Data Shepherding in Nanotechnology: An Antimicrobial Functionality Data Capture Template
Abstract
:1. Introduction
2. Materials and Methods
3. Results
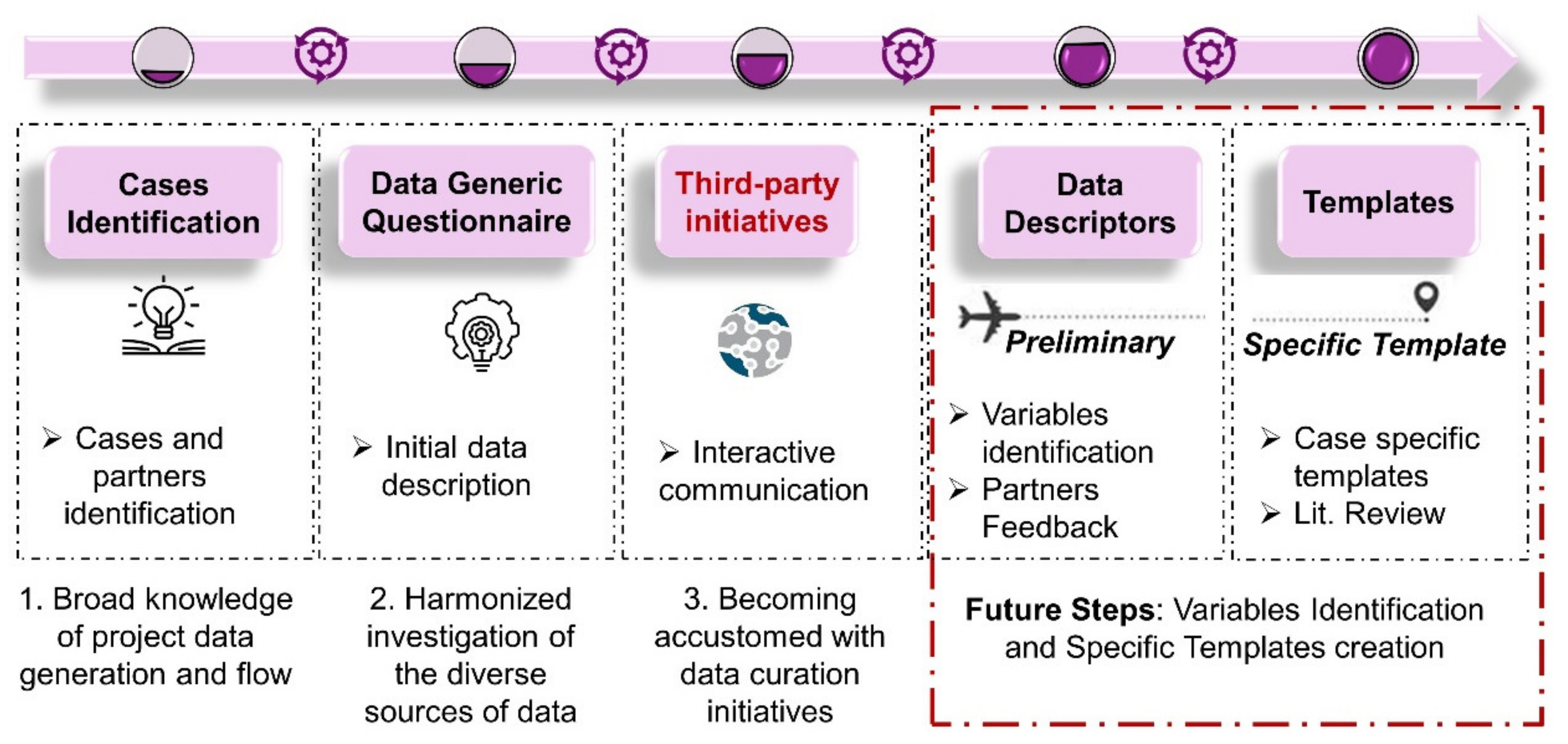
3.1. Initial Data Description—Questionnaire
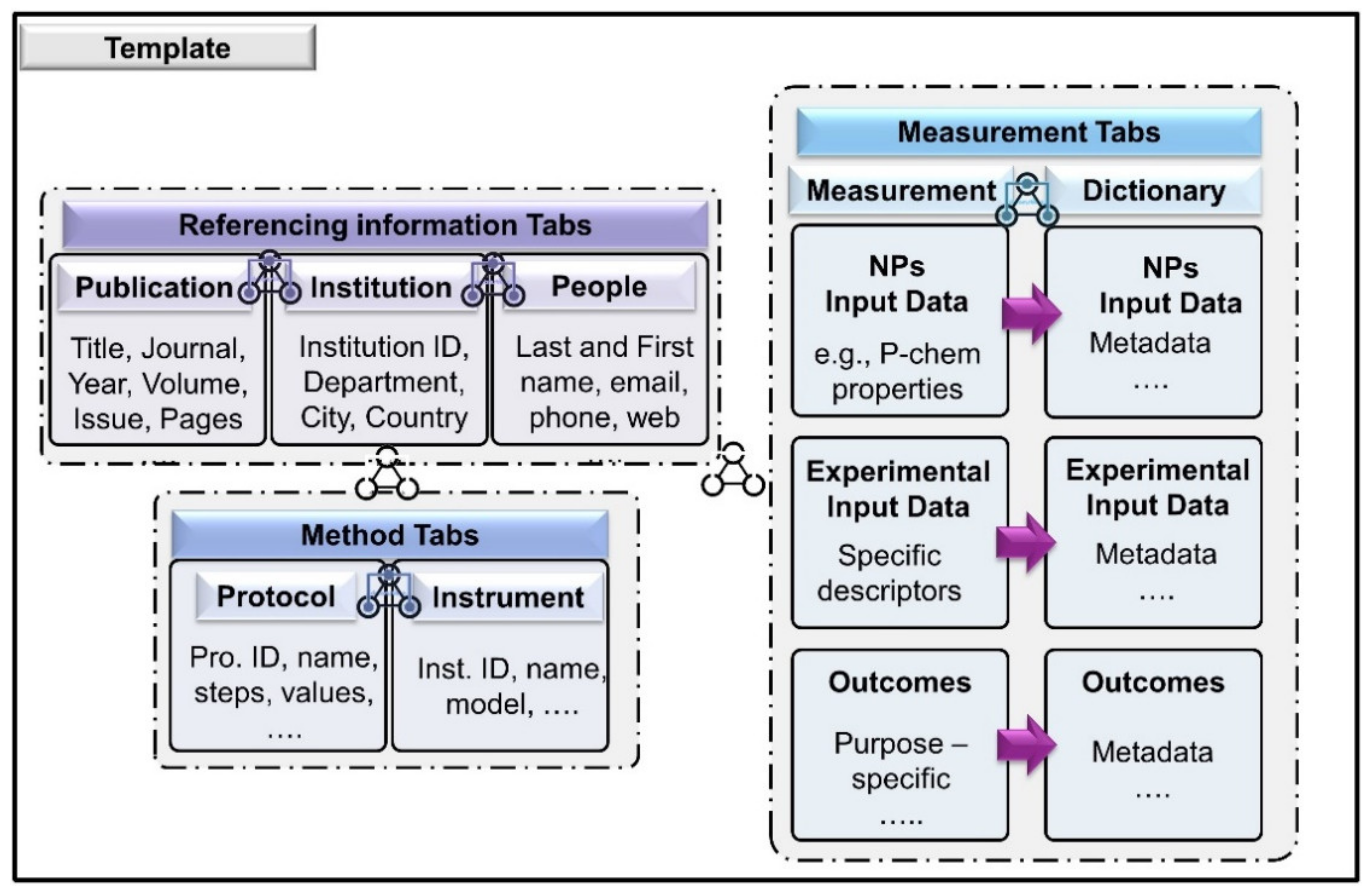
3.2. Preliminary Template from NIKC
3.3. Literature, Internal Communication and Descriptors Identification
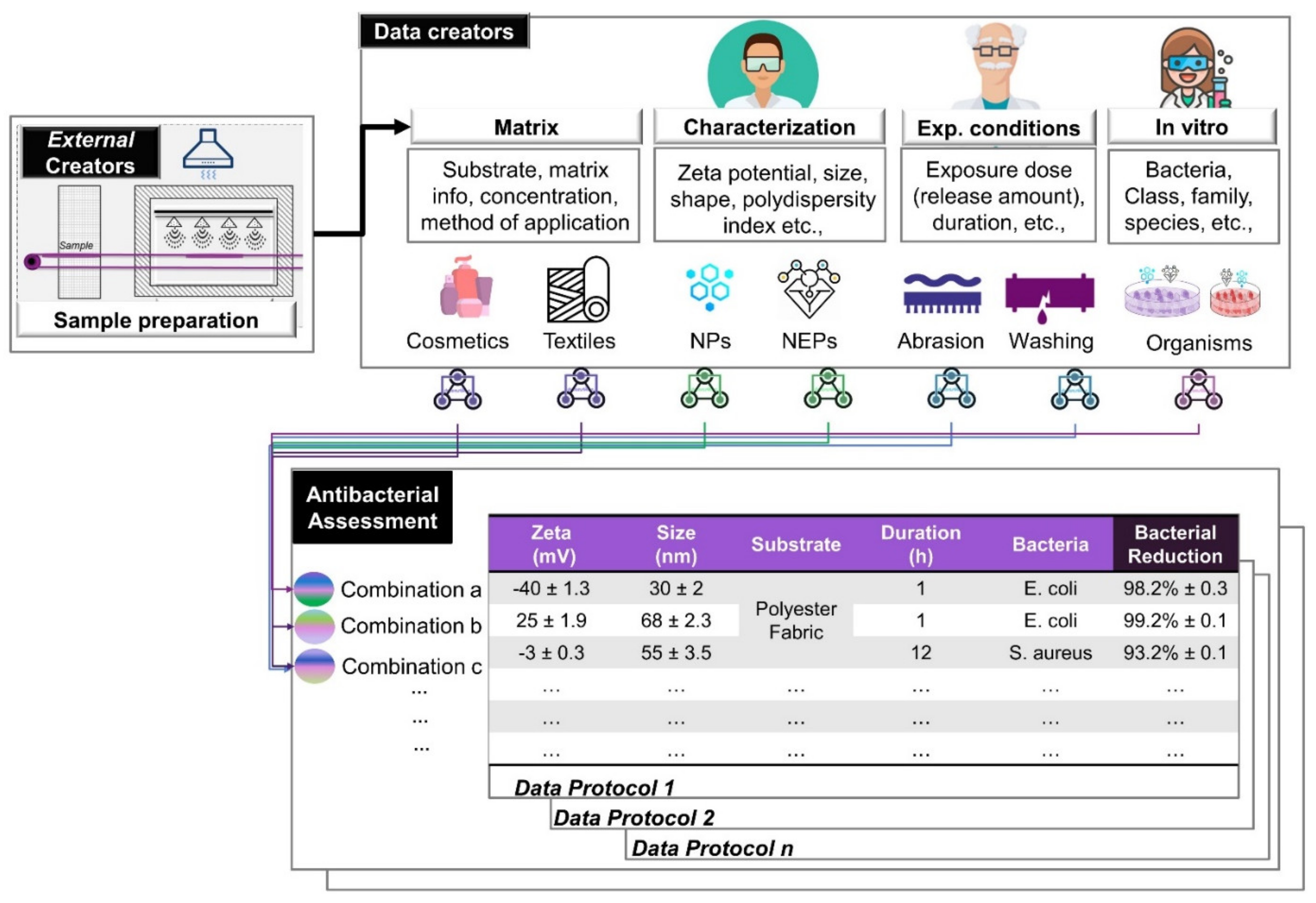
3.3.1. Nanostructured Materials Descriptors
3.3.2. Experimental Setup Descriptors
3.3.3. Antimicrobial Capacity
3.4. Annotation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Yang, X.; Chung, E.; Johnston, I.; Ren, G.; Cheong, Y.K. Exploitation of Antimicrobial Nanoparticles and Their Applications in Biomedical Engineering. Appl. Sci. 2021, 11, 4520. [Google Scholar] [CrossRef]
- Davey, M.E.; O’Toole, G.A. Microbial biofilms: From ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, K.S.; Patwardhan, D.; Jayan, G. Biofilms, medical devices, and antibiofilm technology: Key messages from a recent public workshop. Am. J. Infect. Control. 2015, 43, 2–3. [Google Scholar] [CrossRef]
- Gajdács, M.; Urbán, E.; Stájer, A.; Baráth, Z. Antimicrobial Resistance in the Context of the Sustainable Development Goals: A Brief Review. Eur. J. Investig. Health Psychol. Educ. 2021, 11, 71–82. [Google Scholar] [CrossRef]
- Ruparelia, J.P.; Chatterjee, A.K.; Duttagupta, S.P.; Mukherji, S. Strain specificity in antimicrobial activity of silver and copper nanoparticles. Acta Biomater. 2008, 4, 707–716. [Google Scholar] [CrossRef]
- Alavi, M.; Rai, M. Recent advances in antibacterial applications of metal nanoparticles (MNPs) and metal nanocomposites (MNCs) against multidrug-resistant (MDR) bacteria. Expert Rev. Anti-Infect. Ther. 2019, 17, 419–428. [Google Scholar] [CrossRef]
- Hariharan, H.; Al-Harbi, N.; Karuppiah, P.; Rajaram, S. Microbial synthesis of selenium nanocomposite using Saccharomyces cerevisiae and its antimicrobial activity against pathogens causing nosocomial infection. Chalcogenide Lett. 2012, 9, 509–515. [Google Scholar]
- Saqib, S.; Zaman, W.; Ullah, F.; Majeed, I.; Ayaz, A.; Hussain Munis, M.F. Organometallic assembling of chitosan-Iron oxide nanoparticles with their antifungal evaluation against Rhizopus oryzae. Appl. Organomet. Chem. 2019, 33, e5190. [Google Scholar] [CrossRef]
- Asghar, M.; Habib, S.; Zaman, W.; Hussain, S.; Ali, H.; Saqib, S. Synthesis and characterization of microbial mediated cadmium oxide nanoparticles. Microsc. Res. Tech. 2020, 83, 1574–1584. [Google Scholar] [CrossRef]
- Bhati-Kushwaha, H.; Malik, C. Assessment of Antibacterial and Antifungal Activities of Silver Nanoparticles Obtained from the Callus Extracts (Stem and Leaf) of Tridax procumbens L. Indian J. Biotechnol. 2014, 13, 114–120. [Google Scholar]
- Bhavnani, S.M.; Krause, K.M.; Ambrose, P.G. A Broken Antibiotic Market: Review of Strategies to Incentivize Drug Development; Oxford University Press US: Oxford, England, 2020. [Google Scholar]
- Bush, K.; Pucci, M.J. New antimicrobial agents on the horizon. Biochem. Pharmacol. 2011, 82, 1528–1539. [Google Scholar] [CrossRef]
- Cheesman, M.J.; Ilanko, A.; Blonk, B.; Cock, I.E. Developing new antimicrobial therapies: Are synergistic combinations of plant extracts/compounds with conventional antibiotics the solution? Pharmacogn. Rev. 2017, 11, 57. [Google Scholar]
- Furxhi, I.; Murphy, F.; Mullins, M.; Arvanitis, A.; Poland, C.A. Practices and Trends of Machine Learning Application in Nanotoxicology. Nanomaterials 2020, 10, 116. [Google Scholar] [CrossRef] [Green Version]
