Beta-Lactam Sensitive Bacteria Can Acquire ESBL-Resistance via Conjugation after Long-Term Exposure to Lethal Antibiotic Concentration
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Bioinformatic Analyses
3.2. Evolutionary Rescue Experiments
3.3. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef]
- Cairns, J.; Koskinen, K.; Penttinen, R.; Patinen, T.; Hartikainen, A.; Jokela, R.; Ruusulehto, L.; Viitamäki, S.; Mattila, S.; Hiltunen, T.; et al. Black Queen evolution and trophic interactions determine plasmid survival after the disruption of conjugation network. mSystems 2018, 3, e00104-18. [Google Scholar] [CrossRef] [Green Version]
- Lopatkin, A.J.; Meredith, H.R.; Srimani, J.K.; Pfeiffer, C.; Durrett, R.; You, L. Persistence and reversal of plasmid-mediated antibiotic resistance. Nat. Commun. 2017, 8, 1689. [Google Scholar] [CrossRef]
- Worthington, R.J.; Melander, C. Overcoming resistance to β-lactam antibiotics. J. Org. Chem. 2013, 78, 4207–4213. [Google Scholar] [CrossRef] [Green Version]
- Rawat, D.; Nair, D. Extended-spectrum β-lactamases in Gram Negative Bacteria. J. Glob. Infect Dis. 2010, 2, 263–274. [Google Scholar] [CrossRef]
- Yurtsev, E.A.; Chao, H.X.; Datta, M.S.; Artemova, T.; Gore, J. Bacterial cheating drives the population dynamics of cooperative antibiotic resistance plasmids. Mol. Syst. Biol. 2013, 9, 683. [Google Scholar] [CrossRef] [Green Version]
- Ojala, V.; Mattila, S.; Hoikkala, V.; Bamford, J.K.; Jalasvuori, M. Evolutionary rescue of bacteria via horizontal gene transfer under a lethal β-lactam concentration. J. Glob. Antimicrob. Resist. 2014, 2, 198–200. [Google Scholar] [CrossRef]
