Tiamulin-Resistant Mutants of the Thermophilic Bacterium Thermus thermophilus
Abstract
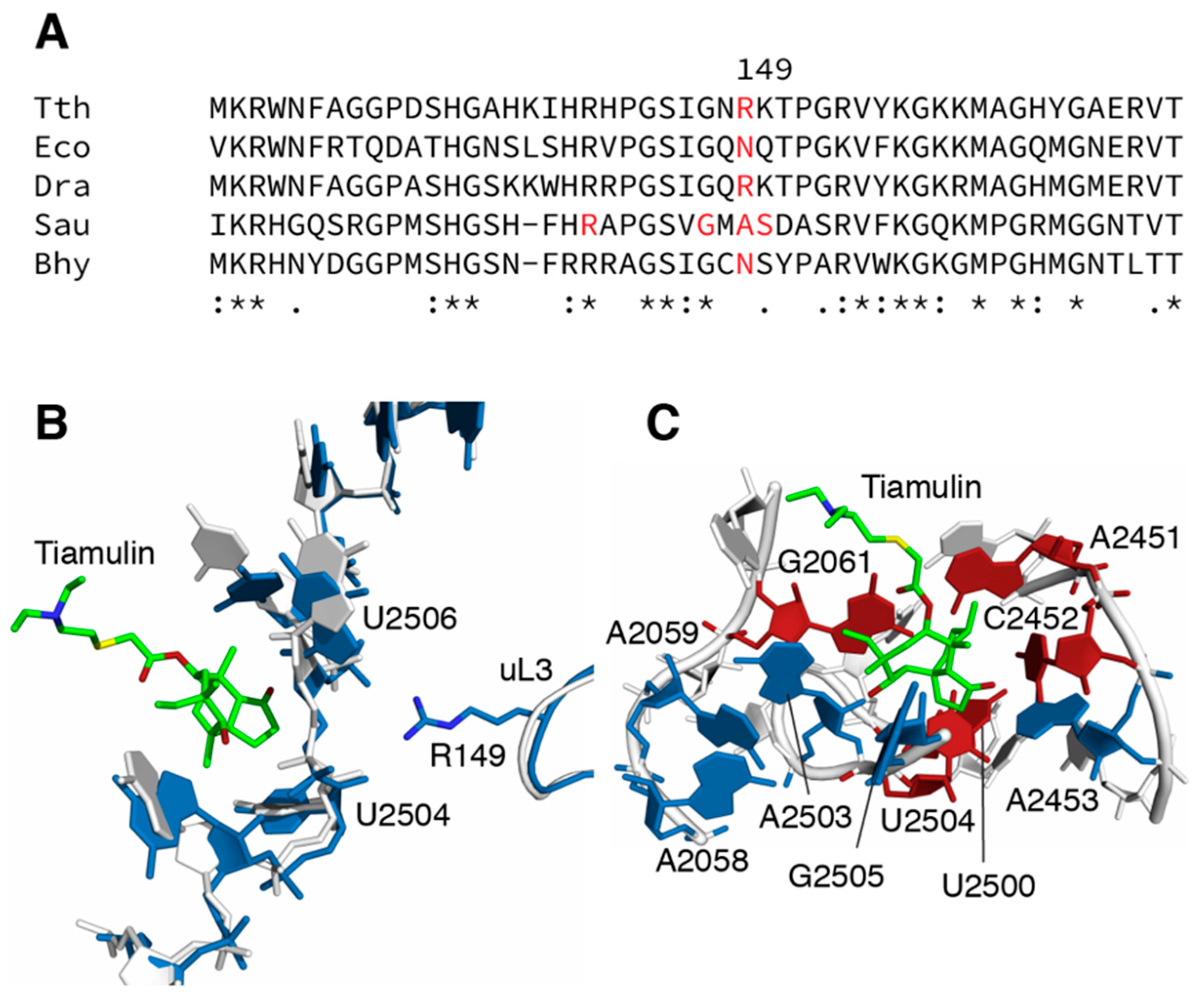
:1. Introduction
2. Results
2.1. Isolation of Spontaneous Tiamulin-Resistant Mutants
2.2. Resistance Phenotypes
2.3. Resistance to Other PTC-Binding Antibiotics
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Cultivation
4.2. Isolation of Spontaneous Mutants
4.3. Analysis of Mutants
4.4. Disc Diffusion Assays
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wilson, D.N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014, 12, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Matzov, D.; Bashan, A.; Yonath, A. A bright future for antibiotics? Annu. Rev. Biochem. 2017, 86, 567–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Zhou, D.; Steitz, T.A.; Polikanov, Y.S.; Gagnon, M.G. Ribosome-targeting antibiotics: Modes of action, mechanisms of resistance, and implications for drug design. Annu. Rev. Biochem. 2018, 87, 451–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavanagh, F.; Hervey, A.; Robbins, W.J. Antibiotic substances from Basidiomycetes: VIII Pleurotus mutilus (Fr.) Sacc. and Pleurotus passeckerianus pilat. Proc. Natl. Acad. Sci. USA 1951, 37, 570–574. [Google Scholar] [CrossRef] [Green Version]
- Paukner, S.; Riedl, R. Pleuromutilins: Potent drugs for resistant bugs-mode of action and resistance. Cold Spring Harb. Perspect. Med. 2017, 7, 27–110. [Google Scholar] [CrossRef]
- Poulsen, S.M.; Karlsson, M.; Johansson, L.B.; Vester, B. The pleuromutilin drugs tiamulin and valnemulin bind to the RNA at the peptidyl transferases centre on the ribosome. Mol. Microbiol. 2001, 41, 1091–1099. [Google Scholar] [CrossRef]
- Hodgin, L.A.; Högenauer, G. The mode of action of pleuromutilin derivatives. Effect on cell-free polypeptide synthesis. Eur. J. Biochem. 1974, 47, 527–533. [Google Scholar] [CrossRef]
- Schlünzen, F.; Pyetan, E.; Fucini, P.; Yonath, A.; Harms, J.M. Inhibition of peptide bond formation by pleuromutilins: The structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin. Mol. Microbiol. 2004, 54, 1287–1294. [Google Scholar] [CrossRef]
- Davidovich, C.; Bashan, A.; Auerbach-Nevo, T.; Yaggie, R.D.; Gontarek, R.R.; Yonath, A. Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity. Proc. Natl. Acad. Sci. USA 2007, 104, 4291–4296. [Google Scholar] [CrossRef] [Green Version]