- Adir, O.; Poley, M.; Chen, G.; Froim, S.; Krinsky, N.; Shklover, J.; Shainsky-Roitman, J.; Lammers, T.; Schroeder, A. Integrating Artificial Intelligence and Nanotechnology for Precision Cancer Medicine. Adv. Mater. 2020, 32, 1901989. [Google Scholar] [CrossRef]
- Winkler, D.A. Role of Artificial Intelligence and Machine Learning in Nanosafety. Small 2020, 16, 2001883. [Google Scholar] [CrossRef]
- Jeliazkova, N.; Apostolova, M.D.; Andreoli, C.; Barone, F.; Barrick, A.; Battistelli, C.; Bossa, C.; Botea-Petcu, A.; Châtel, A.; De Angelis, I.; et al. Towards FAIR nanosafety data. Nat. Nanotechnol. 2021, 16, 644–654. [Google Scholar] [CrossRef]
- Furxhi, I.; Murphy, F.; Mullins, M.; Arvanitis, A.; Poland, C.A. Nanotoxicology data for in silico tools: A literature review. Nanotoxicology 2020, 14, 612–637. [Google Scholar] [CrossRef]
- Dunning, A.; Smaele, M.; Bohmer, J. Are the FAIR Data Principles fair? Int. J. Digit. Curation 2017, 12, 177–195. [Google Scholar] [CrossRef]
- Boeckhout, M.; Zielhuis, G.A.; Bredenoord, A.L. The FAIR guiding principles for data stewardship: Fair enough? Eur. J. Hum. Genet. 2018, 26, 931–936. [Google Scholar] [CrossRef] [Green Version]
- Tropsha, A.; Mills, K.C.; Hickey, A.J. Reproducibility, sharing and progress in nanomaterial databases. Nat. Nanotechnol. 2017, 12, 1111–1114. [Google Scholar] [CrossRef]
- Mirzaei, M.; Furxhi, I.; Murphy, F.; Mullins, M. A Machine Learning Tool to Predict the Antibacterial Capacity of Nanoparticles. Nanomaterials 2021, 11, 1774. [Google Scholar] [CrossRef]
- van Vlijmen, H.; Mons, A.; Waalkens, A.; Franke, W.; Baak, A.; Ruiter, G.; Kirkpatrick, C.; da Silva Santos, L.O.B.; Meerman, B.; Jellema, R.; et al. The Need of Industry to Go FAIR. Data Intell. 2020, 2, 276–284. [Google Scholar] [CrossRef]
- Wise, J.; de Barron, A.G.; Splendiani, A.; Balali-Mood, B.; Vasant, D.; Little, E.; Mellino, G.; Harrow, I.; Smith, I.; Taubert, J.; et al. Implementation and relevance of FAIR data principles in biopharmaceutical R&D. Drug Discov. Today 2019, 24, 933–938. [Google Scholar]
- European Commission. Communication from the Commission to the European Parliament, the European Council, the Council, the European Economic and Social Committee and the Committee of the Regions a New Industrial Strategy for Europe; Secretariat-General: Brussels, Belgium, 2020. [Google Scholar]
- European Commission. Communication from the Commission to the European Parliament, the Council, the European Economic and Social Committee and the Committee of the Regions Chemicals Strategy for Sustainability towards a Toxic-Free Environment; Secretariat-General: Brussels, Belgium, 2020. [Google Scholar]
- European Commission. H2020 Programme: Guidelines on FAIR Data Management in Horizon 2020; Directorate-General for Research & Innovation: Brussels, Belgium, 2016. [Google Scholar]
- Romanos, N.; Kalogerini, M.; Koumoulos, E.P.; Morozinis, A.K.; Sebastiani, M.; Charitidis, C. Innovative Data Management in advanced characterization: Implications for materials design. Mater. Today Commun. 2019, 20, 100541. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [Green Version]
- Jacobsen, A.; de Miranda Azevedo, R.; Juty, N.; Batista, D.; Coles, S.; Cornet, R.; Courtot, M.; Crosas, M.; Dumontier, M.; Evelo, C.T.; et al. FAIR Principles: Interpretations and Implementation Considerations. Data Intell. 2020, 2, 10–29. [Google Scholar] [CrossRef]
- Bloemers, M.; Montesanti, A. The FAIR Funding Model: Providing a Framework for Research Funders to Drive the Transition toward FAIR Data Management and Stewardship Practices. Data Intell. 2020, 2, 171–180. [Google Scholar] [CrossRef]