- Vega, N.M.; Gore, J. Collective antibiotic resistance: Mechanisms and implications. Curr. Opin. Microbiol. 2014, 21, 28–34. [Google Scholar] [CrossRef] [Green Version]
- Bottery, M.J.; Wood, A.J.; Brockhurst, M.A. Selective conditions for a multidrug resistance plasmid depend on the sociality of antibiotic resistance. Antimicrob. Agents Chemother. 2016, 60, 2524–2527. [Google Scholar] [CrossRef] [Green Version]
- Woerther, P.; Burdet, C.; Chachaty, E.; Andremont, A. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: Toward the globalization of CTX-M. Clin. Microbiol. Rev. 2013, 26, 744–758. [Google Scholar] [CrossRef] [Green Version]
- Mattila, S.; Ruotsalainen, P.; Ojala, V.; Tuononen, T.; Hiltunen, T.; Jalasvuori, M. Conjugative ESBL plasmids differ in their potential to rescue susceptible bacteria via horizontal gene transfer in lethal antibiotic concentrations. J. Antibiot. 2017, 70, 805–808. [Google Scholar] [CrossRef]
- Cross, T.; Ransegnola, B.; Shin, J.H.; Weaver, A.; Fauntleroy, K.; VanNieuwenhze, M.; Westblade, L.F.; Dörr, T. Spheroplast-mediated carbapenem tolerance in Gram-negative pathogens. bioRxiv 2019, 578559. [Google Scholar] [CrossRef] [Green Version]
- Medaney, F.; Dimitriu, T.; Ellis, R.J.; Raymond, B. Live to cheat another day: Bacterial dormancy facilitates the social exploitation of β-lactamases. ISME J. 2015, 10, 778–787. [Google Scholar] [CrossRef] [Green Version]
