Antibacterial and Anti-Inflammatory Effects of Novel Peptide Toxin from the Spider Pardosa astrigera
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Bacterial Strains and Cell Lines
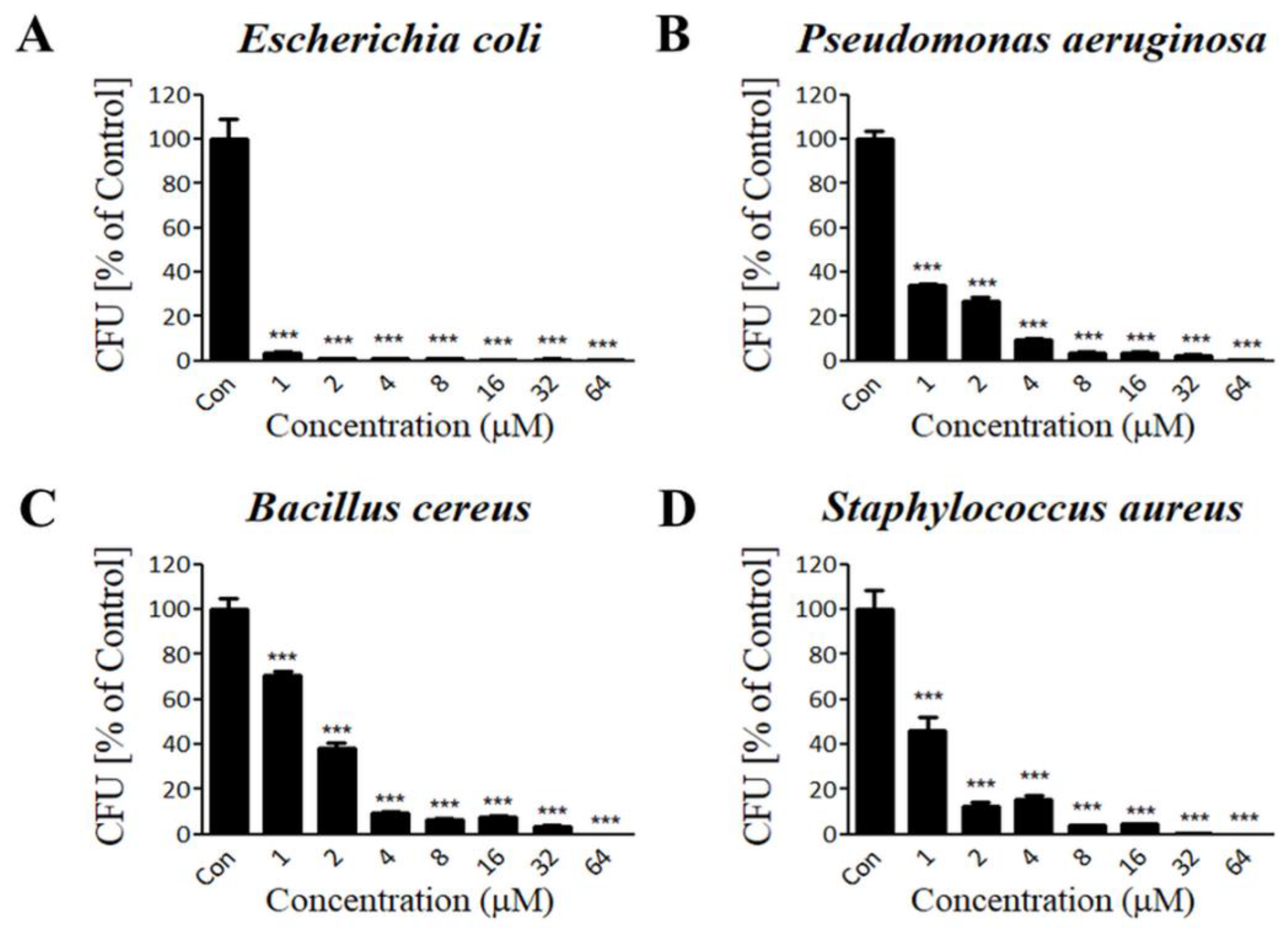
2.3. Antibacterial Activity Assay
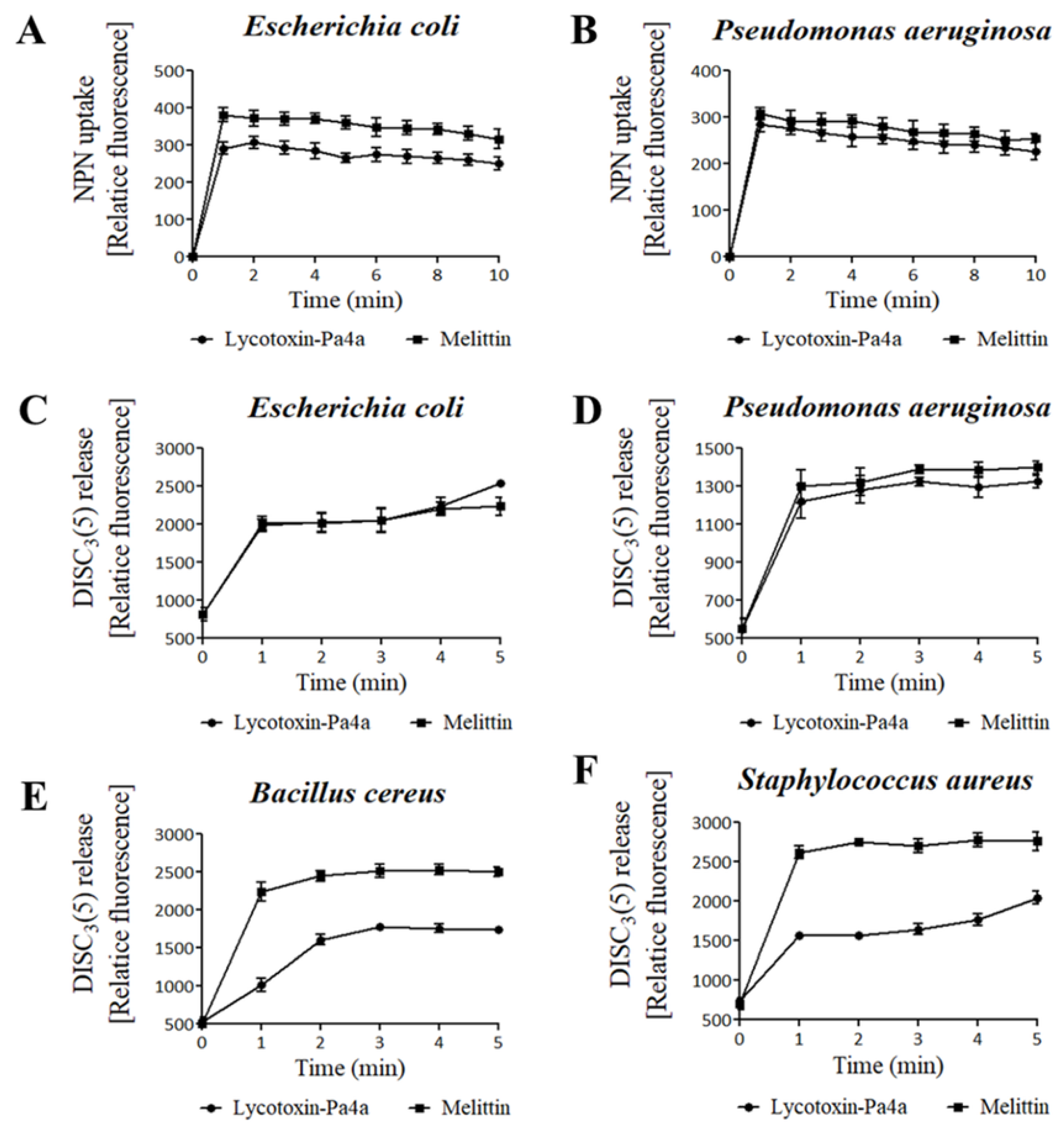
2.4. Membrane Permeability Test
2.5. Nitric oxide (NO) Measurement and Cell Viability Assay
2.6. Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
2.7. Western Blot Analysis
2.8. Statistical Analysis
3. Results
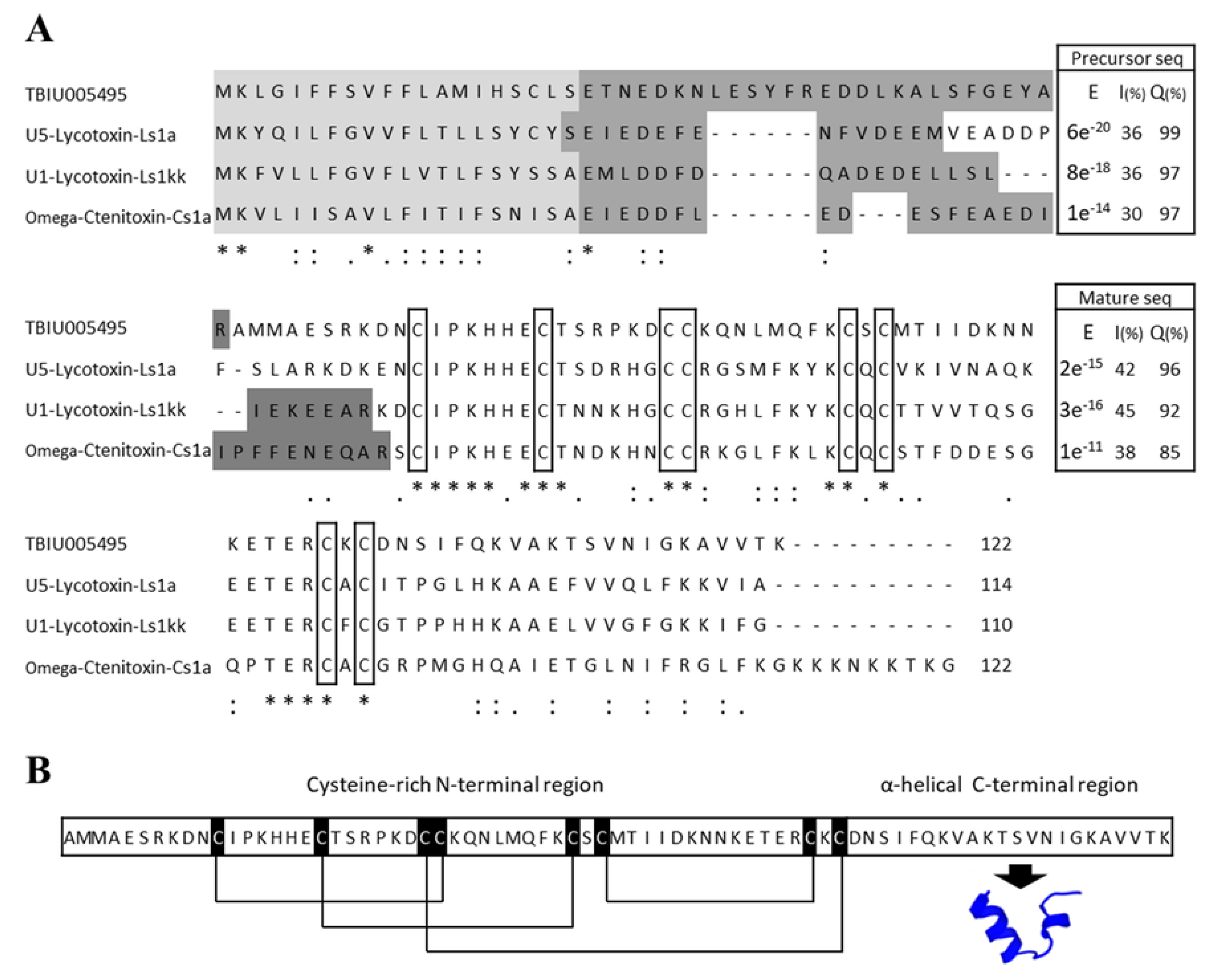
3.1. Identification of Toxin Peptide from the Transcriptome of the Venom Gland of P. astrigera
3.2. Structural Characterization of the Peptide Lycotoxin-Pa4a via in Silico Analysis
3.3. Lycotoxin-Pa4a Shows Antibacterial Activity against Both Gram-Negative and Gram-Positive Bacteria
3.4. Bacteria Membrane Permeabilization by Lycotoxin-Pa4a
3.5. Lycotoxin-Pa4a Exhibits Anti-Inflammatory Effects on LPS-stimulated RAW 264.7
3.6. Inhibition of the MAPK Pathway by Lycotoxin-Pa4a
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Basak, S.; Singh, P.; Rajurkar, M. Multidrug Resistant and Extensively Drug Resistant Bacteria: A Study. J. Pathog. 2016, 2016, 4065603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowler, P.G. Antibiotic resistance and biofilm tolerance: A combined threat in the treatment of chronic infections. J. Wound Care 2018, 27, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Delpech, G.; Bistoletti, M.; Ceci, M.; Lissarrague, S.; Bruni, S.S.; Sparo, M. Bactericidal Activity and Synergy Studies of Peptide AP-CECT7121 Against Multi-resistant Bacteria Isolated from Human and Animal Soft Tissue Infections. Probiotics Antimicrob. Proteins 2017, 9, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.-F.; Jie, M.-M.; Li, B.-S.; Hu, C.-J.; Xie, R.; Tang, B.; Yang, S.-M. Peptide-Based Treatment: A Promising Cancer Therapy. J. Immunol. Res. 2015, 2015, 761820. [Google Scholar] [CrossRef] [Green Version]
