Naphthoquinones and Their Derivatives: Emerging Trends in Combating Microbial Pathogens
Abstract
:1. Introduction
2. Structural Diversity of Naphthoquinones Entities
3. 1,4-Naphthoquinone Derivatives
3.1. 1,4-Naphthoquinone Derivatives of Natural Origin
3.2. Chemically Synthesized 1,4-Naphthoquinone Derivatives
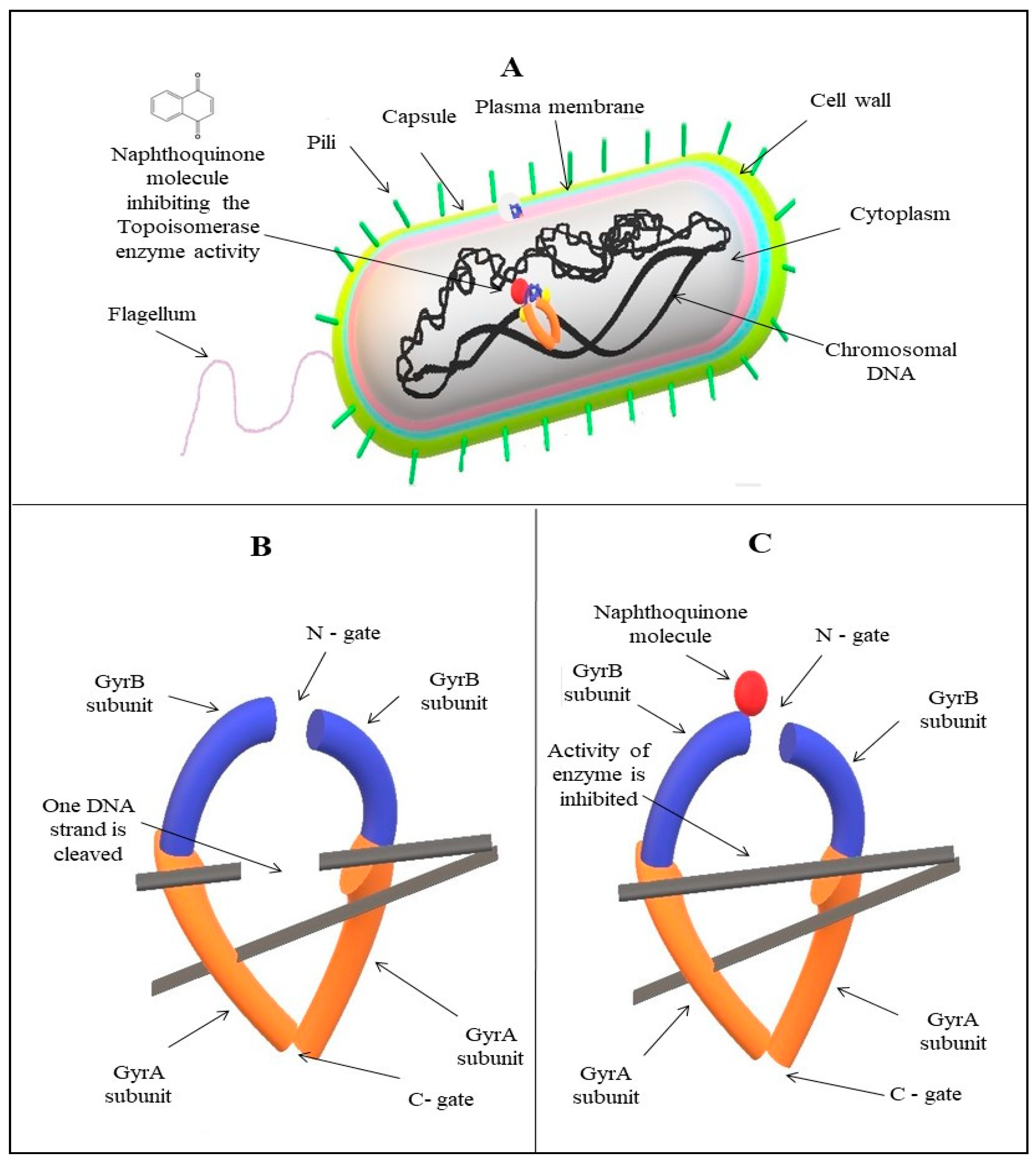
4. Structure–Activity Relationship and Bioinformatics Approach: Determining the Possible Mechanistic Role of 1,4-Naphthoquinones at the Molecular Level in a Cell
5. Possible Mechanistic Roles of Napthoquinones
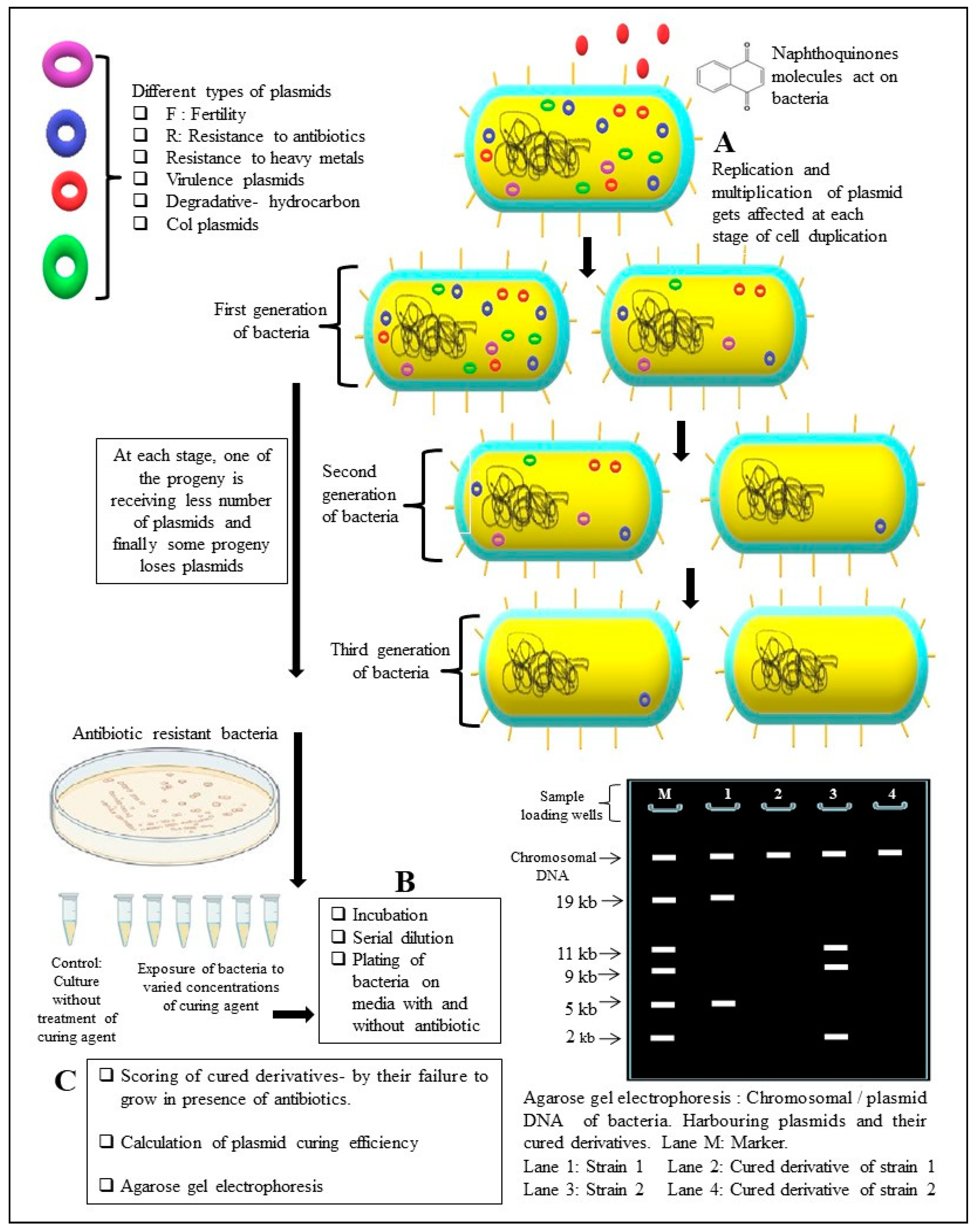
5.1. Efficiency to Cure Plasmids from Bacteria
5.2. Inhibition of Efflux Pumps (EPs)
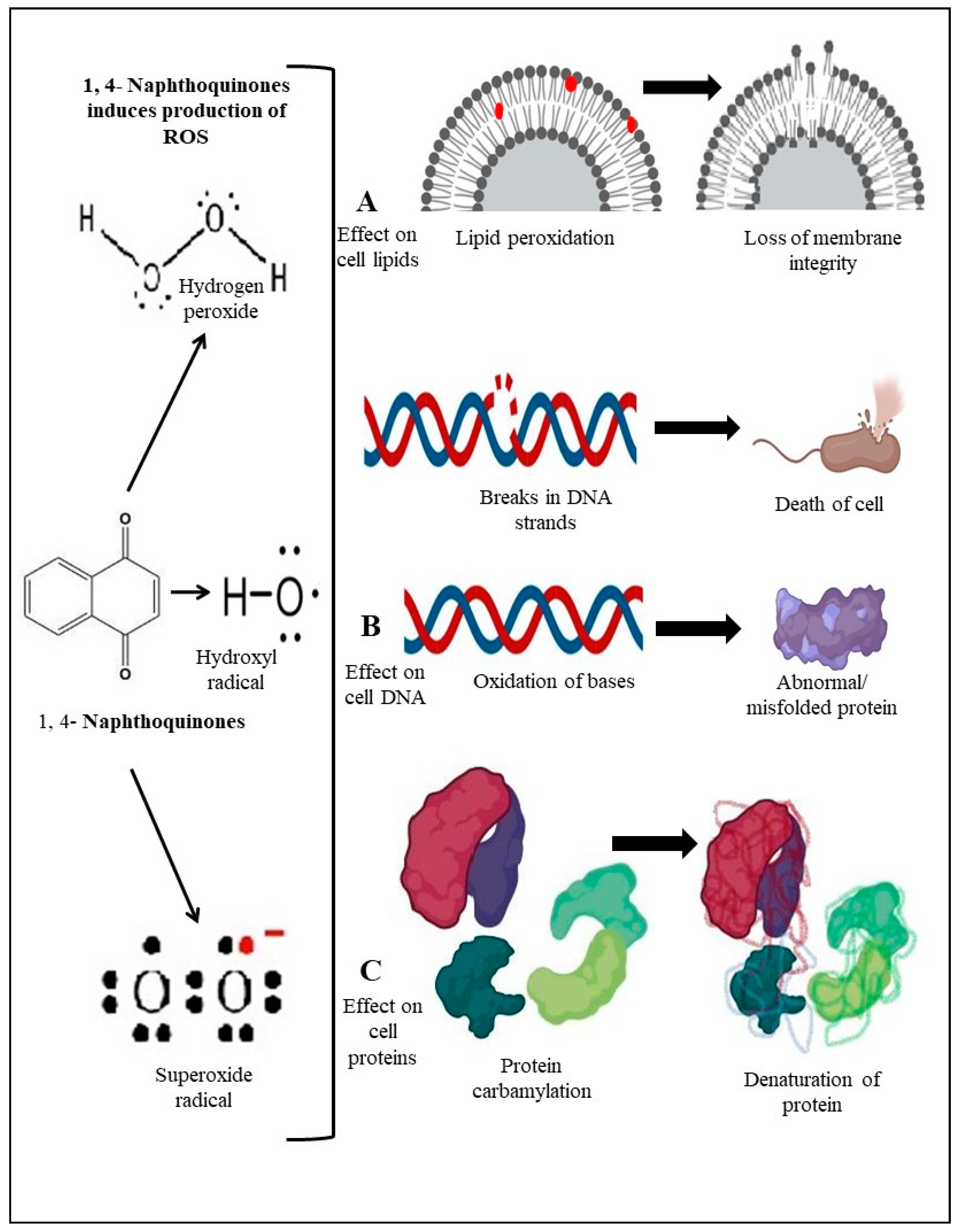
5.3. Generation of Reactive Oxygen Species (ROS) in Bacteria
5.4. Inhibition of Topoisomerase Enzyme
6. Antifungal Potential of Naphthoquinones
7. Antiparasitic Potential of Naphthoquinones
8. Future Prospects
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Gaynes, R. The discovery of Penicillin—New insights after more than 75 years of clinical use. Emerg. Infect. Dis. 2017, 23, 849–853. [Google Scholar] [CrossRef]
- Horner, W.H. Streptomycin. In Biosynthesis; Springer: Berlin/Heidelberg, Germany, 1967; pp. 373–399. [Google Scholar]
- Global Pharma News and Resources. Available online: https://www.pharmiweb.com/press-release/2020-07-02/global-antibiotics-market-is-expected-to-reach-at-a-cagr-of-47-during-the-forecast-period-2019-202 (accessed on 8 September 2020).
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Tanwar, J.; Das, S.; Fatima, Z.; Hameed, S. Multidrug resistance: An emerging crisis. Interdiscip. Perspect. Infect. Dis. 2014, 2014, 541340. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua=1 (accessed on 10 November 2020).
- World Health Organization. Available online: https://www.who.int/news-room/events/detail/2020/11/18/default-calendar/world-antimicrobial-awareness-week-2020 (accessed on 16 November 2020).
- Nikaido, H. Multidrug resistance in bacteria. Annu. Rev. Biochem. 2009, 78, 119–146. [Google Scholar] [CrossRef] [PubMed]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed Res. Int. 2016, 2016, 475067. [Google Scholar] [CrossRef] [PubMed]
- Sharifi-Rad, J.; Salehi, B.; Stojanović-Radić, Z.Z.; Fokou, P.V.T.; Sharifi-Rad, M.; Mahady, G.B.; Sharifi-Rad, M.; Masjedi, M.-R.; Lawal, T.O.; Ayatollahi, S.A.; et al. Medicinal plants used in the treatment of tuberculosis-ethnobotanical and ethnopharmacological approaches. Biotechnol. Adv. 2020, 44, 107629. [Google Scholar] [CrossRef] [PubMed]
- Tiwari Pandey, A.; Pandey, I.; Hachenberger, Y.; Krause, B.C.; Haidar, R.; Laux, P.; Luch, A.; Singh, M.P.; Singh, A.V. Emerging paradigm against global antimicrobial resistance via bioprospecting of mushroom into novel nanotherapeutics development. Trends Food Sci. Technol. 2020, 106, 333–344. [Google Scholar] [CrossRef]
- Tiwari Pandey, A.; Pandey, I.; Zamboni, P.; Gemmati, D.; Kanase, A.; Singh, A.V.; Singh, M.P. Traditional herbal remedies with a multifunctional therapeutic approach as an implication in COVID-19 associated co-infections. Coatings 2020, 10, 761. [Google Scholar] [CrossRef]
- Ansari, M.H.D.; Lavhale, S.; Kalunke, R.M.; Srivastava, P.L.; Pandit, V.; Gade, S.; Yadav, S.; Laux, P.; Luch, A.; Gemmati, D.; et al. Recent advances in plant nanobionics and nanobiosensors for toxicology applications. Curr. Nanosci. 2020, 16, 27–41. [Google Scholar] [CrossRef]
- Singh, V.; Kumar, V.; Kashyap, S.; Singh, A.V.; Kishore, V.; Sitti, M.; Saxena, P.S.; Srivastava, A. Graphene oxide synergistically enhances antibiotic efficacy in vancomycin-resistant Staphylococcus aureus. ACS Appl. Bio Mater. 2019, 2, 1148–1157. [Google Scholar] [CrossRef]
- Kurban, S.; Deniz, N.G.; Sayil, C.; Ozyurek, M.; Guclu, K.; Stasevych, M.; Zvarych, V.; Komarovska-Porokhnyavet, O.; Novikov, V. Synthesis, antimicrobial properties, and inhibition of catalase activity of 1,4-naphtho-and benzoquinone derivatives containing N-, S-, O-substituted. Heteroat. Chem. 2019, 2019, 1658417. [Google Scholar] [CrossRef]
- Sánchez-Calvo, J.M.; Barbero, G.R.; Guerrero-Vásquez, G.; Durán, A.G.; Macías, M.; Rodríguez-Iglesias, M.A.; Molinillo, J.M.; Macías, F.A. Synthesis, antibacterial and antifungal activities of naphthoquinone derivatives: A structure–activity relationship study. Med. Chem. Res. 2016, 25, 1274–1285. [Google Scholar] [CrossRef]
- Mathiyazhagan, K.; Kumaran, A.; Arjun, P. Isolation of natural naphthoquinones from Juglans regia and in vitro antioxidant and cytotoxic studies of naphthoquinones and the synthetic naphthofuran derivatives. Russ. J. Bioorganic Chem. 2018, 44, 346–353. [Google Scholar] [CrossRef]
- Salas, C.O.; Faúndez, M.; Morello, A.; Diego Maya, J.A.; Tapia, R. Natural and synthetic naphthoquinones active against Trypanosoma Cruzi: An initial step towards new drugs for Chagas disease. Curr. Med. Chem. 2011, 18, 144–161. [Google Scholar] [CrossRef] [PubMed]
- Jentzsch, J.; Koko, W.S.; Al Nasr, I.S.; Khan, T.A.; Schobert, R.; Ersfeld, K.; Biersack, B. New antiparasitic bis-naphthoquinone derivatives. Chem. Biodivers. 2020, 17, e1900597. [Google Scholar] [CrossRef] [PubMed]
- Lall, N.; Weiganand, O.; Hussein, A.A.; Meyer, J.J.M. Antifungal activity of naphthoquinones and triterpenes isolated from the root bark of Euclea natalensis. South Afr. J. Bot. 2006, 72, 579–583. [Google Scholar] [CrossRef]