- Gürel, G.; Blaha, G.; Moore, P.B.; Steitz, T.A. U2504 determines the species specificity of the A-site cleft antibiotics: The structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J. Mol. Biol. 2009, 389, 146–156. [Google Scholar] [CrossRef] [Green Version]
- Böck, A.; Turnowsky, F.; Högenauer, G. Tiamulin resistance mutations in Escherichia coli. J. Bacteriol. 1982, 151, 1253–1260. [Google Scholar] [CrossRef] [Green Version]
- Bøsling, J.; Poulsen, S.M.; Vester, B.; Long, K.S. Resistance to the peptidyl transferase inhibitor tiamulin caused by mutation of ribosomal protein L3. Antimicrob. Agents Chemother. 2003, 47, 2892–2896. [Google Scholar] [CrossRef] [Green Version]
- Klitgaard, R.N.; Ntokou, E.; Nørgaard, K.; Biltoft, D.; Hansen, L.H.; Traedholm, N.M.; Kongsted, J.; Vester, B. Mutations in the bacterial ribosomal protein L3 and their association with antibiotic resistance. Antimicrob. Agents Chemother. 2015, 59, 3518–3528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pringle, M.; Poehlsgaard, J.; Vester, B.; Long, K.S. Mutations in ribosomal protein L3 and 23S ribosomal RNA at the peptidyl transferase centre are associated with reduced susceptibility to tiamulin in Brachyspira spp. isolates. Mol. Microbiol. 2004, 54, 1295–1306. [Google Scholar] [CrossRef]
- Gentry, D.R.; Rittenhouse, S.F.; McCloskey, L.; Holmes, D.J. Stepwise exposure of Staphylococcus aureus to pleuromutilins is associated with stepwise acquisition of mutations in rplC and minimally affects susceptibility to retapamulin. Antimicrob. Agents Chemother. 2007, 51, 2048–2052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, K.; Dunsmore, C.J.; Fishwick, C.W.; Chopra, I. Linezolid and tiamulin cross-resistance in Staphylococcus aureus mediated by point mutations in the peptidyl transferase center. Antimicrob. Agents Chemother. 2008, 52, 1737–1742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ban, N.; Nissen, P.; Hansen, J.; Moore, P.B.; Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 2000, 289, 905–920. [Google Scholar] [CrossRef]
- Harms, J.; Schluenzen, F.; Zarivach, R.; Bashan, A.; Gat, S.; Agmon, I.; Bartels, H.; Franceschi, F.; Yonath, A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 2001, 107, 679–688. [Google Scholar] [CrossRef]
- Yusupov, M.M.; Yusupova, G.Z.; Baucom, A.; Lieberman, K.; Earnest, T.N.; Cate, J.H.; Noller, H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science 2001, 292, 883–896. [Google Scholar] [CrossRef]
