayes, A.J.; Feng, J.; Marshall, A.C.; Noll, B.; Milne, R.; et al. Sulfonamide-Based Inhibitors of Biotin Protein Ligase as New Antibiotic Leads. ACS Chem. Boil. 2019, 14, 1990–1997. [Google Scholar] [CrossRef]
- Sternicki, L.; Wegener, K.L.; Bruning, J.B.; Booker, G.W.; Polyak, S.W. Mechanisms Governing Precise Protein Biotinylation. Trends Biochem. Sci. 2017, 42, 383–394. [Google Scholar] [CrossRef]
- Chaudhuri, R.; Allen, A.; Owen, P.J.; Shalom, G.; Stone, K.; Harrison, M.; A Burgis, T.; Lockyer, M.; Garcia-Lara, J.; Foster, S.J.; et al. Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH). BMC Genom. 2009, 10, 291. [Google Scholar] [CrossRef]
- Forsyth, R.A.; Haselbeck, R.J.; Ohlsen, K.L.; Yamamoto, R.T.; Xu, H.; Trawick, J.D.; Wall, D.; Wang, L.; Brown-Driver, V.; Froelich, J.M.; et al. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 2002, 43, 1387–1400. [Google Scholar] [CrossRef]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2006, 6, 29–40. [Google Scholar] [CrossRef]
- Tommasi, R.; Brown, D.G.; Walkup, G.K.; Manchester, J.I.; Miller, A.A. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 2015, 14, 529–542. [Google Scholar] [CrossRef]
- Polyak, S.W.; Abell, A.D.; Wilce, M.C.J.; Zhang, L.; Booker, G.W. Structure, function and selective inhibition of bacterial acetyl-coa carboxylase. Appl. Microbiol. Biotechnol. 2011, 93, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Rock, C. Bacterial fatty acid metabolism in modern antibiotic discovery. Biochim. et Biophys. Acta (BBA) - Mol. Cell Boil. Lipids 2016, 1862, 1300–1309. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, T.P.S.; Tieu, W.; Yap, M.Y.; Pendini, N.R.; Polyak, S.W.; Pedersen, D.S.; Morona, R.; Turnidge, J.D.; Wallace, J.C.; Wilce, M.C.J.; et al. Selective inhibition of Biotin Protein Ligase from Staphylococcus aureus*. J. Boil. Chem. 2012, 287, 17823–17832. [Google Scholar] [CrossRef] [PubMed]
- Duckworth, B.P.; Geders, T.; Tiwari, D.; Boshoff, H.I.; Sibbald, P.A.; Barry, C.E.; Schnappinger, D.; Finzel, B.; Aldrich, C.C. Bisubstrate Adenylation Inhibitors of Biotin Protein Ligase from Mycobacterium tuberculosis. Chem. Boil. 2011, 18, 1432–1441. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Tiwari, D.; Wilson, D.J.; Seiler, C.L.; Schnappinger, D.; Aldrich, C.C. Bisubstrate Inhibitors of Biotin Protein Ligase in Mycobacterium tuberculosis Resistant to Cyclonucleoside Formation. ACS Med. Chem. Lett. 2013, 4, 1213–1217. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, T.P.S.; Tieu, W.; Yap, M.Y.; Zvarec, O.; Bell, J.M.; Turnidge, J.D.; Wallace, J.C.; Booker, G.W.; Wilce, J.A.; Abell, A.D.; et al. Biotin Analogues with Antibacterial Activity Are Potent Inhibitors of Biotin Protein Ligase. ACS Med. Chem. Lett. 2012, 3, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Paparella, A.; Tieu, W.; Heim, D.; Clark, S.; Hayes, A.J.; Booker, G.W.; Polyak, S.W.; Abell, A.D. New Series of BPL Inhibitors To Probe the Ribose-Binding Pocket of Staphylococcus aureus Biotin Protein Ligase. ACS Med. Chem. Lett. 2016, 7, 1068–1072. [Google Scholar] [CrossRef]