- Wellington, K.W.; Nyoka, N.B.; McGaw, L.J. Investigation of the antibacterial and antifungal activity of thiolated naphthoquinones. Drug Dev. Res. 2019, 80, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Tandon, V.K.; Singh, R.V.; Yadav, D.B. Synthesis and evaluation of novel 1,4-naphthoquinone derivatives as antiviral, antifungal and anticancer agents. Bioorg. Med. Chem. Lett. 2004, 14, 2901–2904. [Google Scholar] [CrossRef] [PubMed]
- Schuck, D.C.; Ferreira, S.B.; Cruz, L.N.; Da Rocha, D.R.; Moraes, M.S.; Nakabashi, M.; Rosenthal, P.J.; Ferreira, V.F.; Garcia, C.R. Biological evaluation of hydroxynaphthoquinones as anti-malarials. Malar. J. 2013, 12, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Klaus, V.; Hartmann, T.; Gambini, J.; Graf, P.; Stahl, W.; Hartwig, A.; Klotz, L.-O. 1,4-Naphthoquinones as inducers of oxidative damage and stress signaling in HaCaT human keratinocytes. Arch. Biochem. Biophys. 2010, 496, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Cai, A.; Chen, G.; Xi, H.; Wu, X.; Cui, J.; Zhang, K.; Zhao, X.; Yu, J.; Wei, B.; et al. Shikonin induces mitochondria-mediated apoptosis and enhances chemotherapeutic sensitivity of gastric cancer through reactive oxygen species. Sci. Rep. 2016, 6, 38267. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shen, G.N.; Luo, Y.H.; Piao, X.J.; Jiang, X.Y.; Meng, L.Q.; Wang, Y.; Zhang, Y.; Wang, J.R.; Wang, H.; et al. Novel 1,4-naphthoquinone derivatives induce apoptosis via ROS-mediated P38/MAPK, Akt and STAT3 signaling in human hepatoma Hep3B cells. Int. J. Biochem. Cell Biol. 2018, 96, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.-Y.; Jeon, J.-H.; Jeong, E.-Y.; Lee, C.-H.; Lee, H.-S. Antimicrobial activity of 5-hydroxy-1,4-naphthoquinone isolated from Caesalpinia sappan toward intestinal bacteria. Food Chem. 2007, 100, 1254–1258. [Google Scholar] [CrossRef]
- Periasamy, H.; Iswarya, S.; Pavithra, N.; Senthilnathan, S.; Gnanamani, A. In vitro antibacterial activity of plumbagin isolated from Plumbago zeylanica L. against methicillin-resistant Staphylococcus aureus. Lett. Appl. Microbiol. 2019, 69, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Patwardhan, R.B.; Shinde, P.S.; Chavan, K.R.; Devale, A. Reversal of plasmid encoded antibiotic resistance from nosocomial pathogens by using Plumbago auriculata root extracts. Int. J. Curr. Microbiol. Appl. Sci. 2015, 2, 187–198. [Google Scholar]
- Kaewbumrung, S.; Panichayupakaranant, P. Antibacterial activity of plumbagin derivative-rich Plumbago indica root extracts and chemical stability. Nat. Prod. Res. 2014, 28, 835–837. [Google Scholar] [CrossRef] [PubMed]
- Patwardhan, R.B.; Dhakephalkar, P.K.; Chopade, B.A.; Dhavale, D.D.; Bhonde, R.R. Purification and characterization of an active principle, lawsone, responsible for the plasmid curing activity of Plumbago zeylanica root extracts. Front. Microbiol. 2018, 9, 2618. [Google Scholar] [CrossRef] [PubMed]
- Tekin, V.; Muftuler, F.Z.B.; Guldu, O.K.; Kilcar, A.Y.; Medine, E.I.; Yavuz, M.; Unak, P.; Timur, S. Biological affinity evaluation of Lawsonia inermis origin lawsone compound and its radioiodinated form via in vitro methods. J. Radioanal. Nucl. Chem. 2015, 303, 701–708. [Google Scholar] [CrossRef]
- Huang, X.Y.; Fu, H.L.; Tang, H.Q.; Yin, Z.Q.; Zhang, W.; Shu, G.; Yin, L.Z.; Zhao, L.; Yan, X.R.; Lin, J.C. Optimization extraction of shikonin using ultrasound-assisted response surface methodology and antibacterial studies. Evid. Based Complement Alternat. Med. 2020, 2020, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Lee, D.Y.; Kim, Y.B.; Lee, S.W.; Cha, S.W.; Park, H.W.; Kim, G.S.; Kwon, D.Y.; Lee, M.H.; Han, S.H. The mechanism underlying the antibacterial activity of shikonin against methicillin-resistant Staphylococcus aureus. Evid. Based Complement Alternat. Med. 2015, 2015, 520578. [Google Scholar] [CrossRef]
- Zani, C.L.; De Oliveira, A.B.; De Oliviera, G.G. Furanonaphthoquinones from Tabebuia Ochracea. Phytochemistry 1991, 30, 2379–2381. [Google Scholar] [CrossRef]
- Souza, M.A.; Johann, S.; Lima, L.A.R.d.S.; Campos, F.F.; Mendes, I.C.; Beraldo, H.; Souza-Fagundes, E.M.d.; Cisalpino, P.S.; Rosa, C.A.; Alves, T.M.d.A.; et al. The antimicrobial activity of lapachol and its thiosemicarbazone and semicarbazone derivatives. Mem. Inst. Oswaldo Cruz. 2013, 108, 342–351. [Google Scholar] [CrossRef]
- Widhalm, J.R.; Rhodes, D. Biosynthesis and molecular actions of specialized 1,4-naphthoquinone natural products produced by horticultural plants. Hortic. Res. 2016, 3, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.-Y.; Zhu, B.-W.; Wang, X.-D.; Qin, L.; Li, D.-M.; Miao, L.; Murata, Y. Stability of polyhydroxylated 1,4-naphthoquinone pigment recovered from spines of sea urchin Strongylocentrotus nudus. Int. J. Food Sci. Technol. 2012, 47, 1479–1486. [Google Scholar] [CrossRef]
- Carriço, M.d.P.S.B.; do Carmo Cardoso, M.F.; Da Silva, F.D.C.; Ferreira, V.F.; Lima, E.S.; Souza, J.V.B. Antifungal activity of synthetic naphthoquinones against dermatophytes and opportunistic fungi: Preliminary mechanism-of-action tests. Ann. Clin. Microbiol. Antimicrob. 2014, 13, 26. [Google Scholar]