- Baig, M.H.; Ahmad, K.; Saeed, M.; Alharbi, A.M.; Barreto, G.E.; Ashraf, G.M.; Choi, I. Peptide based therapeutics and their use for the treatment of neurodegenerative and other diseases. Biomed. Pharmacother. 2018, 103, 574–581. [Google Scholar] [CrossRef]
- Bowdish, D.M.E.; Davidson, D.J.; Scott, M.G.; Hancock, R.E.M. Immunomodulatory Activities of Small Host Defense Peptides. Antimicrob. Agents Chemother. 2005, 49, 1727–1732. [Google Scholar] [CrossRef] [Green Version]
- Perumal Samy, R.; Stiles, B.G.; Franco, O.L.; Sethi, G.; Lim, L.H.K. Animal venoms as antimicrobial agents. Biochem. Pharmacol. 2017, 134, 127–138. [Google Scholar] [CrossRef]
- Tan, H.; Ding, X.; Meng, S.; Liu, C.; Wang, H.; Xia, L.; Liu, Z.; Liang, S. Antimicrobial potential of lycosin-I, a cationic and amphiphilic peptide from the venom of the spider Lycosa singorensis. Curr. Mol. Med. 2013, 13, 900–910. [Google Scholar] [CrossRef]
- Tang, Y.; Hou, S.; Li, X.; Wu, M.; Ma, B.; Wang, Z.; Jiang, J.; Deng, M.; Duan, Z.; Tang, X.; et al. Anti-parasitic effect on Toxoplasma gondii induced by a spider peptide lycosin-I. Exp. Parasitol. 2019, 198, 17–25. [Google Scholar] [CrossRef]
- Kozlov, S.A.; Vassilevski, A.A.; Feofanov, A.V.; Surovoy, A.Y.; Karpunin, D.Y.; Grishin, E.V. Latarcins, antimicrobial and cytolytic peptides from the venom of the spider Lachesana tarabaevi (Zodariidae) that exemplify biomolecular diversity. J. Biol. Chem. 2006, 281, 20983–20992. [Google Scholar] [CrossRef] [Green Version]