- Papadiamantis, A.G.; Klaessig, F.C.; Exner, T.E.; Hofer, S.; Hofstaetter, N.; Himly, M.; Williams, M.A.; Doganis, P.; Hoover, M.D.; Afantitis, A.; et al. Metadata Stewardship in Nanosafety Research: Community-Driven Organisation of Metadata Schemas to Support FAIR Nanoscience Data. Nanomaterials 2020, 10, 2033. [Google Scholar] [CrossRef]
- Eleraky, N.E.; Allam, A.; Hassan, S.B.; Omar, M.M. Nanomedicine Fight against Antibacterial Resistance: An Overview of the Recent Pharmaceutical Innovations. Pharmaceutics 2020, 12, 142. [Google Scholar] [CrossRef] [Green Version]
- Furxhi, I.; Arvanitis, A.; Murphy, F.; Costa, A.; Blosi, M. Data Shepherding in Nanotechnology. The Initiation. Nanomaterials 2021, 11, 1520. [Google Scholar] [CrossRef] [PubMed]
- Furxhi, I.; Koivisto, A.J.; Murphy, F.; Trabucco, S.; Del Secco, B.; Arvanitis, A. Data Shepherding in Nanotechnology. The Exposure Field Campaign Template. Nanomaterials 2021, 11, 1818. [Google Scholar] [CrossRef] [PubMed]
- Furxhi, I.; Massimo, P.; Murphy, F.; Costa, A. A Data Driven Methodological Sustainable and Safe by Design Approach and Tool. Front. Bioeng. Biotechnol. Nanobiotechnol. 2021. paper under Review: Perspective. [Google Scholar]
- Powers, C.M.; Mills, K.A.; Morris, S.A.; Klaessig, F.; Gaheen, S.; Lewinski, N.; Ogilvie Hendren, C. Nanocuration workflows: Establishing best practices for identifying, inputting, and sharing data to inform decisions on nanomaterials. Beilstein J. Nanotechnol. 2015, 6, 1860–1871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holec, D.; Dumitraschkewitz, P.; Vollath, D.; Fischer, F.D. Surface energy of Au nanoparticles depending on their size and shape. Nanomaterials 2020, 10, 484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gholami, A.; Habibi, B.; Matin, A.A.; Samadi, N. Controlled release of anticancer drugs via the magnetic magnesium iron nanoparticles modified by graphene oxide and polyvinyl alcohol: Paclitaxel and docetaxel. Nanomed. J. 2021, 8, 200–210. [Google Scholar]
- Yi, Z.; Loosli, F.; Wang, J.; Berti, D.; Baalousha, M. How to distinguish natural versus engineered nanomaterials: Insights from the analysis of TiO2 and CeO2 in soils. Environ. Chem. Lett. 2020, 18, 215–227. [Google Scholar] [CrossRef]
- Sarker, N.C.; Ray, P.; Pfau, C.; Kalavacharla, V.; Hossain, K.; Quadir, M. Development of functional nanomaterials from wheat bran derived arabinoxylan for nucleic acid delivery. J. Agric. Food Chem. 2020, 68, 4367–4373. [Google Scholar] [CrossRef]
- Shen, X.; Wang, Z.; Gao, X.; Zhao, Y. Density Functional Theory-Based Method to Predict the Activities of Nanomaterials as Peroxidase Mimics. ACS Catal. 2020, 10, 12657–12665. [Google Scholar] [CrossRef]
- Durán, N.; Durán, M.; De Jesus, M.B.; Seabra, A.B.; Fávaro, W.J.; Nakazato, G. Silver nanoparticles: A new view on mechanistic aspects on antimicrobial activity. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 789–799. [Google Scholar] [CrossRef]
- Hajipour, M.J.; Fromm, K.M.; Ashkarran, A.A.; de Aberasturi, D.J.; de Larramendi, I.R.; Rojo, T.; Serpooshan, V.; Parak, W.J.; Mahmoudi, M. Antibacterial properties of nanoparticles. Trends Biotechnol. 2012, 30, 499–511. [Google Scholar] [CrossRef] [Green Version]
- Nel, A. Atmosphere. Air pollution-related illness: Effects of particles. Science 2005, 308, 804–806. [Google Scholar] [CrossRef]
- Soenen, S.J.; Rivera-Gil, P.; Montenegro, J.-M.; Parak, W.J.; De Smedt, S.C.; Braeckmans, K. Cellular toxicity of inorganic nanoparticles: Common aspects and guidelines for improved nanotoxicity evaluation. Nano Today 2011, 6, 446–465. [Google Scholar] [CrossRef]
- Nel, A.E.; Mädler, L.; Velegol, D.; Xia, T.; Hoek, E.M.; Somasundaran, P.; Klaessig, F.; Castranova, V.; Thompson, M. Understanding biophysicochemical interactions at the nano–bio interface. Nat. Mater. 2009, 8, 543–557. [Google Scholar] [CrossRef]