- Adusei, E.; Adosraku, R.K.; Oppong-Kyekyeku, J.; Amengor, C.D.; Jibira, Y. Resistance modulation action, time-kill kinetics assay, and inhibition of biofilm formation effects of plumbagin from Plumbago zeylanica Linn. J. Trop. Med. 2019, 2019, 31. [Google Scholar] [CrossRef]
- Vukic, M.D.; Vukovic, N.L.; Djelic, G.T.; Popovic, S.L.; Zaric, M.M.; Baskic, D.D.; Krstic, G.B.; Tesevic, V.V.; Kacaniova, M.M. Antibacterial and cytotoxic activities of naphthoquinone pigments from Onosma visianii Clem. EXCLI J. 2017, 16, 73–88. [Google Scholar] [PubMed]
- Petrosyan, M.; Shcherbakova, Y.; Sahakyan, N.; Vardanyan, Z.; Poladyan, A.; Popov, Y.; Trchounian, A. Alkanna orientalis (L.) Boiss. Plant isolated cultures and antimicrobial activity of their extracts: Phenomenon, dependence on different factors and effects on some membrane-associated properties of bacteria. Plant Cell Tissue Organ Cult. 2015, 122, 727–738. [Google Scholar] [CrossRef]
- Wang, J.; Cheng, Y.; Wu, R.; Jiang, D.; Bai, B.; Tan, D.; Yan, T.; Sun, X.; Zhang, Q.; Wu, Z. Antibacterial activity of juglone against Staphylococcus aureus: From apparent to proteomic. Int. J. Mol. Sci. 2016, 17, 965. [Google Scholar] [CrossRef] [PubMed]
- Padhye, S.; Dandawate, P.; Yusufi, M.; Ahmad, A.; Sarkar, F.H. Perspectives on medicinal properties of plumbagin and its analogs. Med. Res. Rev. 2012, 32, 1131–1158. [Google Scholar] [CrossRef] [PubMed]
- Al-Mussawi, A.A. Isolation and identification of shikonin from Arnebia Decumbens L. and its antibacterial activity. Res. J. Appl. Sci. 2010, 6, 1452–1456. [Google Scholar]
- Andújar, I.; Ríos, J.L.; Giner, R.M.; Recio, M.C. Pharmacological properties of shikonin-a review of literature since 2002. Planta Med. 2013, 79, 1685–1697. [Google Scholar] [CrossRef]
- Aljanaby, A.J. Antibacterial Activity of an aqueous extracts of Alkanna tinctoria roots against drug resistant aerobic pathogenic bacteria isolated from patients with burns infections. Russ. Open Med. J. 2018, 7, e0104. [Google Scholar] [CrossRef]
- De Almeida, E.R. Preclinical and clinical studies of lapachol and beta-lapachone. Open Nat. Prod. J. 2009, 2, 42–47. [Google Scholar] [CrossRef]
- Balachandran, C.; Al-Dhabi, N.A.; Duraipandiyan, V.; Ignacimuthu, S. Bluemomycin, a new naphthoquinone derivative from Streptomyces sp. with antimicrobial and cytotoxic properties. Biotechnol. Lett. 2021, 43, 1–4. [Google Scholar]
- Ravichandiran, P.; Sheet, S.; Premnath, D.; Kim, A.R.; Yoo, D.J. 1,4-Naphthoquinone analogues: Potent antibacterial agents and mode of action evaluation. Molecules 2019, 24, 1437. [Google Scholar] [CrossRef] [PubMed]
- Tandon, V.K.; Yadav, D.B.; Singh, R.V.; Chaturvedi, A.K.; Shukla, P.K. Synthesis and biological evaluation of novel (L)-α-amino acid methyl ester, heteroalkyl, and aryl substituted 1,4-naphthoquinone derivatives as antifungal and antibacterial agents. Bioorg. Med. Chem. Lett. 2005, 15, 5324–5328. [Google Scholar] [CrossRef] [PubMed]
- Yap, J.K.Y.; Tan, S.Y.Y.; Tang, S.Q.; Thien, V.K.; Chan, E.W.L. Synergistic antibacterial activity between 1,4-naphthoquinone and β-lactam antibiotics against methicillin-resistant Staphylococcus aureus. Microb. Drug Resist. 2021, 27, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Ghosh, S.; Shaw, R.; Patra, M.M.; Calcuttawala, F.; Mukherjee, N.; Gupta, S.K.D. Mycobacterium tuberculosis thymidylate synthase (ThyX) is a target for plumbagin, a natural product with antimycobacterial activity. PLoS ONE 2020, 15, e0228657. [Google Scholar] [CrossRef] [PubMed]
- Meah, M.S.; Lertcanawanichakul, M.; Pedpradab, P.; Lin, W.; Zhu, K.; Li, G.; Panichayupakaranant, P. Synergistic effect on anti-methicillin-resistant Staphylococcus aureus among combinations of α-mangostin-rich extract, lawsone methyl ether and ampicillin. Lett. Appl. Microbiol. 2020, 71, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Yu, B.; Friedrich, D.; Li, J.; Shen, H.; Krautscheid, H.; Huang, S.D.; Kim, M.-H. Naphthoquinone-derivative as a synthetic compound to overcome the antibiotic resistance of methicillin-resistant S. aureus. Commun. Biol. 2020, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Linzner, N.; Fritsch, V.N.; Busche, T.; Tung, Q.N.; Loi, V.V.; Bernhardt, J.; Kalinowski, J.; Antelmann, H. The plant-derived naphthoquinone lapachol causes an oxidative stress response in Staphylococcus aureus. Free Radic. Biol. Med. 2020, 158, 126–136. [Google Scholar] [CrossRef]
- Figueredo, F.G.; Ramos, I.T.L.; Paz, J.A.; Silva, T.M.S.; Camara, C.A.; Oliveira-Tintino, C.D.d.M.; Relison Tintino, S.; de Farias, P.A.M.; Coutinho, H.D.M.; Fonteles, M.M.d.F. In silico evaluation of the antibacterial and modulatory activity of lapachol and nor-lapachol derivates. Microb. Pathog. 2020, 144, 104181. [Google Scholar] [CrossRef]
- Choudhari, D.; Salunke-Gawali, S.; Chakravarty, D.; Shaikh, S.R.; Lande, D.N.; Gejji, S.P.; Rao, P.K.; Satpute, S.; Puranik, V.G.; Gonnade, R. Synthesis and biological activity of imidazole based 1,4-naphthoquinones. New J. Chem. 2020, 44, 6889–6901. [Google Scholar] [CrossRef]
- Choudhari, D.; Chakravarty, D.; Lande, D.N.; Parveen, S.; Gejji, S.P.; Kodam, K.M.; Salunke-Gawali, S. Crystal structures and biological activity of homologated (N)-n-alkylammonium salts of 2-bromo-3-oxido-1,4-naphthoquinone. Struct. Chem. 2019, 30, 2257–2270. [Google Scholar] [CrossRef]