- Mutz, K.-O.; Heilkenbrinker, A.; Lönne, M.; Walter, J.-G.; Stahl, F. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 2013, 24, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Sigurdardottir, T.; Andersson, P.; Davoudi, M.; Malmsten, M.; Schmidtchen, A.; Bodelsson, M. In Silico Identification and Biological Evaluation of Antimicrobial Peptides Based on Human Cathelicidin LL-37. Antimicrob. Agents Chemother. 2006, 50, 2983–2989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, S. Insilico Studies on Antimicrobial Peptides (AMPs) from Earthworm. Int. J. Pept. Res. Ther. 2019. Available online: https://link.springer.com/article/10.1007/s10989-019-09970-9 (accessed on 15 November 2019). [CrossRef]
- Helander, I.M.; Mattila-Sandholm, T. Fluorometric assessment of Gram-negative bacterial permeabilization. J. Appl. Microbiol. 2000, 88, 213–219. [Google Scholar] [CrossRef]
- Te Winkel, J.D.; Gray, D.A.; Seistrup, K.H.; Hamoen, L.W.; Strahl, H. Analysis of Antimicrobial-Triggered Membrane Depolarization Using Voltage Sensitive Dyes. Front Cell Dev. Biol. 2016, 4, 29. [Google Scholar] [CrossRef] [Green Version]
- Kuhn-Nentwig, L.; Fedorova, I.M.; Lüscher, B.P.; Kopp, L.S.; Trachsel, C.; Schaller, J.; Vu, X.L.; Seebeck, T.; Streitberger, K.; Nentwig, W.; et al. A venom-derived neurotoxin, CsTx-1, from the spider Cupiennius salei exhibits cytolytic activities. J. Biol. Chem. 2012, 287, 25640–25649. [Google Scholar] [CrossRef] [Green Version]
- Dennison, S.; Wallace, J.; Harris, F.; Phoenix, D. Amphiphilic α-Helical Antimicrobial Peptides and Their Structure / Function Relationships. Protein Pept. Let. 2005, 12, 31–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Huang, J.; Chen, Y. Alpha-helical cationic antimicrobial peptides: Relationships of structure and function. Protein Cell 2010, 1, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Yang, H.; Xiao, H.; Farooq, A.; Liu, Z.; Hu, M.; Shi, X. The Spider Venom Peptide Lycosin-II Has Potent Antimicrobial Activity against Clinically Isolated Bacteria. Toxins (Basel) 2016, 8, 119. [Google Scholar] [CrossRef]