- Lallo da Silva, B.; Caetano, B.L.; Chiari-Andréo, B.G.; Pietro, R.C.L.R.; Chiavacci, L.A. Increased antibacterial activity of ZnO nanoparticles: Influence of size and surface modification. Colloids Surf. B Biointerfaces 2019, 177, 440–447. [Google Scholar] [CrossRef]
- Pal, S.; Tak, Y.K.; Song, J.M. Does the Antibacterial Activity of Silver Nanoparticles Depend on the Shape of the Nanoparticle? A Study of the Gram-Negative Bacterium Escherichia coli. Appl. Environ. Microbiol. 2007, 73, 1712–1720. [Google Scholar] [CrossRef] [Green Version]
- Suttiponparnit, K.; Jiang, J.; Sahu, M.; Suvachittanont, S.; Charinpanitkul, T.; Biswas, P. Role of surface area, primary particle size, and crystal phase on titanium dioxide nanoparticle dispersion properties. Nanoscale Res. Lett. 2011, 6, 27. [Google Scholar] [CrossRef] [Green Version]
- Tang, S.; Zheng, J. Antibacterial Activity of Silver Nanoparticles: Structural Effects. Adv. Healthc. Mater. 2018, 7, 1701503. [Google Scholar] [CrossRef]
- Simon-Deckers, A.; Loo, S.; Mayne-L’hermite, M.; Herlin-Boime, N.; Menguy, N.; Reynaud, C.; Gouget, B.; Carrière, M. Size-, Composition- and Shape-Dependent Toxicological Impact of Metal Oxide Nanoparticles and Carbon Nanotubes toward Bacteria. Environ. Sci. Technol. 2009, 43, 8423–8429. [Google Scholar] [CrossRef]
- Panáček, A.; Kvitek, L.; Prucek, R.; Kolář, M.; Večeřová, R.; Pizúrová, N.; Sharma, V.K.; Nevěčná, T.j.; Zbořil, R. Silver colloid nanoparticles: Synthesis, characterization, and their antibacterial activity. J. Phys. Chem. B 2006, 110, 16248–16253. [Google Scholar] [CrossRef]
- Pan, X.; Wang, Y.; Chen, Z.; Pan, D.; Cheng, Y.; Liu, Z.; Lin, Z.; Guan, X. Investigation of antibacterial activity and related mechanism of a series of nano-Mg(OH)2. ACS Appl. Mater. Interfaces 2013, 5, 1137–1142. [Google Scholar] [CrossRef]
- Chen, J.; Li, S.; Luo, J.; Wang, R.; Ding, W. Enhancement of the Antibacterial Activity of Silver Nanoparticles against Phytopathogenic Bacterium Ralstonia solanacearum by Stabilization. J. Nanomater. 2016, 2016, 7135852. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227–1249. [Google Scholar] [CrossRef] [Green Version]
- Armijo, L.M.; Wawrzyniec, S.J.; Kopciuch, M.; Brandt, Y.I.; Rivera, A.C.; Withers, N.J.; Cook, N.C.; Huber, D.L.; Monson, T.C.; Smyth, H.D.C.; et al. Antibacterial activity of iron oxide, iron nitride, and tobramycin conjugated nanoparticles against Pseudomonas aeruginosa biofilms. J. Nanobiotechnol. 2020, 18, 35. [Google Scholar] [CrossRef] [Green Version]
- Daly, C.A.; Hernandez, R. Learning from the Machine: Uncovering Sustainable Nanoparticle Design Rules. J. Phys. Chem. C 2020, 124, 13409–13420. [Google Scholar] [CrossRef]
- Thatikayala, D.; Jayarambabu, N.; Banothu, V.; Ballipalli, C.B.; Park, J.; Rao, K.V. Biogenic synthesis of silver nanoparticles mediated by Theobroma cacao extract: Enhanced antibacterial and photocatalytic activities. J. Mater. Sci. Mater. Electron. 2019, 30, 17303–17313. [Google Scholar] [CrossRef]
- Wu, T.; Wu, C.; Fu, S.; Wang, L.; Yuan, C.; Chen, S.; Hu, Y. Integration of lysozyme into chitosan nanoparticles for improving antibacterial activity. Carbohydr. Polym. 2017, 155, 192–200. [Google Scholar] [CrossRef]
- Budama, L.; Çakır, B.A.; Topel, Ö.; Hoda, N. A new strategy for producing antibacterial textile surfaces using silver nanoparticles. Chem. Eng. J. 2013, 228, 489–495. [Google Scholar] [CrossRef]
- Slavin, Y.N.; Asnis, J.; Häfeli, U.O.; Bach, H. Metal nanoparticles: Understanding the mechanisms behind antibacterial activity. J. Nanobiotechnol. 2017, 15, 65. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, V.P.; Rajput, K.; Mondal Roy, S.; Chaudhuri, T.K.; Roy, D.R. Experimental and first-principles investigation on the structural, electronic and antimicrobial properties of nickel hydroxide nanoparticles. J. Phys. Chem. Solids 2022, 160, 110367. [Google Scholar] [CrossRef]