- Andrade, J.C.; Morais Braga, M.F.B.; Guedes, G.M.M.; Tintino, S.R.; Freitas, M.A.; Quintans, L.J.; Menezes, I.R.A.; Coutinho, H.D.M. Menadione (vitamin K) enhances the antibiotic activity of drugs by cell membrane permeabilization mechanism. Saudi J. Biol. Sci. 2017, 24, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Moreira, C.S.; Silva, A.; Novais, J.S.; Sá Figueiredo, A.M.; Ferreira, V.F.; da Rocha, D.R.; Castro, H.C. Searching for a potential antibacterial lead structure against bacterial biofilms among new naphthoquinone compounds. J. Appl. Microbiol. 2017, 122, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Janeczko, M.; Demchuk, O.M.; Strzelecka, D.; Kubiński, K.; Masłyk, M. New family of antimicrobial agents derived from 1,4-naphthoquinone. Eur. J. Med. Chem. 2016, 124, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.Y.; Wang, P.F.; Wang, Z.Z.; Luo, Y.L.; Hu, D.Q.; Qi, J.L.; Lu, G.H.; Pang, Y.J.; Yang, R.W.; Zhu, H.L.; et al. Shikonin derivatives as inhibitors of tyrosyl-TRNA synthetase: Design, synthesis and biological evaluation. RSC Adv. 2016, 6, 83003–83010. [Google Scholar] [CrossRef]
- Zmantar, T.; Miladi, H.; Kouidhi, B.; Chaabouni, Y.; Slama, R.B.; Bakhrouf, A.; Mahdouani, K.; Chaieb, K. Use of juglone as antibacterial and potential efflux pump inhibitors in Staphylococcus aureus isolated from the oral cavity. Microb. Pathog. 2016, 101, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Ravichandiran, P.; Premnath, D.; Vasanthkumar, S. Synthesis, molecular docking and antibacterial evaluation of 2-(4-(4-aminophenylsulfonyl) phenylamino)-3-(thiophen-2-ylthio) naphthalene-1,4-dione derivatives. Front. Chem. Sci. Eng. 2015, 9, 46–56. [Google Scholar] [CrossRef]
- Sreelatha, T.; Kandhasamy, S.; Dinesh, R.; Shruthy, S.; Shweta, S.; Mukesh, D.; Karunagaran, D.; Balaji, R.; Mathivanan, N.; Perumal, P.T. Synthesis and sar study of novel anticancer and antimicrobial naphthoquinone amide derivatives. Bioorg. Med. Chem. Lett. 2014, 24, 3647–3651. [Google Scholar] [CrossRef]
- Schlievert, P.M.; Merriman, J.A.; Salgado-Pabón, W.; Mueller, E.A.; Spaulding, A.R.; Vu, B.G.; Chuang-Smith, O.N.; Kohler, P.L.; Kirby, J.R. Menaquinone analogs inhibit growth of bacterial pathogens. Antimicrob. Agents Chemother. 2013, 57, 5432–5437. [Google Scholar] [CrossRef] [PubMed]
- Rahmoun, N.M.; Boucherit-Otmani, Z.; Boucherit, K.; Benabdallah, M.; Villemin, D.; Choukchou-Braham, N. Antibacterial and antifungal activity of lawsone and novel naphthoquinone derivatives. Med. Mal. Infect. 2012, 42, 270–275. [Google Scholar] [CrossRef]
- Mathew, R.; Kruthiventi, A.K.; Prasad, J.V.; Kumar, S.P.; Srinu, G.; Chatterji, D. Inhibition of mycobacterial growth by plumbagin derivatives. Chem. Biol. Drug Des. 2010, 76, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Tandon, V.K.; Maurya, H.K.; Mishra, N.N.; Shukla, P.K. Design, synthesis and biological evaluation of novel nitrogen and sulfur containing hetero-1,4-naphthoquinones as potent antifungal and antibacterial agents. Eur. J. Med. Chem. 2009, 44, 3130–3137. [Google Scholar] [CrossRef] [PubMed]
- Medina, L.F.C.; Hertz, P.F.; Stefani, V.; Henriques, J.A.P.; Zanotto-Filho, A.; Brandelli, A. Aminonaphthoquinone induces oxidative stress in Staphylococcus aureus. Biochem. Cell Biol. 2006, 84, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.V.; Ansari, M.H.D.; Rosenkranz, D.; Maharjan, R.S.; Kriegel, F.L.; Gandhi, K.; Kanase, A.; Singh, R.; Laux, P.; Luch, A. Artificial intelligence and machine learning in computational nanotoxicology: Unlocking and empowering nanomedicine. Adv. Healthc. Mater. 2020, 9, 1901862. [Google Scholar] [CrossRef]
- Singh, A.V.; Rosenkranz, D.; Ansari, M.H.D.; Singh, R.; Kanase, A.; Singh, S.P.; Blair, J.; Jutta, T.; Laux, P.; Luch, A. Artificial intelligence and machine learning empower advanced biomedical material design to toxicity prediction. Adv. Intell. Syst. 2020, 2, 2000084. [Google Scholar] [CrossRef]
- Singh, A.V.; Maharjan, R.-S.; Kanase, A.; Siewert, K.; Rosenkranz, D.; Singh, R.; Laux, P.; Luch, A. Machine-learning-based approach to decode the influence of nanomaterial properties on their interaction with cells. ACS Appl. Mater. Interfaces 2021, 13, 1943–1955. [Google Scholar] [CrossRef] [PubMed]
- Figueredo, F.G.; Ramos, I.T.; Paz, J.A.; Silva, T.M.; Câmara, C.A.; de Morais Oliveira-Tintino, C.D.; Tintino, S.R.; de Farias, P.A.M.; de Menezes, I.R.A.; Coutinho, H.D.M.; et al. Effect of hydroxyamines derived from lapachol and norlachol against Staphylococcus aureus strains carrying the NorA efflux pump. Infect. Genet. Evol. 2020, 84, 104370. [Google Scholar] [CrossRef]
- Klotz, L.O.; Hou, X.; Jacob, C. 1,4-Naphthoquinones: From oxidative damage to cellular and inter-cellular signaling. Molecules 2014, 19, 14902–14918. [Google Scholar] [CrossRef]
- Lee, M.H.; Lapidus, R.G.; Ferraris, D.; Emadi, A. Analysis of the mechanisms of action of naphthoquinone-based anti-acute myeloid leukemia chemotherapeutics. Molecules 2019, 24, 3121. [Google Scholar] [CrossRef] [PubMed]
- Pereyra, C.E.; Dantas, R.F.; Ferreira, S.B.; Gomes, L.P.; Silva, F.P., Jr. The diverse mechanisms and anticancer potential of naphthoquinones. Cancer Cell Int. 2019, 19, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Shintani, M.; Sanchez, Z.K.; Kimbara, K. Genomics of microbial plasmids: Classification and identification based on replication and transfer systems and host taxonomy. Front. Microbiol. 2015, 6, 242. [Google Scholar] [CrossRef] [PubMed]