- Benton, B.M.; Zhang, J.P.; Bond, S.; Pope, C.; Christian, T.; Lee, L.; Winterberg, K.M.; Schmid, M.B.; Buysse, J.M. Large-Scale Identification of Genes Required for Full Virulence of Staphylococcus aureus. J. Bacteriol. 2004, 186, 8478–8489. [Google Scholar] [CrossRef]
- Bae, T.; Banger, A.K.; Wallace, A.; Glass, E.M.; Åslund, F.; Schneewind, O.; Missiakas, M.M. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. USA 2004, 101, 12312–12317. [Google Scholar] [CrossRef]
- Rodionov, D.A.; Mironov, A.A.; Gelfand, M. Conservation of the Biotin Regulon and the BirA Regulatory Signal in Eubacteria and Archaea. Genome Res. 2002, 12, 1507–1516. [Google Scholar] [CrossRef]
- Satiaputra, J.; Eijkelkamp, B.; McDevitt, C.A.; Shearwin, K.; Booker, G.W.; Polyak, S.W. Biotin-mediated growth and gene expression in Staphylococcus aureus is highly responsive to environmental biotin. Appl. Microbiol. Biotechnol. 2018, 102, 3793–3803. [Google Scholar] [CrossRef] [PubMed]
- Satiaputra, J.; Shearwin, K.E.; Booker, G.; Polyak, S.W. Mechanisms of biotin-regulated gene expression in microbes. Synth. Syst. Biotechnol. 2016, 1, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Satiaputra, J.; Sternicki, L.M.; Hayes, A.J.; Pukala, T.L.; Booker, G.W.; Shearwin, K.; Polyak, S.W. Native mass spectrometry identifies an alternative DNA-binding pathway for BirA from Staphylococcus aureus. Sci. Rep. 2019, 9, 2767. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, T.P.S.; Yap, M.Y.; Perugini, M.; Wallace, J.C.; Abell, A.D.; Wilce, M.C.J.; Polyak, S.W.; Booker, G.W. Dual roles of F123 in protein homodimerization and inhibitor binding to biotin protein ligase from Staphylococcus aureus. Mol. Microbiol. 2013, 91, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Pendini, N.R.; Yap, M.Y.; Polyak, S.W.; Cowieson, N.P.; Abell, A.; Booker, G.W.; Wallace, J.C.; Wilce, J.A.; Wilce, M.C.J. Structural characterization of Staphylococcus aureus biotin protein ligase and interaction partners: An antibiotic target. Protein Sci. 2013, 22, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.H.; Cronan, J.E.; Grøtli, M.; Beckett, D. The Biotin Repressor: Modulation of Allostery by Corepressor Analogs. J. Mol. Boil. 2004, 337, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Wood, Z.A.; Weaver, L.H.; Brown, P.H.; Beckett, D.; Matthews, B.W. Co-repressor Induced Order and Biotin Repressor Dimerization: A Case for Divergent Followed by Convergent Evolution. J. Mol. Boil. 2006, 357, 509–523. [Google Scholar] [CrossRef]
- Paparella, A.; Lee, K.J.; Hayes, A.J.; Feng, J.; Feng, Z.; Cini, D.; Deshmukh, S.; Booker, G.W.; Wilce, M.C.J.; Polyak, S.W.; et al. Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus. ACS Infect. Dis. 2017, 4, 175–184. [Google Scholar] [CrossRef]
- Tieu, W.; Jarrad, A.; Paparella, A.; Keeling, K.A.; Da Costa, T.P.S.; Wallace, J.C.; Booker, G.W.; Polyak, S.W.; Abell, A.D. Heterocyclic acyl-phosphate bioisostere-based inhibitors of Staphylococcus aureus biotin protein ligase. Bioorganic Med. Chem. Lett. 2014, 24, 4689–4693. [Google Scholar] [CrossRef]