- Mohammadi, A.; Hashemi, M.; Masoud Hosseini, S. Effect of chitosan molecular weight as micro and nanoparticles on antibacterial activity against some soft rot pathogenic bacteria. LWT Food Sci. Technol. 2016, 71, 347–355. [Google Scholar] [CrossRef]
- Yalcinkaya, F.; Komarek, M.; Lubasova, D.; Sanetrnik, F.; Maryska, J. Preparation of Antibacterial Nanofibre/Nanoparticle Covered Composite Yarns. J. Nanomater. 2016, 2016, 7565972. [Google Scholar] [CrossRef]
- Textor, T.; Fouda, M.M.; Mahltig, B. Deposition of durable thin silver layers onto polyamides employing a heterogeneous Tollens’ reaction. Appl. Surf. Sci. 2010, 256, 2337–2342. [Google Scholar] [CrossRef]
- Montazer, M.; Shamei, A.; Alimohammadi, F. Synthesizing and stabilizing silver nanoparticles on polyamide fabric using silver-ammonia/PVP/UVC. Prog. Org. Coat. 2012, 75, 379–385. [Google Scholar] [CrossRef]
- Mao, J.W.; Murphy, L. Durable freshness for textiles. AATCC Rev. 2001, 1, 28–31. [Google Scholar]
- Hady, A.; Farouk, A.; Sharaf, S. Flame retardancy and UV protection of cotton based fabrics using nano ZnO and polycarboxylic acid. Carbohydr. Polym. 2013, 92, 400–406. [Google Scholar] [CrossRef]
- Sasaki, K.; Tenjimbayashi, M.; Manabe, K.; Shiratori, S. Asymmetric superhydrophobic/superhydrophilic cotton fabrics designed by spraying polymer and nanoparticles. ACS Appl. Mater. Interfaces 2016, 8, 651–659. [Google Scholar] [CrossRef]
- Sun, Y.; Sun, G. Durable and regenerable antimicrobial textile materials prepared by a continuous grafting process. J. Appl. Polym. Sci. 2002, 84, 1592–1599. [Google Scholar] [CrossRef]
- Catauro, M.; Barrino, F.; Blanco, I.; Piccolella, S.; Pacifico, S. Use of the Sol–Gel Method for the Preparation of Coatings of Titanium Substrates with Hydroxyapatite for Biomedical Application. Coatings 2020, 10, 203. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, A.; Jarrais, B.; Pereira, C.; Morgado, J.; Freire, C.; Pereira, M. Functionalization of textiles with multi-walled carbon nanotubes by a novel dyeing-like process. J. Mater. Sci. 2012, 47, 5263–5275. [Google Scholar] [CrossRef]
- Petkova, P.; Francesko, A.; Perelshtein, I.; Gedanken, A.; Tzanov, T. Simultaneous sonochemical-enzymatic coating of medical textiles with antibacterial ZnO nanoparticles. Ultrason. Sonochem. 2016, 29, 244–250. [Google Scholar] [CrossRef]
- Perelshtein, I.; Applerot, G.; Perkas, N.; Wehrschetz-Sigl, E.; Hasmann, A.; Guebitz, G.; Gedanken, A. Antibacterial properties of an in situ generated and simultaneously deposited nanocrystalline ZnO on fabrics. ACS Appl. Mater. Interfaces 2009, 1, 361–366. [Google Scholar] [CrossRef]
- Sun, G.; Hong, K.H. Photo-induced antimicrobial and decontaminating agents: Recent progresses in polymer and textile applications. Text. Res. J. 2013, 83, 532–542. [Google Scholar] [CrossRef]
- Klemenčič, D.; Tomšič, B.; Kovač, F.; Žerjav, M.; Simončič, A.; Simončič, B. Preparation of novel fibre–silica–Ag composites: The influence of fibre structure on sorption capacity and antimicrobial activity. J. Mater. Sci. 2014, 49, 3785–3794. [Google Scholar] [CrossRef]
- Jana, T.K.; Jana, S.K.; Kumar, A.; De, K.; Maiti, R.; Mandal, A.K.; Chatterjee, T.; Chatterjee, B.K.; Chakrabarti, P.; Chatterjee, K. The antibacterial and anticancer properties of zinc oxide coated iron oxide nanotextured composites. Colloids Surf. B Biointerfaces 2019, 177, 512–519. [Google Scholar] [CrossRef]