- King, G.F.; Gentz, M.C.; Escoubas, P.; Nicholson, G.M. A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon 2008, 52, 264–276. [Google Scholar] [CrossRef] [Green Version]
- Raghuraman, H.; Chattopadhyay, A. Melittin: A Membrane-active Peptide with Diverse Functions. Biosci. Rep. 2007, 27, 187–223. [Google Scholar] [CrossRef]
- Brandenburg, L.-O.; Merres, J.; Albrecht, L.-J.; Varoga, D.; Pufe, T. Antimicrobial Peptides: Multifunctional Drugs for Different Applications. Polymers 2012, 4, 539–560. [Google Scholar] [CrossRef] [Green Version]
- Sonyot, W.; Lamlertthon, S.; Luangsa-ard, J.J.; Mongkolsamrit, S.; Usuwanthim, K.; Ingkaninan, K.; Waranuch, N.; Suphrom, N. In Vitro Antibacterial and Anti-Inflammatory Effects of Novel Insect Fungus Polycephalomyces phaothaiensis Extract and Its Constituents against Propionibacterium acnes. Antibiotics 2020, 9, 274. [Google Scholar] [CrossRef] [PubMed]
- Annadurai, R.S.; Neethiraj, R.; Jayakumar, V.; Damodaran, A.C.; Rao, S.N.; Katta, M.A.V.S.K.; Gopinathan, S.; Sarma, S.P.; Senthilkumar, V.; Niranjan, V.; et al. De Novo Transcriptome Assembly (NGS) of Curcuma longa L. Rhizome Reveals Novel Transcripts Related to Anticancer and Antimalarial Terpenoids. PLoS ONE 2013, 8, e56217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, C.-S.; Lee, S.B.; Jung, M.-P.; Lee, J.-H.; Lee, S.; Lee, S.H. Accumulated Heavy Metal Content in Wolf Spider, Pardosa astrigera (Araneae: Lycosidae), as a Bioindicator of Exposure. J. Asia-Pac. Entomol. 2005, 8, 185–192. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Diamond, G.; Beckloff, N.; Weinberg, A.; Kisich, K.O. The roles of antimicrobial peptides in innate host defense. Curr. Pharm. Des. 2009, 15, 2377–2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Yuan, S.; Gao, B.; Zhu, S. Molecular diversity of fungal inhibitor cystine knot peptides evolved by domain repeat and fusion. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Zhang, S.-K.; Song, J.-W.; Gong, F.; Li, S.-B.; Chang, H.-Y.; Xie, H.-M.; Gao, H.-W.; Tan, Y.-X.; Ji, S.-P. Design of an α-helical antimicrobial peptide with improved cell-selective and potent anti-biofilm activity. Sci. Rep. 2016, 6, 27394. [Google Scholar] [CrossRef] [Green Version]
- Idiong, G.; Won, A.; Ruscito, A.; Leung, B.O.; Hitchcock, A.P.; Ianoul, A. Investigating the effect of a single glycine to alanine substitution on interactions of antimicrobial peptide latarcin 2a with a lipid membrane. Eur. Biophys. J. 2011, 40, 1087–1100. [Google Scholar] [CrossRef]
- Park, H.G.; Deng, Y.; Lee, K.S.; Kim, B.Y.; Yoon, H.J.; Lee, K.Y.; Jin, B.R. Molecular cloning and antifungal activity of an inhibitor cysteine knot peptide from the bumblebee Bombus ignites. J. Asia-Pac. Entomol. 2016, 19, 59–64. [Google Scholar] [CrossRef]
- Sun, Y.; Shang, D. Inhibitory Effects of Antimicrobial Peptides on Lipopolysaccharide-Induced Inflammation. Mediat. Inflamm. 2015, 2015, 167572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanjeewa, K.K.A.; Nagahawatta, D.P.; Yang, H.-Y.; Oh, J.Y.; Jayawardena, T.U.; Jeon, Y.-J.; de Zoysa, M.; Whang, I.; Ryu, B. Octominin Inhibits LPS-Induced Chemokine and Pro-inflammatory Cytokine Secretion from RAW 264.7 Macrophages via Blocking TLRs/NF-κB Signal Transduction. Biomolecules 2020, 10, 511. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.; LEE, M.W.; Wong, G.C.L. Modulation of toll-like receptor signaling by antimicrobial peptides. Semin. Cell Dev. Biol. 2019, 88, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Bode, J.G.; Ehlting, C.; Häussinger, D. The macrophage response towards LPS and its control through the p38MAPK–STAT3 axis. Cell. Signal. 2012, 24, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Rajaiah, R.; Perkins, D.J.; Ireland, D.D.C.; Vogel, S.N. CD14 dependence of TLR4 endocytosis and TRIF signaling displays ligand specificity and is dissociable in endotoxin tolerance. Proc. Natl. Acad. Sci. USA 2015, 112, 8391–8396. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, Z.; Chen, L.; Guang, H.; Li, Z.; Yang, H.; Li, J.; You, D.; Yu, H.; Lai, R. Cathelicidin-BF, a Snake Cathelicidin-Derived Antimicrobial Peptide, Could Be an Excellent Therapeutic Agent for Acne Vulgaris. PLoS ONE 2011, 6, e22120. [Google Scholar] [CrossRef] [Green Version]
- Ko, S.J.; Park, E.; Asandei, A.; Choi, J.-Y.; Lee, S.-C.; Seo, C.H.; Luchian, T.; Park, Y. Bee venom-derived antimicrobial peptide melectin has broad-spectrum potency, cell selectivity, and salt-resistant properties. Sci. Rep. 2020, 10, 10145. [Google Scholar] [CrossRef]
- Woodburn, K.W.; Jaynes, J.; Clemens, L.E. Designed Antimicrobial Peptides for Topical Treatment of Antibiotic Resistant Acne Vulgaris. Antibiotics 2020, 9, 23. [Google Scholar] [CrossRef] [Green Version]
- Kwon, J.Y.; Kim, M.K.; Mereuta, L.; Seo, C.H.; Luchian, T.; Park, Y. Mechanism of action of antimicrobial peptide P5 truncations against Pseudomonas aeruginosa and Staphylococcus aureus. AMB Express 2019, 9, 122. [Google Scholar] [CrossRef] [Green Version]