- Okpokwasili, G.C.; Somerville, C.C.; Sullivan, M.; Grimes, D.J.; Colwell, R.R. Plasmid mediated degradation of hydrocarbons in estuarine bacteria. Oil Chem. Pollut. 1986, 3, 117–129. [Google Scholar] [CrossRef]
- Campelo, A.B.; Gaspar, P.; Roces, C.; Rodríguez, A.; Kok, J.; Kuipers, O.P.; Neves, A.R.; Martínez, B. The Lcn972 bacteriocin-encoding plasmid PBL1 impairs cellobiose metabolism in Lactococcus lactis. Appl. Environ. Microbiol. 2011, 77, 7576–7585. [Google Scholar] [CrossRef] [PubMed]
- Buckner, M.M.; Ciusa, M.L.; Piddock, L.J. Strategies to combat antimicrobial resistance: Anti-plasmid and plasmid curing. FEMS Microbiol. Rev. 2018, 42, 781–804. [Google Scholar] [CrossRef] [PubMed]
- Letchumanan, V.; Chan, K.-G.; Lee, L.-H. An insight of traditional plasmid curing in vibrio species. Front. Microbiol. 2015, 6, 735. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, V.V.; Thomas, C.M. Curing of F-like plasmid TP181 by plumbagin is due to interference with both replication and maintenance functions. Microbiology 1996, 142, 2399–2406. [Google Scholar] [CrossRef]
- Beg, A.Z.; Ahmad, I. Effect of Plumbago zeylanica extract and certain curing agents on multidrug resistant bacteria of clinical origin. World J. Microbiol. Biotechnol. 2000, 16, 841–844. [Google Scholar] [CrossRef]
- Masi, M.; Réfregiers, M.; Pos, K.M.; Pagès, J.-M. Mechanisms of envelope permeability and antibiotic influx and efflux in gram-negative bacteria. Nat. Microbiol. 2017, 2, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Shriram, V.; Khare, T.; Bhagwat, R.; Shukla, R.; Kumar, V. Inhibiting bacterial drug efflux pumps via phyto-therapeutics to combat threatening antimicrobial resistance. Front. Microbiol. 2018, 9, 2990. [Google Scholar] [CrossRef] [PubMed]
- Blanco, P.; Hernando-Amado, S.; Reales-Calderon, J.A.; Corona, F.; Lira, F.; Alcalde-Rico, M.; Bernardini, A.; Sanchez, M.B.; Martinez, J.L. Bacterial multidrug efflux pumps: Much more than antibiotic resistance determinants. Microorganisms 2016, 4, 14. [Google Scholar] [CrossRef]
- Kumar, A.; Khan, I.A.; Koul, S.; Koul, J.L.; Taneja, S.C.; Ali, I.; Ali, F.; Sharma, S.; Mirza, Z.M.; Kumar, M.; et al. Novel structural analogues of piperine as inhibitors of the NorA efflux pump of Staphylococcus aureus. J. Antimicrob. Chemother. 2008, 61, 1270–1276. [Google Scholar] [CrossRef]
- Stermitz, F.R.; Tawara-Matsuda, J.; Lorenz, P.; Mueller, P.; Zenewicz, L.; Lewis, K. 5’-Methoxyhydnocarpin-D and pheophorbide A: Berberis species components that potentiate berberine growth inhibition of resistant Staphylococcus aureus. J. Nat. Prod. 2000, 63, 1146–1149. [Google Scholar] [CrossRef]
- Chevalier, J.; Atifi, S.; Eyraud, A.; Mahamoud, A.; Barbe, J.; Pagès, J.-M. New pyridoquinoline derivatives as potential inhibitors of the fluoroquinolone efflux pump in resistant Enterobacter aerogenes strains. J. Med. Chem. 2001, 44, 4023–4026. [Google Scholar] [CrossRef]
- Ohene-Agyei, T.; Mowla, R.; Rahman, T.; Venter, H. Phytochemicals increase the antibacterial activity of antibiotics by acting on a drug efflux pump. Microbiologyopen 2014, 3, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Drlica, K. Reactive oxygen species and the bacterial response to lethal stress. Curr. Opin. Microbiol. 2014, 21, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Acker, H.V.; Coenye, T. The role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol. 2017, 25, 456–466. [Google Scholar] [CrossRef]
- Dharmaraja, A.T.; Alvala, M.; Sriram, D.; Yogeeswari, P.; Chakrapani, H. Design, synthesis and evaluation of small molecule reactive oxygen species generators as selective Mycobacterium tuberculosis inhibitors. Chem. Commun. 2012, 48, 10325–10327. [Google Scholar] [CrossRef]
- Memar, M.Y.; Ghotaslou, R.; Samiei, M.; Adibkia, K. Antimicrobial use of reactive oxygen therapy: Current insights. Infect. Drug Resist. 2018, 11, 567–576. [Google Scholar] [CrossRef]
- Singh, A.V.; Jungnickel, H.; Leibrock, L.; Tentschert, J.; Reichardt, P.; Katz, A.; Laux, P.; Luch, A. ToF-SIMS 3D imaging unveils important insights on the cellular microenvironment during biomineralization of gold nanostructures. Sci. Rep. 2020, 10, 261. [Google Scholar] [CrossRef]
- Singh, A.V. Commentary on “Peptide-conjugated nanoparticles as targeted anti-angiogenesis therapeutic and diagnostic in Cancer” by Shaker, A. Mousa, Pharmaceutical Research Institute, Albany College of Pharmacy and Health Sciences, Rensselaer, NY 12144, United States-Peptide-Conjugated Nanoparticles for Multimodal Nanomedicine. Curr. Med. Chem. 2020, 27, 2927–2928. [Google Scholar] [PubMed]
- Bush, N.G.; Evans-Roberts, K.; Maxwell, A. DNA topoisomerases. EcoSal Plus 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- D’Atanasio, N.; Capezzone de Joannon, A.; Di Sante, L.; Mangano, G.; Ombrato, R.; Vitiello, M.; Bartella, C.; Magarò, G.; Prati, F.; Milanese, C. Antibacterial activity of novel dual bacterial DNA type II topoisomerase inhibitors. PLoS ONE 2020, 15, e0228509. [Google Scholar] [CrossRef] [PubMed]
- Karkare, S.; Chung, T.T.; Collin, F.; Mitchenall, L.A.; McKay, A.R.; Greive, S.J.; Meyer, J.J.; Lall, N.; Maxwell, A. The naphthoquinone diospyrin is an inhibitor of DNA gyrase with a novel mechanism of action. J. Biol. Chem. 2013, 288, 5149–5156. [Google Scholar] [CrossRef] [PubMed]