- El-Sheikh, M.A.; El Gabry, L.K.; Ibrahim, H.M. Photosynthesis of Carboxymethyl Starch-Stabilized Silver Nanoparticles and Utilization to Impart Antibacterial Finishing for Wool and Acrylic Fabrics. J. Polym. 2013, 2013, 792035. [Google Scholar] [CrossRef]
- Bajpai, V.; Dey, A.; Ghosh, S.; Bajpai, S.; Jha, M.K. Quantification of bacterial adherence on different textile fabrics. Int. Biodeterior. Biodegrad. 2011, 65, 1169–1174. [Google Scholar] [CrossRef]
- Hsieh, Y.-L.; Timm, D.A.; Merry, J. Bacterial adherence on fabrics by a radioisotope labeling method. Text. Res. J. 1987, 57, 20–28. [Google Scholar] [CrossRef]
- Saleem, H.; Zaidi, S.J. Sustainable Use of Nanomaterials in Textiles and Their Environmental Impact. Materials 2020, 13, 5134. [Google Scholar] [CrossRef]
- Rehman, A.; Houshyar, S.; Wang, X. Nanodiamond-Based Fibrous Composites: A Review of Fabrication Methods, Properties, and Applications. ACS Appl. Nano Mater. 2021, 4, 2317–2332. [Google Scholar] [CrossRef]
- Som, C.; Wick, P.; Krug, H.; Nowack, B. Environmental and health effects of nanomaterials in nanotextiles and façade coatings. Environ. Int. 2011, 37, 1131–1142. [Google Scholar] [CrossRef]
- Limpiteeprakan, P.; Babel, S.; Lohwacharin, J.; Takizawa, S. Release of silver nanoparticles from fabrics during the course of sequential washing. Environ. Sci. Pollut. Res. 2016, 23, 22810–22818. [Google Scholar] [CrossRef] [PubMed]
- Mitrano, D.M.; Limpiteeprakan, P.; Babel, S.; Nowack, B. Durability of nano-enhanced textiles through the life cycle: Releases from landfilling after washing. Environ. Sci. Nano 2016, 3, 375–387. [Google Scholar] [CrossRef]
- Mitrano, D.M.; Rimmele, E.; Wichser, A.; Erni, R.; Height, M.; Nowack, B. Presence of nanoparticles in wash water from conventional silver and nano-silver textiles. ACS Nano 2014, 8, 7208–7219. [Google Scholar] [CrossRef]
- Gabrielyan, L.; Hovhannisyan, A.; Gevorgyan, V.; Ananyan, M.; Trchounian, A. Antibacterial effects of iron oxide (Fe3O4) nanoparticles: Distinguishing concentration-dependent effects with different bacterial cells growth and membrane-associated mechanisms. Appl. Microbiol. Biotechnol. 2019, 103, 2773–2782. [Google Scholar] [CrossRef] [PubMed]
- Pazos-Ortiz, E.; Roque-Ruiz, J.H.; Hinojos-Márquez, E.A.; López-Esparza, J.; Donohué-Cornejo, A.; Cuevas-González, J.C.; Espinosa-Cristóbal, L.F.; Reyes-López, S.Y. Dose-dependent antimicrobial activity of silver nanoparticles on polycaprolactone fibers against gram-positive and gram-negative bacteria. J. Nanomater. 2017, 2017, 4752314. [Google Scholar] [CrossRef] [Green Version]
- Haase, H.; Jordan, L.; Keitel, L.; Keil, C.; Mahltig, B. Comparison of methods for determining the effectiveness of antibacterial functionalized textiles. PLoS ONE 2017, 12, e0188304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joyce, M.; Woods, C.W. Antibacterial susceptibility testing in the clinical laboratory. Infect. Dis. Clin. N. Am. 2004, 18, 401–434. [Google Scholar] [CrossRef] [PubMed]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Blosi, M.; Costa, A.L.; Ortelli, S.; Belosi, F.; Ravegnani, F.; Varesano, A.; Tonetti, C.; Zanoni, I.; Vineis, C. Polyvinyl alcohol/silver electrospun nanofibers: Biocidal filter media capturing virus-size particles. J. Appl. Polym. Sci. 2021, 138, 51380. [Google Scholar] [CrossRef]
- Joiner, B.G. Determining Antimicrobial Efficacy and Biocompatibility of Treated Articles Using Standard Test Methods. In Bioactive Fibers and Polymers; ACS Publications: Washington, DC, USA, 2001; Available online: https://www.researchgate.net/publication/286910803_Determining_Antimicrobial_Efficacy_and_Biocompatibility_of_Treated_Articles_Using_Standard_Test_Methods (accessed on 29 November 2021).