- Jia, F.; Wang, J.; Peng, J.; Zhao, P.; Kong, Z.; Wang, K.; Yan, W.; Wang, R. D-amino acid substitution enhances the stability of antimicrobial peptide polybia-CP. Acta. Biochim. Biophys. Sin. 2017, 49, 916–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irazazabal, L.N.; Porto, W.F.; Ribeiro, S.M.; Casale, S.; Humblot, V.; Ladram, A.; Franco, O.L. Selective amino acid substitution reduces cytotoxicity of the antimicrobial peptide mastoparan. Biochim. Biophys. Acta 2016, 1858, 2699–2708. [Google Scholar] [CrossRef] [PubMed]
- Chung, P.Y.; Khanum, R. Antimicrobial peptides as potential anti-biofilm agents against multidrug-resistant bacteria, Journal of Microbiology. J. Microbiol. Immunol. Infect. 2017, 50, 405–410. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, M.K.; Hwang, I.-W.; Kim, Y.; Kim, S.T.; Jang, W.; Lee, S.; Bang, W.Y.; Bae, C.-H.; Sung, J.-S. Antibacterial and Anti-Inflammatory Effects of Novel Peptide Toxin from the Spider Pardosa astrigera. Antibiotics 2020, 9, 422. https://doi.org/10.3390/antibiotics9070422
Shin MK, Hwang I-W, Kim Y, Kim ST, Jang W, Lee S, Bang WY, Bae C-H, Sung J-S. Antibacterial and Anti-Inflammatory Effects of Novel Peptide Toxin from the Spider Pardosa astrigera. Antibiotics. 2020; 9(7):422. https://doi.org/10.3390/antibiotics9070422
Chicago/Turabian StyleShin, Min Kyoung, In-Wook Hwang, Yunkyung Kim, Seung Tae Kim, Wonhee Jang, Seungki Lee, Woo Young Bang, Chang-Hwan Bae, and Jung-Suk Sung. 2020. "Antibacterial and Anti-Inflammatory Effects of Novel Peptide Toxin from the Spider Pardosa astrigera" Antibiotics 9, no. 7: 422. https://doi.org/10.3390/antibiotics9070422
APA StyleShin, M. K., Hwang, I.-W., Kim, Y., Kim, S. T., Jang, W., Lee, S., Bang, W. Y., Bae, C.-H., & Sung, J.-S. (2020). Antibacterial and Anti-Inflammatory Effects of Novel Peptide Toxin from the Spider Pardosa astrigera. Antibiotics, 9(7), 422. https://doi.org/10.3390/antibiotics9070422