- Mollaei, S.; Khanehbarndaz, O.; Gerami-Khashal, Z.; Ebadi, M. Molecular identification and phytochemical screening of endophytic fungi isolated from Lithospermum officinale L. roots: A new source of shikonin. Phytochemistry 2019, 168, 112116. [Google Scholar] [CrossRef] [PubMed]
- Hughes, L.M.; Lanteri, C.A.; O’Neil, M.T.; Johnson, J.D.; Gribble, G.W.; Trumpower, B.L. Design of anti-parasitic and anti-fungal hydroxy-naphthoquinones that are less susceptible to drug resistance. Mol. Biochem. Parasitol. 2011, 177, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Altharawi, A.; Gut, J.; Rosenthal, P.J.; Long, T.E. 1,4-Naphthoquinone cations as antiplasmodial agents: Hydroxy, acyloxy-, and alkoxy-substituted analogues. ACS Med. Chem. Lett. 2012, 3, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Moreira, D.R.M.; de Sá, M.S.; Macedo, T.S.; Menezes, M.N.; Reys, J.R.M.; Santana, A.E.G.; Silva, T.L.; Maia, G.L.A.; Filho, J.M.B.; de Amorim Camara, C.; et al. Evaluation of naphthoquinones identified the acetylated isolapachol as a potent and selective antiplasmodium agent. J. Enzym. Inhib. Med. Chem. 2015, 30, 615–621. [Google Scholar] [CrossRef] [PubMed]
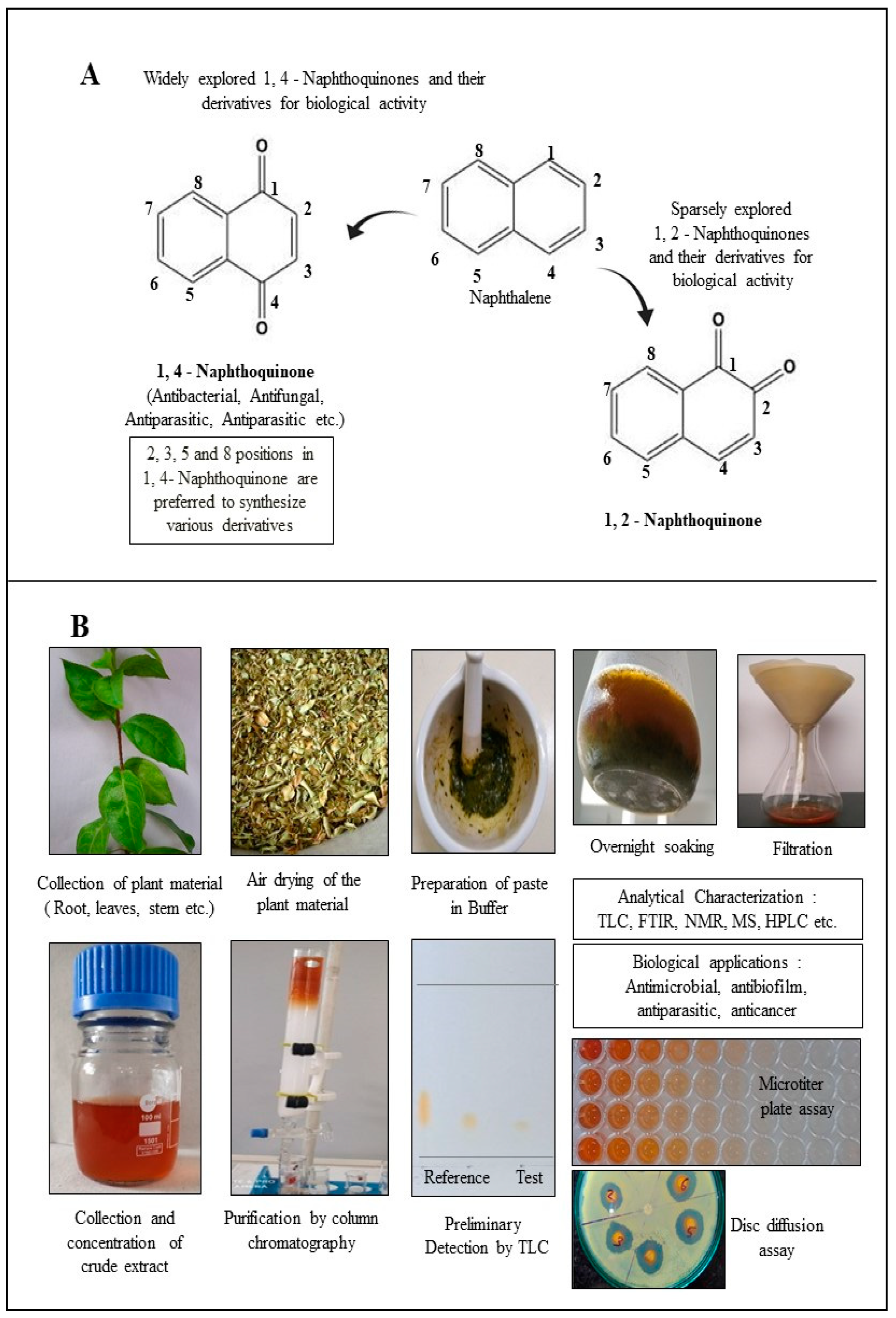

| Name and Structure of the Naphthoquinone Derivative | Source of Plant Material | Reference |
|---|---|---|
(A) Juglone: 5-Hydroxy-1,4-naphthalenedione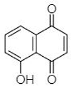 | Caesalpinia sappan | [30] |
(B) Plumbagin: 5-Hydroxy-2-methyl-naphthalene-1,4-dione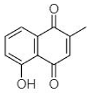 | Plumbago zeylanica | [31] |
| Plumbago auriculata | [32] | |
| Plumbago indica | [33] | |
(C) Lawsone: 2-Hydroxy-1,4-naphthoquinone | Plumbago zeylanica | [34] |
| Lawsonia inermis | [35] | |
(D) Shikonin: 5,8-Dihydroxy-2-[(1R)-1-hydroxy-4-methyl-3-penten-1-yl]-1,4-naphthoquinone | Arnebia euchroma | [36] |
| Lithospermum erythrorhizon | [37] | |
(E) Lapachol: 2-Hydroxy-3-(3-methyl-2-butenyl)-1,4-Naphthalenedione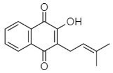 | Tabebuia ochracea | [38,39] |
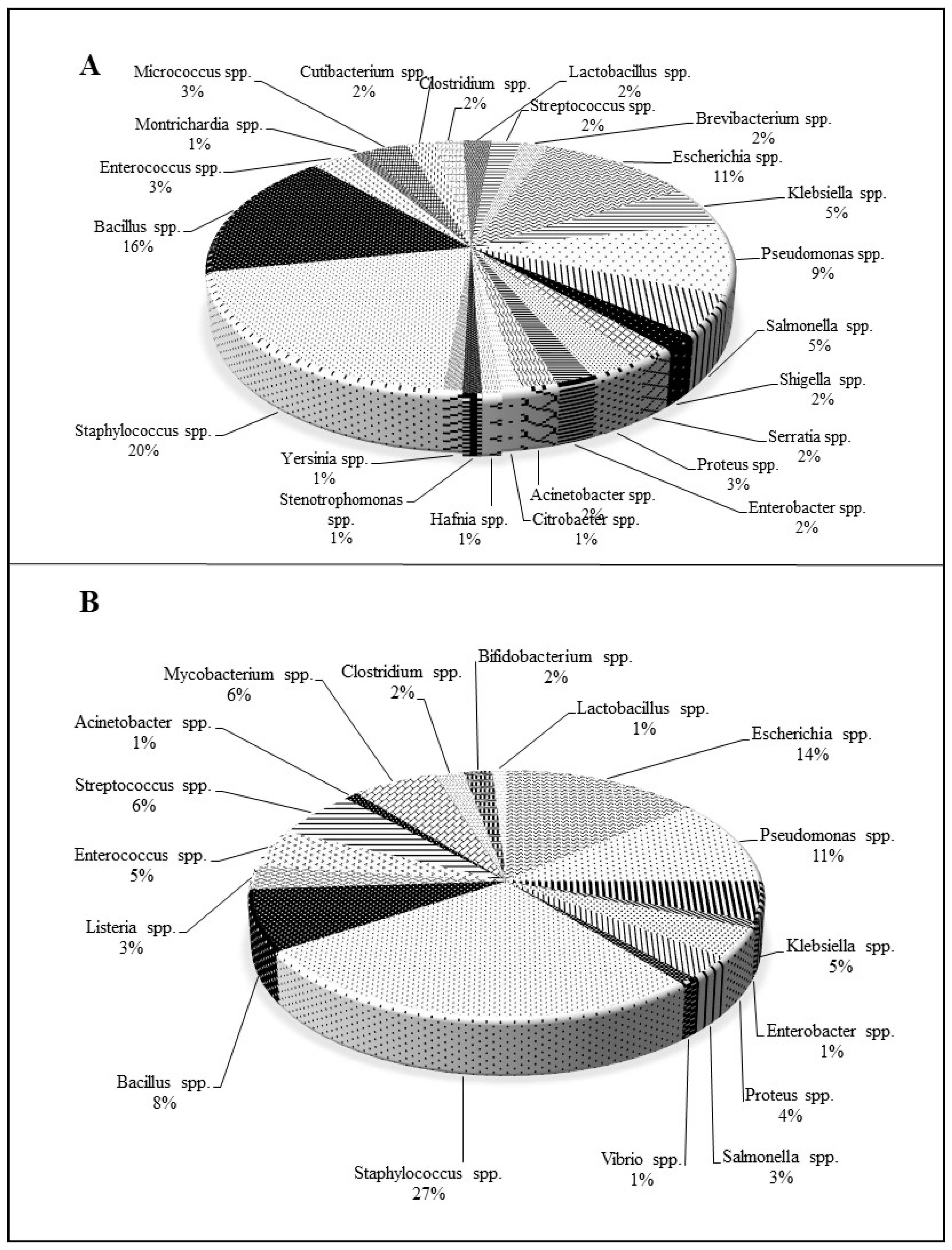
| Type of Naphthoquinone | Active against Bacterial Strains | MIC (µg/mL) | Reference |
|---|---|---|---|
| Shikonin from Arnebia euchroma root extract | Staphylococcus aureus | 128 | [36] |
| Streptococcus agalactiae | 128 | ||
| Escherichia coli | 256 | ||
| Salmonella isolates | 256 | ||
| Pseudomonas aeruginosa | 512 | ||
| Plumbagin from Plumbago zeylanica | Staphylococcus aureus | 0.5 | [43] |
| Escherichia coli | 8 | ||
| Klebsiella pneumoniae | 2 | ||
| Pseudomonas aeruginosa | 8 | ||
| Plumbagin from Plumbago zeylanica | S. aureus (MRSA) | 4–8 | [31] |
| Lawsone from Plumbago zeylanica root extract | Staphylococcus aureus | 400 | [34] |
| Salmonella typhi | 200 | ||
| Bacillus cereus | 200 | ||
| Bacillus subtilis | 200 | ||
| Pseudomonas aeruginosa | 800 | ||
| Escherichia coli | 800 | ||
| Shigella dysenteriae | 400 | ||
| Serratia marcescens | >1600 | ||
| Proteus mirabilis | >1600 | ||
| Klebsiellapneumoniae | 800 | ||
| Enterobacter | 800 | ||
| Acinetobacter baumannii | 800 | ||
| Naphthoquinone pigments from Onosma visianii | Bacillus megaterium | 9.54–54.28 | [44] |
| Montrichardia arborescens | 6.82–54.28 | ||
| Micrococcus luteus | 9.54–76.20 | ||
| Staphylococcus epidermidis | 9.54–54.28 | ||
| Enterococcus faecalis | 6.82–38.10 | ||
| Citrobacter koseri | 6.82–76.20 | ||
| Hafnia alvei | 6.82–54.28 | ||
| Pseudomonas proteolytica | 4.77–76.20 | ||
| Stenotrophomonas maltophilia | 4.77–76.20 | ||
| Yersinia intermedia | 6.82–76.20 | ||
| - | (values depict MIC90) | ||
| Shikonin from Lithospermum erythrorhizon | Staphylococcus aureus (MRSA) | 7.8–31.2 | [37] |
| Staphylococcus aureus (MSSA) | 7.8 | ||
| Plumbago auriculata root extracts | Proteus vulgaris | Strong activity | [32] |
| Klebsiella. pneumoniae | Strong activity | ||
| Escherichia coli | Moderate activity | ||
| Pseudomonas aeruginosa | Less activity | ||
| Naphthoquinone from Alkanna orientalis extracts | Bacillus megaterium | 125 | [45] |
| Bacillus mesentericus | 125 | ||
| Bacillus mycoides | 125 | ||
| Bacillus subtilis | 125 | ||
| Brevibacterium flavum | 250 | ||
| Enterococcus hirae | 250 | ||
| Micrococcus luteus | 250 | ||
| Staphylococcus aureus | 125 | ||
| Staphylococcus citreus | 500 | ||
| Staphylococcus roseus | 125 | ||
| Escherichia. coli | 500 | ||
| Salmonella typhimurium | 750 | ||
| Lawsone from Lawsonia inermis | Bacillus subtilis | Activity at 1000 µg/disc | [35] |
| Staphylococcus aureus | No activity | ||
| Escherichia coli | No activity | ||
| Pseudomonas aeruginosa | No activity | ||
| Plumbago indica from root extracts | Cutibacterium acnes (formerly Propionibacterium acnes) | 12.5 | [33] |
| Staphylococcus aureus | 12.5 | ||
| Staphylococcus epidermidis | 0.78 | ||
| Naphthoquinones from Caesalpinia sappan | Clostridium perfringens | Activity observed | [30] |
| Bacillus bifidum | Activity observed | ||
| Bacillus. breve | No significant activity | ||
| Lactobacillus casei | Minute activity | ||
| Escherichia coli | Minute activity |
| Type of Naphthoquinone | Antibacterial Activity Against | MIC (µg/mL) | Reference |
|---|---|---|---|
| Plumbagin | Mycobacterium tuberculosis | 4 | [56] |
| Mycobacterium smegmatis | 4 | ||