- Noeske, J.; Wasserman, M.R.; Terry, D.S.; Altman, R.B.; Blanchard, S.C.; Cate, J.H. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 2015, 22, 336–341. [Google Scholar] [CrossRef]
- Long, K.S.; Poehlsgaard, J.; Hansen, L.H.; Hobbie, S.N.; Bottger, E.C.; Vester, B. Single 23S rRNA mutations at the ribosomal peptidyl transferase centre confer resistance to valnemulin and other antibiotics in Mycobacterium smegmatis by perturbation of the drug binding pocket. Mol. Microbiol. 2009, 71, 1218–1227. [Google Scholar] [CrossRef]
- Li, B.-B.; Shen, J.-Z.; Cao, X.-Y.; Wang, Y.; Dai, L.; Huang, S.-Y.; Wu, C.-M. Mutations in 23S rRNA gene associated with decreased susceptibility to tiamulin and valnemulin in Mycoplasma gallisepticum. Fems Microbiol. Lett. 2010, 308, 144–149. [Google Scholar] [CrossRef] [Green Version]
- Gregory, S.T.; Carr, J.F.; Rodriguez–Correa, D.; Dahlberg, A.E. 2005. Mutational analysis of 16S and 23S rRNA genes of Thermus thermophilus. J. Bacteriol. 2005, 187, 4804–4812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demirci, H.; Wang, L.; Murphy, F.V.; Murphy, E.L.; Carr, J.F.; Blanchard, S.C.; Jogl, G.; Dahlberg, A.E.; Gregory, S.T. The central role of protein S12 in organizing the structure of the decoding site of the ribosome. RNA 2013, 19, 1791–1801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oshima, T.; Imahori, K. Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int. J. Syst. Evol. Microbiol. 1974, 24, 102–112. [Google Scholar] [CrossRef] [Green Version]
- Kristjansson, J.K.; Hreggvidsson, G.O.; Alfredsson, G.A. 1986. Isolation of halotolerant Thermus spp. from submarine hot springs in Iceland. Appl. Environ. Microbiol. 1986, 52, 1313–1316. [Google Scholar] [CrossRef] [Green Version]
- Kearsey, S.E.; Craig, I.W. Altered ribosomal RNA genes in mitochondria from mammalian cells with chloramphenicol resistance. Nature 1981, 290, 607–608. [Google Scholar] [CrossRef]
- Beringer, M.; Bruell, C.; Xiong, L.; Pfister, P.; Bieling, P.; Katunin, V.I.; Mankin, A.S.; Böttger, E.C.; Rodnina, M.V. Essential mechanisms in the catalysis of peptide bond formation on the ribosome. J. Biol. Chem. 2005, 280, 36065–36072. [Google Scholar] [CrossRef] [Green Version]
- Selmer, M.; Dunham, C.M.; Murphy, F.V.; Weixlbaumer, A.; Petry, S.; Kelly, A.C.; Weir, J.R.; Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 2006, 313, 1935–1942. [Google Scholar] [CrossRef] [Green Version]