- Ojala, V.; Mattila, S.; Hoikkala, V.; Bamford, J.K.H.; Hiltunen, T.; Jalasvuori, M. Scoping the effectiveness and evolutionary obstacles in utilizing plasmid-dependent phages to fight antibiotic resistance. Future Microbiol. 2016, 11, 999–1009. [Google Scholar] [CrossRef]
- Mercier, R.; Yoshikazu, K.; Errington, J. General principles for the formation and proliferation of a wall-free (L-form) state in bacteria. eLife 2014, 30, 3. [Google Scholar] [CrossRef]
- Cushnie, T.P.; O’Driscoll, N.H.; Lamb, A.J. Morphological and ultrastructural changes in bacterial cells as an indicator of antibacterial mechanism of action. Cell. Mol. Life. Sci. 2016, 73, 4471–4492. [Google Scholar] [CrossRef]
- Martinac, B.; Buechner, M.; Delcour, A.H.; Adler, J.; Kung, C. Pressure-sensitive ion channel in Escherichia coli. Proc. Natl. Acad. Sci. USA 1987, 84, 2297–2301. [Google Scholar] [CrossRef] [Green Version]
- Yosef, I.; Manor, M.; Kiro, R.; Qimron, U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 7267–7272. [Google Scholar] [CrossRef] [Green Version]
- Ruotsalainen, P.; Penttinen, R.; Mattila, S.; Jalasvuori, M. Midbiotics: Conjugative plasmids for genetic engineering of natural gut flora. Gut Microbes 2019, 10, 643–653. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Xie, Y.; Liu, M.; Tai, C.; Sun, J.; Deng, Z.; Ou, H.Y. oriTfinder: A web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 2018, 46, W229–W234. [Google Scholar] [CrossRef]
- Luo, H.; Zhang, C.T.; Gao, F. Ori-Finder 2, an integrated tool to predict replication origins in the archaeal genomes. Front. Microbiol. 2014, 5, 482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Lennox, E.S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1955, 1, 190–206. [Google Scholar] [CrossRef]