| Lawsone | Mycobacterium tuberculosis | >16 | |
| Mycobacterium smegmatis | >32 | ||
| Lawsone methyl ether | Staphylococcus aureus (MRSA) | 62.5–125 | [57] |
| Lawsone derivatives | Staphylococcus aureus (MRSA) | 32–128 & >128 | [58] |
| Staphylococcusaureus (MSSA) | 0.6 –128 & >128 | ||
| Lapachol | Staphylococcus aureus | 1.25 mM | [59] |
| Lapachol, nor lapachol | Staphylococcus aureus (MRSA) | 30–500 & >500 | [60] |
| Imidazole derivatives of 1,4-naphthoquinone | Staphylococcus aureus | 8–512 | [61] |
| Bacillus subtilis | 32–512 | ||
| Pseudomonas aeruginosa | 8–256 | ||
| Proteus vulgaris | 16–256 | ||
| Escherichia coli | 32–256 | ||
| 1,4-naphthoquinone derivatives | Staphylococcus aureus | 4–256 | [62] |
| Bacillus subtilis | 128–512 | ||
| Pseudomonas aeruginosa | 32–512 | ||
| Proteus vulgaris | 128–512 | ||
| Escherichia coli | 256–512 | ||
| Phenylamino-phenylthio derivatives of 1,4-naphthoquinone (no. of derivatives synthesized) | Staphylococcus aureus | 31.25–500 | [53] |
| Listeria monocytogenes | 62.5–500 | ||
| Pseudomonas aeruginosa | 62.5–500 | ||
| Escherichia coli | 15.6–500 | ||
| Klebsiella pneumoniae | 62.5–500 | ||
| Sulfide derivatives of 1,4-naphthoquinone | Staphylococcus aureus | 7.8–250 & >250 | [24] |
| Escherichia coli | 31.3–250 & >250 | ||
| Menadione | Staphylococcus aureus | 128 | [63] |
| Pseudomonas aeruginosa | 64 | ||
| Escherichia coli | 128 | ||
| Klebsiella pneumoniae | 128 | ||
| Arylsulfanylmethyl-[1,4]-naphthoquinone derivatives | Staphylococcus aureus | 32–256 | [64] |
| Staphylococcus epidermidis | 16–256 | ||
| Staphylococcus simulans | 32–256 | ||
| Escherichia coli | 256 (less activity) | ||
| Enterococcus faecalis | 32–256 | ||
| 1,4-naphthoquinone substituted at positions 2 and 3 | Staphylococcus aureus | 7.8–500 | [65] |
| Salmonella bongori | 125–500 | ||
| Pseudomonas aeruginosa | 125–500 | ||
| Proteus vulgaris | 250–500 | ||
| Escherichia coli | 125–500 | ||
| Klebsiella pneumoniae | 62.5–500 | ||
| Enterococcus faecalis | 125–500 | ||
| Enterobacter cloacae | 250–500 | ||
| Shikonin derivatives | Staphylococcus aureus | 1.1–45.4 | [66] |
| Bacillus subtilis | 2–50 & >50 | ||
| Escherichia coli | 3.1–50 & >50 | ||
| Pseudomonas aeruginosa | 4–50 & >50 | ||
| Naphthoquinone derivatives | Staphylococcus aureus | 16–256 | [19] |
| Pseudomonas aeruginosa | 64–256 | ||
| Escherichia coli | 64–256 | ||
| Enterococcusfaecalis | 64–256 | ||
| 2-bromo-5-hydroxy-1,4-NQ | Staphylococcus aureus | 16 | |
| Juglone | Staphylococcus aureus | 32–256 | [67] |
| Staphylococcus epidermidis | 128 | ||
| Bacillus cereus | 256 | ||
| Salmonella enterica | 256 | ||
| Listeria monocytogenes | 256 | ||
| Pseudomonas aeruginosa | 128 | ||
| Escherichia coli | 128 | ||
| Enterococcus feacalis | 256 | ||
| Vibrio alginolyticus | 256 | ||
| Nitrogen and sulfur derivatives of 1,4-naphthoquinone | Bacillus subtilis | 1.4–19.3 | [68] |
| Proteus vulgaris | 2.7–39 | ||
| Plumbagin, juglone, lawsone, menadione and their analogues | Staphylococcusaureus (MRSA) | 3.9–125 | [69] |
| Pseudomonas aeruginosa | No significant activity | ||
| Menadione | Staphylococcus aureus | 3.1 | [70] |
| Bacillus anthracis | 6.25 | ||
| Streptococcus pyogenes | 25 | ||
| Streptococcus agalactiae | 6.25 | ||
| 1,4-naphthoquinone | Staphylococcus aureus | 6.25 | |
| Bacillus anthracis | 12.5 | ||
| Streptococcus pyogenes | 50 | ||
| Streptococcus agalactiae | 12.5 | ||
| 2-hydroxy, 1,4 naphthoquinone (Lawsone) and its 2-hydroxynaphthoquinone derivatives | Staphylococcus aureus | 16–512 & >512 | [71] |
| Listeria monocytogenes | 512 & >512 | ||
| Pseudomonas aeruginosa | 512 & >512 | ||
| Escherichia coli | 512 & >512 | ||
| Enterococcus faecalis | 256–512 | ||
| Acinetobacter baumannii | 512 & >512 | ||
| Salmonella typhimurium | 512 & >512 | ||
| Plumbagin derivatives | Mycobacterium smegmatis | 13.3–30.4 | [72] |
| Mycobacterium tuberculosis | 15.6–77.4 | ||
| Nitrogen, sulfur groups substitution at 2, 3 positions of 1,4-naphthoquinone | Staphylococcus aureus | 6.25–50 & >50 | [73] |
| Escherichia coli | 6.25–50 & >50 | ||
| Klebsiella pneumoniae | 1.56–50 & >50 | ||
| 5-Hydroxy-2-methyl-1,4-NQ | Clostridium perfringens | Antibacterial activity observed however MIC values are not mentioned specifically for the test organisms | [30] |
| 1,4-naphthoquinone | Lactobacillus casei | ||
| Bifidobacterium bifidum | |||
| Bifidobacterium breve | |||
| Clostridium perfringens | |||
| Escherichia coli | |||
| 1,2-naphthoquinone | Clostridium perfringens | ||
| Bifidobacterium bifidum | |||
| Bifidobacterium breve | |||
| 5-Amino-8-Hydroxy-1,4-NQ | Staphylococcus aureus | 50 | [74] |
| 1,4-naphthoquinone | Staphylococcus aureus | 10 | |
| (L)-a-amino acid methyl ester, heteroalkyl and aryl substituted1,4-naphthoquinone derivatives | Staphylococcus aureus | 12.5–50 & >50 | [54] |
| Streptococcus faecalis | 12.5–50 & >50 | ||
| Klebsiella pneumoniae | 6.25–50 & >50 | ||
| Escherichia coli | 12.5–50 & >50 | ||
| Pseudomonas aeruginosa | 50 & >50 |
| Test Organism | MIC of Antimicrobial Compound | MIC of NQs/Substances (S)/Derivatives (Individual) | MIC of Antimicrobial Compound with NQs | Reference | |
|---|---|---|---|---|---|
| Name of the Antimicrobial Compound | MIC of AC (Individual) | ||||
| - | MIC in µg/mL | - | |||