- Khusainov, I.; Vicens, Q.; Bochler, A.; Grosse, F.; Myasnikov, A.; Ménétret, J.F.; Chicher, J.; Marzi, S.; Romby, P.; Yusupova, G. Structure of the 70S ribosome from human pathogen Staphylococcus aureus. Nucleic Acids Res. 2016, 44, 10491–10504. [Google Scholar] [CrossRef] [Green Version]
- Nissen, P.; Hansen, J.; Ban, N.; Moore, P.B.; Steitz, T.A. The structural basis of ribosome activity in peptide bond formation. Science 2000, 289, 920–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannone, J.J.; Subramanian, S.; Schnare, M.N.; Collett, J.R.; D’Souza, L.M.; Du, Y.; Feng, B.; Lin, N.; Madabusi, L.V.; Müller, K.M. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron and other RNA. BMC Bioinform. 2002, 3, 2. [Google Scholar]
- Thompson, J.D.; Kim, D.F.; O’Connor, M.; Lieberman, K.R.; Bayfield, M.A.; Gregory, S.T.; Green, R.; Noller, H.F.; Dahlberg, A.E. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc. Natl. Acad. Sci. USA 2001, 98, 9002–9007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaha, G.; Gürel, G.; Schroeder, S.J.; Moore, P.B.; Steitz, T.A. Mutations outside the anisomycin-binding site can make ribosomes drug-resistant. J. Mol. Biol. 2008, 379, 505–519. [Google Scholar] [CrossRef] [Green Version]
- Davidovitch, C.; Bashan, A.; Yonath, A. Structural basis for cross-resistance to ribosomal PTC antibiotics. Proc. Natl. Acad. Sci. USA 2008, 105, 20665–20670. [Google Scholar] [CrossRef] [Green Version]
| Strain | Genotype | Mutation | Tiamulin Selection | Temperature |
|---|---|---|---|---|
| IB-21 | rplC-R149H | uL3-R149H | 50 μg/mL | 72 °C |
| IB-21 | rplC-R149C | uL3-R149C | 100 μg/mL | 72 °C |
| HB27 | rplC-R149C | uL3-R149C | 100 μg/mL | 72 °C |
| IB-21 | rrlAB-G2061A | 23S-G2061A | 50, 100 μg/mL | 72 °C |
| HB27 | rrlAB-G2061A | 23S-G2061A | 100 μg/mL | 65 °C |
| IB-21 | rrlAB-G2061U | 23S-G2061U | 100 μg/mL | 62 °C |
| HB27 | rrlAB-G2061U | 23S-G2061U | 100 μg/mL | 72 °C |
| HB27 | rrlAB-A2451U | 23S-A2451U | 100 μg/mL | 72 °C |
| IB-21 | rrlAB-C2452U | 23S-C2452U | 50 μg/mL | 72 °C |
| IB-21 | rrlAB-U2500A | 23S-U2500A | 100, 200 μg/mL | 72 °C |
| HB27 | rrlAB-U2500A | 23S-U2500A | 50 μg/mL | 65 °C |
| IB-21 | rrlAB-U2504G | 23S-U2504G | 100 μg/mL | 62 °C |
| HB27 | rrlAB-U2504G | 23S-U2504G | 200 μg/mL | 72 °C |
| Mutant | Zone of Inhibition (mm) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tam | Ery | Clr | Rox | Azm | Mid | Tyl | Spi | Lin | Chl | Str | Rif | Amp | Cip | |
| WT (IB-21) | 30 | 27 | 36 | 32 | 22 | 21 | 36 | 16 | 36 | 29 | 14 | 17 | 40 | 22 |