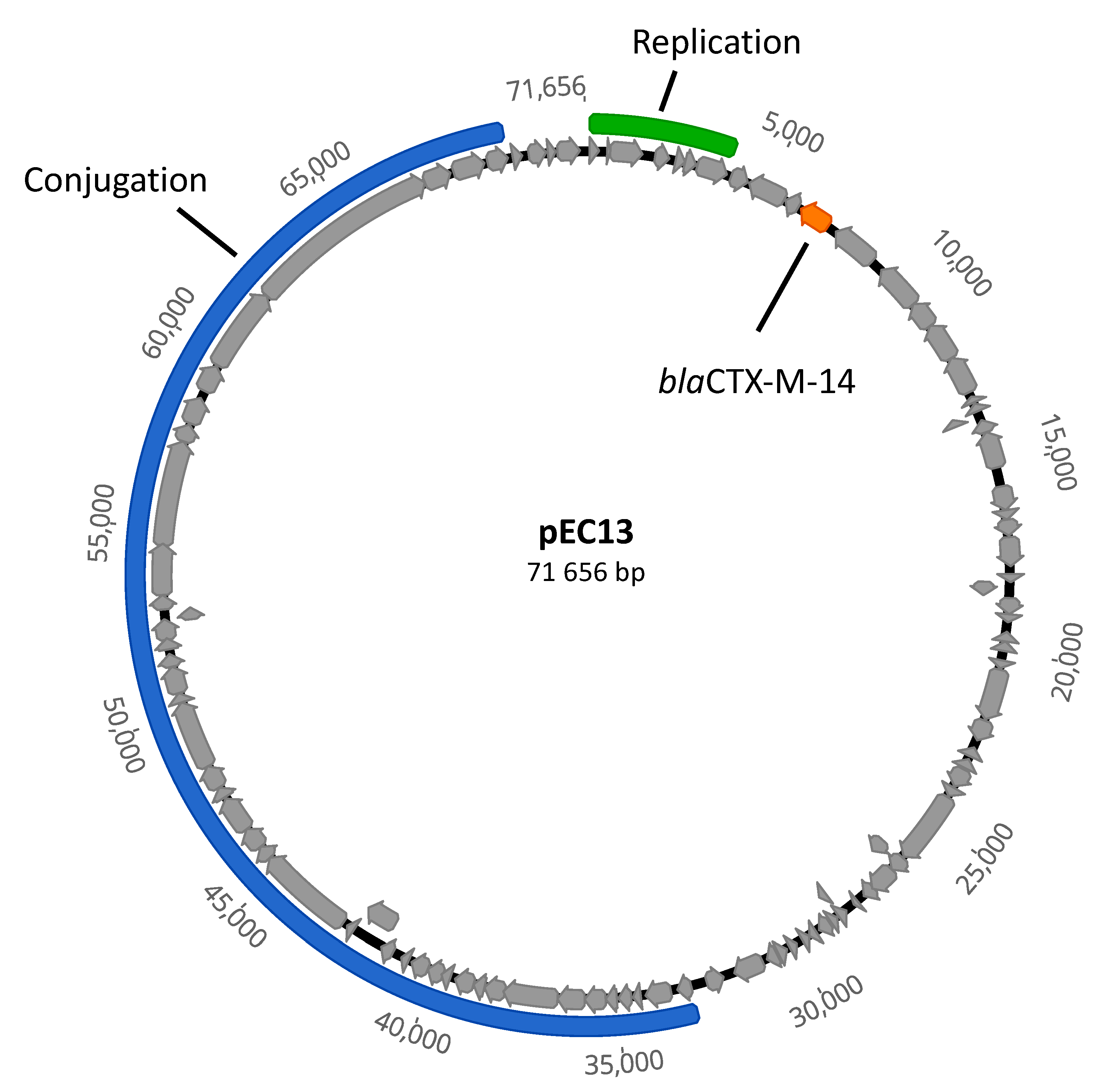
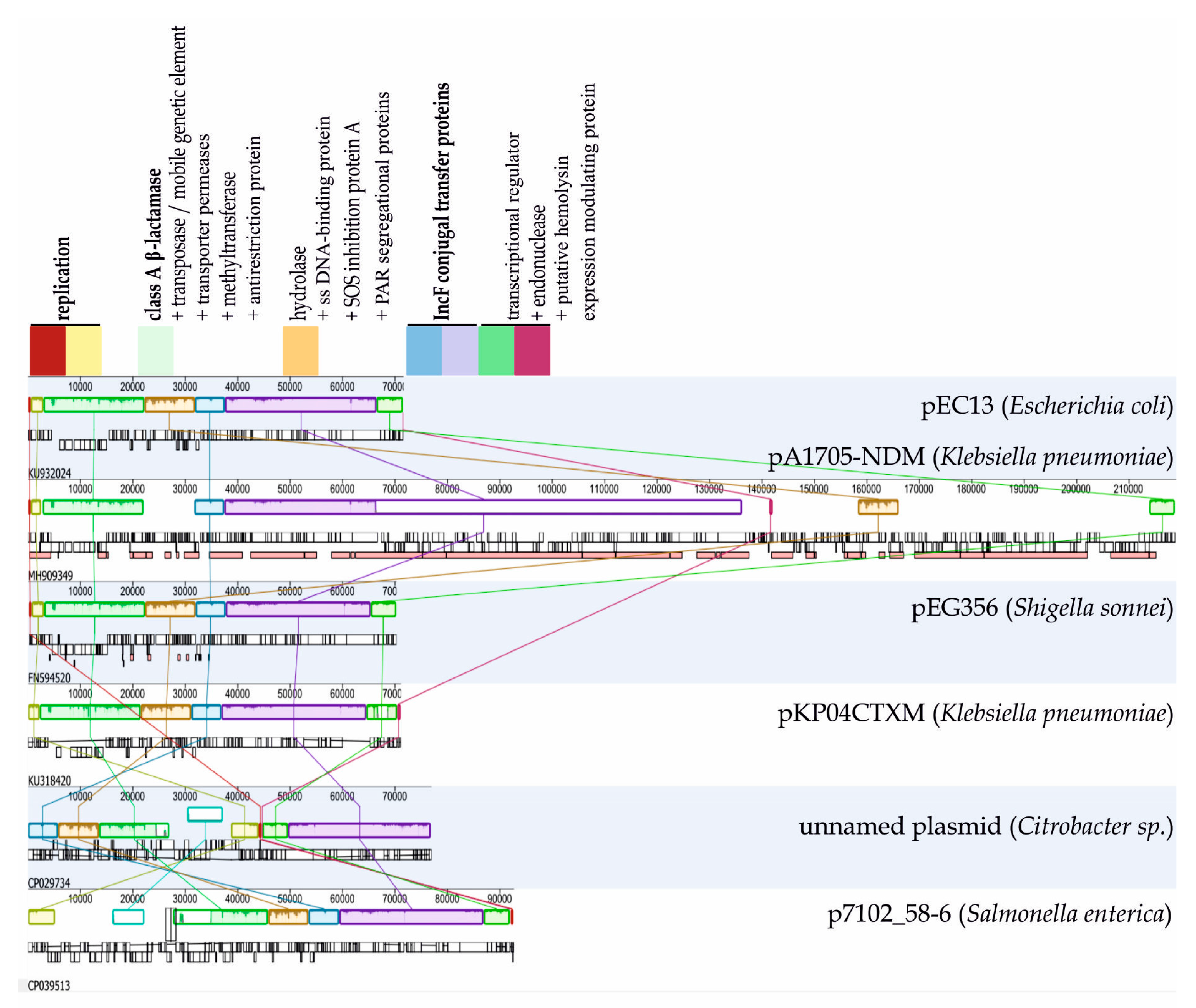

| Beta-Lactam Susceptible Strain | Replicate | Survivors (cfu/mL) |
|---|---|---|
| 1 | <200 | |
| E. coli DH5α (pCas9-gRNA) | 2 | <200 |
| 3 | <200 | |
| 1 | 1.52 × 106 | |
| E. coli DH5α (pCas9-CTRL) | 2 | 1.76 × 106 |
| 3 | 2.12 × 106 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruotsalainen, P.; Given, C.; Penttinen, R.; Jalasvuori, M. Beta-Lactam Sensitive Bacteria Can Acquire ESBL-Resistance via Conjugation after Long-Term Exposure to Lethal Antibiotic Concentration. Antibiotics 2020, 9, 296. https://doi.org/10.3390/antibiotics9060296
Ruotsalainen P, Given C, Penttinen R, Jalasvuori M. Beta-Lactam Sensitive Bacteria Can Acquire ESBL-Resistance via Conjugation after Long-Term Exposure to Lethal Antibiotic Concentration. Antibiotics. 2020; 9(6):296. https://doi.org/10.3390/antibiotics9060296
Chicago/Turabian StyleRuotsalainen, Pilvi, Cindy Given, Reetta Penttinen, and Matti Jalasvuori. 2020. "Beta-Lactam Sensitive Bacteria Can Acquire ESBL-Resistance via Conjugation after Long-Term Exposure to Lethal Antibiotic Concentration" Antibiotics 9, no. 6: 296. https://doi.org/10.3390/antibiotics9060296
APA StyleRuotsalainen, P., Given, C., Penttinen, R., & Jalasvuori, M. (2020). Beta-Lactam Sensitive Bacteria Can Acquire ESBL-Resistance via Conjugation after Long-Term Exposure to Lethal Antibiotic Concentration. Antibiotics, 9(6), 296. https://doi.org/10.3390/antibiotics9060296