- Tieu, W.; Polyak, S.W.; Paparella, A.; Yap, M.Y.; Da Costa, T.P.S.; Ng, B.; Wang, G.; Lumb, R.; Bell, J.M.; Turnidge, J.D.; et al. Improved Synthesis of Biotinol-5′-AMP: Implications for Antibacterial Discovery. ACS Med. Chem. Lett. 2014, 6, 216–220. [Google Scholar] [CrossRef]
- Bockman, M.R.; Engelhart, C.A.; Dawadi, S.; Larson, P.; Tiwari, D.; Ferguson, D.M.; Schnappinger, D.; Aldrich, C.C. Avoiding Antibiotic Inactivation in Mycobacterium tuberculosis by Rv3406 through Strategic Nucleoside Modification. ACS Infect. Dis. 2018, 4, 1102–1113. [Google Scholar] [CrossRef] [PubMed]
- Bockman, M.R.; Kalinda, A.S.; Petrelli, R.; De La Mora-Rey, T.; Tiwari, D.; Liu, F.; Dawadi, S.; Nandakumar, M.; Rhee, K.Y.; Schnappinger, D.; et al. Targeting Mycobacterium tuberculosis Biotin Protein Ligase (MtBPL) with Nucleoside-Based Bisubstrate Adenylation Inhibitors. J. Med. Chem. 2015, 58, 7349–7369. [Google Scholar] [CrossRef] [PubMed]
- Garcia, L. Synergism Testing: Broth Microdilution Checkerboard and Broth Macrodilution Methods. In Clinical Microbiology Procedures Handbook, 3rd ed.; ASM Press: Washington, DC, USA, 2010; pp. 140–162. [Google Scholar]
- Odds, F.C. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 2003, 52, 1. [Google Scholar] [CrossRef] [PubMed]
- Blake, K.L.; Randall, C.; O’Neill, A. In Vitro Studies Indicate a High Resistance Potential for the Lantibiotic Nisin in Staphylococcus aureus and Define a Genetic Basis for Nisin Resistance. Antimicrob. Agents Chemother. 2011, 55, 2362–2368. [Google Scholar] [CrossRef] [PubMed]
- Sims, D.; Sudbery, I.; Ilott, N.; Heger, A.; Ponting, C.P. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef]
- Broeke-Smits, N.J.P.T.; Pronk, T.; Jongerius, I.; Bruning, O.; Wittink, F.R.; Breit, T.M.; Van Strijp, J.A.G.; Fluit, A.C.; Boel, C.H.E. Operon structure of Staphylococcus aureus. Nucleic Acids Res. 2010, 38, 3263–3274. [Google Scholar] [CrossRef]
- Rahman, M.M.; Hunter, H.N.; Prova, S.; Verma, V.; Qamar, A.; Golemi-Kotra, D. The Staphylococcus aureus Methicillin Resistance Factor FmtA Is a d-Amino Esterase That Acts on Teichoic Acids. mBio 2016. [Google Scholar] [CrossRef]
- Corrigan, R.M.; Abbott, J.; Burhenne, H.; Kaever, V.; Gründling, A. c-di-AMP Is a New Second Messenger in Staphylococcus aureus with a Role in Controlling Cell Size and Envelope Stress. PLOS Pathog. 2011, 7, e1002217. [Google Scholar] [CrossRef]
- Engman, J.; Rogstam, A.; Frees, D.; Ingmer, H.; Von Wachenfeldt, C. The YjbH Adaptor Protein Enhances Proteolysis of the Transcriptional Regulator Spx in Staphylococcus aureus. J. Bacteriol. 2011, 194, 1186–1194. [Google Scholar] [CrossRef]
- Fey, P.D.; Endres, J.L.; Yajjala, V.K.; Widhelm, T.J.; Boissy, R.J.; Bose, J.L.; Bayles, K.W. A Genetic Resource for Rapid and Comprehensive Phenotype Screening of Nonessential Staphylococcus aureus Genes. mBio 2013, 4, e00537-12. [Google Scholar] [CrossRef]