| WT (HB27) | 32 | 29 | 36 | 33 | 25 | 24 | 37 | 15 | 41 | 33 | 16 | - | 38 | 21 |
| uL3-R149H | 16 | 23 | 33 | 28 | 18 | 25 | 39 | 10 | 44 | 38 | 15 | 17 | 45 | 20 |
| uL3-R149C | 17 | 25 | 36 | 32 | 22 | 23 | 38 | 12 | 35 | 31 | 17 | 16 | 51 | 25 |
| 23S-G2061A | - | 42 | 63 | 65 | 34 | 46 | 71 | 40 | 34 | - | 20 | 27 | 55 | 28 |
| 23S-G2061U | - | 22 | 35 | 24 | 11 | 24 | 48 | 13 | 7 | 9 | 17 | 17 | 50 | 25 |
| 23S-A2451U | - | 12 | 33 | 28 | 15 | 28 | 64 | 27 | - | - | 30 | 21 | 50 | 26 |
| 23S-C2452U | - | 14 | 25 | 20 | 10 | 20 | 48 | 13 | 20 | 9 | 16 | 15 | 42 | 22 |
| 23S-U2500A | - | 14 | 20 | 19 | 11 | 12 | 36 | 7 | 18 | 15 | 15 | 17 | 40 | 21 |
| 23S-U2504G | - | 18 | 34 | 28 | 13 | 18 | 46 | 16 | 22 | - | 16 | 16 | 42 | 22 |
| Mutation | Tiamulin Phenotype 1 | Conservation | Nearest Neighbor | Distance (Dra, Å) | Distance (Hma, Å) |
|---|---|---|---|---|---|
| A2058G | S | 98.33/42.28 | A2058-N6:Tam-O2 | 7.1 | 7.1 |
| A2059G | S | 99.17/99.41 | A2059-N1:Tam-O2 | 6.7 | 5.8 |
| G2061A,U | R,R | 100.00/99.88 | G2061-N2:Tam-O4 | 3.0 | 3.0 |
| G2447A | R | 96.28/98.20 | G2447-N1:Tam-C8 | 5.3 | 5.0 |
| A2451U | R | 100.00/100.00 | A2451-N6:Tam-C7 | 3.6 | 3.3 |
| C2452U | R | 100.00/100.00 | C2452-C1’:Tam-O1 C2452-O2:Tam-O1 | 3.9 4.0 | 4.7 2.9 |
| A2453G | S | 99.59/82.39 | A2453-N3:Tam-C2 | 6.0 | 6.6 |
| U2500A,C | R,S | 100.00/99.64 | U2500-O2:Tam-C1 U2500-O2:Tam-C2 | 7.2 7.4 | 6.4 6.1 |
| A2503G | S | 99.59/98.31 | A2503-O2’:Tam-C17 A2503-C8:Tam-O2 | 3.0 - | 4.0 3.0 |
| U2504G,C,A | R,S,R | 99.59/98.43 | U2504-C5:Tam-C1 U2504-O2’:Tam-C2 | 3.5 5.0 | 5.6 3.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Killeavy, E.E.; Jogl, G.; Gregory, S.T. Tiamulin-Resistant Mutants of the Thermophilic Bacterium Thermus thermophilus. Antibiotics 2020, 9, 313. https://doi.org/10.3390/antibiotics9060313
Killeavy EE, Jogl G, Gregory ST. Tiamulin-Resistant Mutants of the Thermophilic Bacterium Thermus thermophilus. Antibiotics. 2020; 9(6):313. https://doi.org/10.3390/antibiotics9060313
Chicago/Turabian StyleKilleavy, Erin E., Gerwald Jogl, and Steven T. Gregory. 2020. "Tiamulin-Resistant Mutants of the Thermophilic Bacterium Thermus thermophilus" Antibiotics 9, no. 6: 313. https://doi.org/10.3390/antibiotics9060313
APA StyleKilleavy, E. E., Jogl, G., & Gregory, S. T. (2020). Tiamulin-Resistant Mutants of the Thermophilic Bacterium Thermus thermophilus. Antibiotics, 9(6), 313. https://doi.org/10.3390/antibiotics9060313