- Höfer, D. Antimicrobial textiles—Evaluation of their effectiveness and safety. Biofunct. Text. Ski. 2006, 33, 42–50. [Google Scholar]
- Zille, A.; Almeida, L.; Amorim, T.; Carneiro, N.; Esteves, M.F.; Silva, C.J.; Souto, A.P. Application of nanotechnology in antimicrobial finishing of biomedical textiles. Mater. Res. Express 2014, 1, 032003. [Google Scholar] [CrossRef] [Green Version]
| Antimicrobial Capacity | |||
|---|---|---|---|
| Element | Response—Data creators (STIIMA-CNR) | Response—Data creators (BioNanoPlus) | Response—Data reusers (RedOfView and CENTI) |
| Data Identification | |||
| Dataset description | Antimicrobial tests on coated textiles before and after use simulation tests | Functionality analysis of nanostructured capsules that deliver active phases in cosmetics | Antimicrobial tests of cosmetic formulations |
| Source | Data from a measurement experiment | Data from a measurement experiment | Data from a measurement experiment |
| Partner’s Activities and Responsibilities | |||
| Partner owner of the data, copyright holder | The data that each partner provides to the template is under their responsibility | The data that each partner provides to the template is under their responsibility | The data that each partner provides to the template is under their responsibility |
| Partner in charge of data collection/analysis/storage/related WP(s) | Data collection between data creators and shepherd; construction of a measuring matrix in collaboration with stakeholders, as well as the reporting strategy throughout the project duration | Data collection between data creators and shepherd; construction of a measuring matrix in collaboration with stakeholders, as well as the reporting strategy throughout the project duration | Data creators and shepherd; bridging information for future analysis |
| Expected Input Variables | |||
| Description of the information required (WPs and/or tasks) in order to move forward | - P-chem characterization of NMs deposited into the textiles | Colloidal properties (particle size, zeta potential) of NMs once integrated into the use and testing matrices | Design of the measurement matrix with contextual information to facilitate testing |
| Expected outcomes | |||
| Description of the specific endpoint measurement variables/outcomes | Bacteria reduction (%): number of viable bacterial cells | Microbial log reduction |
|
| Standards | |||
| Detailed description of the methods/protocols |
| Internal protocol involving a disc diffusion test: E.coli, P. aeruginosa and S. aureus are grown in Petri dishes, applying the product to be tested and then measuring the inhibition halo (zone of inhibition) obtained |
|
| Category | Variables | Content | Metadata Description |
|---|---|---|---|
| Testing material | Nanomaterial (NM) | Text | NMs/nanoforms tested., ID code (ie., JRCxxx) or CAS for example. The core nanoform composition element. In the case of ASINA, a codification is used for internal communication purposes. |
| Nanoenabled products (NEPs) | Text | Nanoenabled product tested. In this case, specify the NMs that were used to produce the final NEP (textiles, cosmetics, etc.) first. In the comment section, the data creator can specify whether the antibacterial testing was performed on naked NMs or NEPs. | |
| Provider/ manufacturer | Text | Information related to the provider of NMs/NEPs. In the case of a project, this information allows for traceability among partner information exchanges. | |
| Batch | Text | A code provided by a manufacturer or distributor to a manufactured material or product that is anticipated to be homogeneous after production. In the case of ASINA, the batch can reflect a sample being produced and circulated from different partners for traceability. | |
| P-chem properties | Primary size | Number (nm) + SD | According to the EU definition, NM means a natural, incidental or manufactured material containing particles in an unbound state or as an aggregate or as agglomerate and where, for 50% or more of the particles in the number size distribution, one or more external dimensions is in the size range of 1—100 nm. |
| Shape | Text | The spatial arrangement of something as being distinct from its substance. High aspect ratio NMs include nanotubes and nanowires with diverse shapes. Small aspect ratio morphologies include spherical, oval, cubic and pillar [39]. | |
| Coating/ stabilizer | Text | Indicates the presence of a coating and/or of a stabilizer agent surrounding the particles [40]. Chemical composition of a coating layer on a material surface. | |
| Composition | Text and number | Chemical composition of a coating layer on a material surface. The metrics can be expressed as a percentage or a molar ratio. | |
| Hydrodynamic diameter | Number (nm) + SD | The diameter of a solid sphere that would exhibit the same hydrodynamic friction as the particle of interest [41]. The determined diameter is an indicator of the apparent size of the solvated particle that is approximated as being spherical. | |
| Polydispersity index (PDI) | Number + SD | A measurement of the size distribution, indicating the uniformity of NMs [42]. A measure of the heterogeneity of sizes of molecules or particles in a mixture. | |
| Zeta potential | Number (mV) + SD | The zeta potential is a measure of the electrical potential difference between the bulk fluid in which a particle is dispersed and the layer of fluid containing the oppositely charged ions that are associated with the nanoparticle’s surface. | |
| Medium tested | Number | Medium in which zeta potential was measured, e.g., water or culture. | |
| pH tested | Number | The pH in which the zeta potential was measured. In cases such as for cosmetics, the pH value could represent that of the final NMs solution since the sample is diluted before the zeta measurement, which results in a different pH. | |
| Density | Number (g/mL) | Mass (amount) of a material substance per unit volume [43]. | |