- Pendini, N.R.; Polyak, S.W.; Booker, G.W.; Wallace, J.C.; Wilce, J.A. Purification, crystallization and preliminary crystallographic analysis of biotin protein ligase from Staphylococcus aureus. Acta Crystallogr. Sect. F Struct. Boil. Cryst. Commun. 2008, 64, 520–523. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, F.; Cui, L.; Priest, D.; Endy, D.; Dodd, I.; Shearwin, K. One-Step Cloning and Chromosomal Integration of DNA. ACS Synth. Boil. 2013, 2, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Blount, K.; Breaker, R.R. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 2006, 24, 1558–1564. [Google Scholar] [CrossRef]
- Pedrolli, D.; Matern, A.; Wang, J.; Ester, M.; Siedler, K.; Breaker, R.R.; Mack, M. A highly specialized flavin mononucleotide riboswitch responds differently to similar ligands and confers roseoflavin resistance to Streptomyces davawensis. Nucleic Acids Res. 2012, 40, 8662–8673. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.P.; Shewchuk, L.M.; Brennan, R.G.; Otsuka, A.J.; Matthews, B.W. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc. Natl. Acad. Sci. USA 1992, 89, 9257–9261. [Google Scholar] [CrossRef]
- Buoncristiani, M.R.; Howard, P.K.; Otsuka, A.J. DNA-binding and enzymatic domains of the bifunctional biotin operon repressor (BirA) of Escherichia. Gene 1986, 44, 255–261. [Google Scholar] [CrossRef]
- Hellman, L.; Fried, M. Electrophoretic mobility shift assay (EMSA) for detecting protein–nucleic acid interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef]
- Pope, C.F.; O’Sullivan, D.M.; McHugh, T.D.; Gillespie, S.H. A Practical Guide to Measuring Mutation Rates in Antibiotic Resistance. Antimicrob. Agents Chemother. 2008, 52, 1209–1214. [Google Scholar] [CrossRef]
- Weiner, O.; Se, L.; M, D. Faculty of 1000 evaluation for Mutations of Bacteria from Virus Sensitivity to Virus Resistance. F1000 - Post-publication peer review of the biomedical literature 2010, 28, 491–511. [Google Scholar]
- Young, K. In Vitro Antibacterial Resistance Selection and Quantitation. Curr. Protoc. Pharmacol. 2006, 34, 13A.6.1–13A.6.22. [Google Scholar] [CrossRef]
- Friedman, L.; Alder, J.D.; Silverman, J.A. Genetic Changes That Correlate with Reduced Susceptibility to Daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 2137–2145. [Google Scholar] [CrossRef] [PubMed]
- Deatherage, D.E.; Barrick, J.E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Breast Cancer 2014, 1151, 165–188. [Google Scholar]
- Giachino, P.; Engelmann, S.; Bischoff, M. ςB Activity Depends on RsbU in Staphylococcus aureus. J. Bacteriol. 2001, 183, 1843–1852. [Google Scholar] [CrossRef] [PubMed]
- Brune, I.; Götker, S.; Schneider, J.; Rodionov, D.A.; Tauch, A. Negative transcriptional control of biotin metabolism genes by the TetR-type regulator BioQ in biotin-auxotrophic Corynebacterium glutamicum ATCC 13032. J. Biotechnol. 2012, 159, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Chapman-Smith, A.; Turner, D.L.; E Cronan, J.; Morris, T.W.; Wallace, J.C. Expression, biotinylation and purification of a biotin-domain peptide from the biotin carboxy carrier protein of Escherichia coli acetyl-CoA carboxylase. Biochem. J. 1994, 302, 881–887. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).