| - | Ethidium bromide (EtBr) | EtBr | ≥1024 (S3) ≥1024 (S4) ≥1024 (S5) | With 1/8th MIC of S3, S4 and S5 | [78] |
| Staphylococcus aureus SA-1199 (wildtype -WT) | 64 | S. aureus (WT) | |||
| 16 (EtBr + S3) | |||||
| 32 (EtBr + S4) | |||||
| 32 (EtBr + S5) | |||||
| Staphylococcus aureus SA-1199B strain (expressing norA gene expressing NorA efflux protein) | 256 | S. aureus (NorA) | |||
| 32 (EtBr + S3) | |||||
| 128 (EtBr + S4) | |||||
| 64 (EtBr + S5) | |||||
| - | Norfloxacin (NF) | Norfloxacin | ≥1024 (S3) ≥1024 (S4) ≥1024 (S5) | - | [78] |
| Staphylococcus aureus SA-1199 wildtype | 32 | S. aureus (WT) | |||
| 1 (NF + S3) | |||||
| 2.51 (NF + S4) | |||||
| 4 (NF + S5) | |||||
| Staphylococcus aureus SA-1199B strain (expressing norA gene expressing NorA efflux protein) | 406.3 | S. aureus (NorA) | |||
| 80 (NF + S3) | |||||
| 50.8 (NF + S4) | |||||
| 101.6 (NF + S5) | |||||
| - | MIC in µg/mL | - | |||
| - | Erythromycin | Erythromycin | Juglone | With ½ MIC of Juglone | [67] |
| Bacillus cereus | 128 | 256 | 32 | ||
| Escherichia coli | 64 | 128 | 16 | ||
| Salmonella enterica | 4 | 256 | 2 | ||
| Staphylococcus aureus | 128 | 128 | 16 | ||
| Listeria monocytogenes | 64 | 256 | 16 | ||
| Enterococcus feacalis | 64 | 256 | 32 | ||
| Staphylococcus epidermidis | 32 | 128 | 4 | ||
| Staphylococcus aureus | 16–256 | 32–256 | 4–32 | ||
| (Oral strains = 8) | - | - | (2 to 8-fold decrease in MIC) | ||
| - | Tetracycline | Tetracycline | Juglone | With ½ MIC of Juglone | [67] |
| Bacillus cereus | 2 | 256 | 1 | ||
| Vibrio alginolyticus | 256 | 256 | 64 | ||
| Salmonella enterica | 128 | 256 | 64 | ||
| Staphylococcus aureus | 4 | 128 | 2 | ||
| Enterococcus feacalis | 128 | 256 | 64 | ||
| Staphylococcus epidermidis | 8 | 128 | 2 | ||
| Pseudomonas aeruginosa | 64 | 128 | 32 | ||
| Staphylococcus aureus | 2–32 | 32–256 | 1–16 | ||
| (Oral strains = 8) | - | - | (2 to 8-fold decrease in the MIC) | ||
| - | Benzalkonium-chloride | Benzalkonium-chloride | Juglone | With ½ MIC of Juglone | [67] |
| Bacillus cereus | 16 | 256 | 4 | ||
| Escherichia coli | 16 | 128 | 8 | ||
| Salmonella enterica | 16 | 256 | 4 | ||
| Staphylococcus aureus | 16 | 128 | 8 | ||
| Listeria monocytogenes | 2 | 256 | 1 | ||
| Enterococcus feacalis | 8 | 256 | 2 | ||
| Staphylococcus epidermidis | 4 | 128 | 2 | ||
| Pseudomonas aeruginosa | 32 | 128 | 16 | ||
| Staphylococcus aureus | 4–16 | 32–256 | 1–4 | ||
| (Oral strains = 8) | - | - | (2 to 8-fold decrease in the MIC) | ||
| MIC in mg/L | |||||
| - | - | AC | Plumbagin | Plumbagin (64) + AC | [95] |
| Escherichia coli | Erythromycin | 256 | 128 (Individual MIC) | 64 | |
| Chloramphenicol | 4 | 2 | |||
| Tetraphenyl- phosphonium | 1024 | 256 | |||
| - | AC | Plumbagin | Plumbagin (10) + AC | ||
| Escherichia coli ΔAcrB (Drug sensitive) | Erythromycin | 16 | 10 (Individual MIC) | 16 | |
| Chloramphenicol | 1 | 1 | |||
| Tetraphenyl- phosphonium | 16 | 16 | |||
| - | - | AC | Shikonin | Shikonin (256) | |
| Escherichia coli | Tetraphenyl- phosphonium | 1024 | >256 | 64 | |
| - | - | AC | Shikonin | Shikonin (256) + AC | |
| Escherichia coli DAcrB (Drug sensitive) | Tetraphenyl- phosphonium | 16 | >256 (Individual MIC) | 16 | |
| - | - | AC | Plumbagin nordihydroguaretic acid (NDGA) | Plumbagin NDGA (256) + AC | [95] |
| Escherichia coli | Erythromycin | 256 | 512 (Individual MIC) | 64 | |
| Chloramphenicol | 4 | 1 | |||
| Novobiocin | 512 | 64 | |||
| Tetraphenyl- Phosphonium | 1024 | 64 | |||
| Tetracycline | 1 | 0.25 | |||
| - | - | AC | NDGA | NDGA (25) + AC | |
| Escherichia coli DAcrB (Drug sensitive) | Erythromycin | 16 | 25 (Individual MIC) | 16 | |
| Chloramphenicol | 1 | 1 | |||
| Novobiocin | 16 | 16 | |||
| Tetraphenyl- Phosphonium | 16 | 16 | |||
| Tetracycline | 0.5 | 0.5 | |||
| Test Organism | IC50 (µM) of Naphthoquinone Derivative Inhibiting Supercoiling Activity of DNA Gyrase (Bacterial Topoisomerase II) Enzyme | Reference | |||
|---|---|---|---|---|---|
| Diospyrin | 7-Methyljuglone | Menadione | Shinanolone | ||
| Staphylococcus aureus | 8 | 60 | - | - | [104] |
| Escherichia coli | 4 | 30 | - | - | |
| Mycobacterium tuberculosis | 15 | 30 | >200 | >200 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mone, N.S.; Bhagwat, S.A.; Sharma, D.; Chaskar, M.; Patil, R.H.; Zamboni, P.; Nawani, N.N.; Satpute, S.K. Naphthoquinones and Their Derivatives: Emerging Trends in Combating Microbial Pathogens. Coatings 2021, 11, 434. https://doi.org/10.3390/coatings11040434
Mone NS, Bhagwat SA, Sharma D, Chaskar M, Patil RH, Zamboni P, Nawani NN, Satpute SK. Naphthoquinones and Their Derivatives: Emerging Trends in Combating Microbial Pathogens. Coatings. 2021; 11(4):434. https://doi.org/10.3390/coatings11040434
Chicago/Turabian StyleMone, Nishigandha S., Srushti A. Bhagwat, Deepansh Sharma, Manohar Chaskar, Rajendra H. Patil, Paolo Zamboni, Neelu N. Nawani, and Surekha K. Satpute. 2021. "Naphthoquinones and Their Derivatives: Emerging Trends in Combating Microbial Pathogens" Coatings 11, no. 4: 434. https://doi.org/10.3390/coatings11040434
APA StyleMone, N. S., Bhagwat, S. A., Sharma, D., Chaskar, M., Patil, R. H., Zamboni, P., Nawani, N. N., & Satpute, S. K. (2021). Naphthoquinones and Their Derivatives: Emerging Trends in Combating Microbial Pathogens. Coatings, 11(4), 434. https://doi.org/10.3390/coatings11040434