| Viscosity | Number (cP) | The resistance of a fluid to flow when it is subjected to a force. The result is usually represented in centipoise (cP), which is equal to 1 mPa s (millipascal second). | |
| Ion fraction (equilibrium) | Number (wt%) | Released ion mass fraction of particles. Particles in solution exist in an equilibrium between ions and NMs. Ions released in suspension represent the synthesis conversion of ions into NMs. The higher the ion fraction, the lower the conversion. The value is expressed as a mass percent of the total testing element [44]. | |
| Suspension concentration | Number (NM wt% ± SD) | The elemental concentration of NPs in a medium (water, cell media or biological matrix) | |
| Sample | Application method | Text | The method of NMs deposition: dip coating, spraying, sonochemical deposition, etc. |
| Substrate (matrix type) | Text | Description of the matrix (polyester fibers, cotton, polymethylmethacrylate, clay, epoxy, fil, liquid). | |
| Matrix | Text | Composition of the substrate, e.g., 50% X, 20% Y, 5% Z. | |
| Deposited concentration | Number (mg NM/g product ± SD) | Example: milligrams of active ingredient per gram of product for cosmetics; milligrams per gram of textile or grams per meter squared for textiles. |
| Category | Variables | Content | Metadata Description |
|---|---|---|---|
| Experimental conditions | Abrasion cycles | Number | Number of abrasion cycles. |
| Abrasion cycles to specimen breakdown | Number | Abrasion cycles to the fabric end-of-life. A physical test for the assessment of specimen breakdown [83,84]. Specimens abraded under a test pressure using abrasive cloth for a pre-determined number of cycles or until failure occurs. | |
| Washing cycles | Number | Washing cycles. | |
| Exposure conditions | Exposure dose | Number (e.g., ppm) | The dose exposed to the bacteria/microorganisms. Example: exposure dose for fabrics calculated in parts per million by considering the amount of the element on the fabrics (via ICP analysis) and the weight of fabric (1 g) per volume of inoculum (50 mL). The data creator should provide comments regarding what the dose reflects and how it was derived. |
| Exposure duration | Number (h) | Either exposure duration that an in vitro system is exposed to or the incubation contact time duration, depending on the methodology. | |
| Organisms | Culture medium | Text | Culture medium: information related to the in vitro biological system exposed. |
| Initial bacterium number | Text | Initial population of bacteria. | |
| Class | Text | One of the bacteria classification systems comprises five kingdoms that are further split into phylum, class, order, family, genus and species. Examples of class: Gammaproteobacteria, Sordariomycetes and Bacilli. | |
| Family | Text | Trichocomaceae, Enterobacteriaceae, Listeriaceae, etc. | |
| Species | Text | E. coli, P. aeruginosa, K. pneumoniae, etc. |
| Category | Variables | Content | Metadata Description |
|---|---|---|---|
| Measured outcomes | Bacteria reduction percentage | Number (%) | Bacteria reduction is the percentage ratio of the difference between the reference bacteria concentration (e.g., inoculum, untreated sample) and the bacteria concentration after contact with the sample, and the reference bacteria concentration [94]. |
| Minimum inhibitory concentration (MIC) | Number ( μ g/mL) | The MIC is the lowest concentration of antimicrobial agent that completely inhibits the growth of the organism in tubes or microdilution wells, as detected by the unaided eye [95]. | |
| Colony-forming unit concentration (CFU) | Number (CFU/mL) | Concentration of bacteria that can form a colony. | |
| Minimum bactericidal concentration (MBC) or minimum fungicidal concentration (MFC) | Number (μ g/mL) | The MBC is defined as the lowest concentration of antimicrobial agent that is needed to kill 99.9% of the final inoculum after incubation for 24 h under a standardized set of conditions [93]. It is also known as the minimum lethal concentration (MLC). | |
| Bacteria log reduction | Number | Bacteria log reduction is defined as the common logarithm of the ratio between the bacteria concentration after contact with the sample and the reference bacteria concentration (e.g., inoculum, untreated sample). | |
| Zone of inhibition (ZOI) | Number (mm) | ZOI is the gap without bacteria colonies around the sample on a contaminated solid medium. | |
| Optical density at 600 nm (OD600) | Number (absorbance) | Optical density at a wavelength of 600 nm is used for estimating the concentration of bacteria in a liquid. | |
| Half maximal inhibitory concentration (IC50) | Number (molar concentration (mol/L)) | IC50 is the capacity of a biocidal agent to inhibit the growth of a specific microorganism. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furxhi, I.; Varesano, A.; Salman, H.; Mirzaei, M.; Battistello, V.; Tomasoni, I.T.; Blosi, M. Data Shepherding in Nanotechnology: An Antimicrobial Functionality Data Capture Template. Coatings 2021, 11, 1486. https://doi.org/10.3390/coatings11121486
Furxhi I, Varesano A, Salman H, Mirzaei M, Battistello V, Tomasoni IT, Blosi M. Data Shepherding in Nanotechnology: An Antimicrobial Functionality Data Capture Template. Coatings. 2021; 11(12):1486. https://doi.org/10.3390/coatings11121486
Chicago/Turabian StyleFurxhi, Irini, Alessio Varesano, Hesham Salman, Mahsa Mirzaei, Vittoria Battistello, Ivonne Tonani Tomasoni, and Magda Blosi. 2021. "Data Shepherding in Nanotechnology: An Antimicrobial Functionality Data Capture Template" Coatings 11, no. 12: 1486. https://doi.org/10.3390/coatings11121486
APA StyleFurxhi, I., Varesano, A., Salman, H., Mirzaei, M., Battistello, V., Tomasoni, I. T., & Blosi, M. (2021). Data Shepherding in Nanotechnology: An Antimicrobial Functionality Data Capture Template. Coatings, 11(12), 1486. https://doi.org/10.3390/coatings11121486







