Diphenyl-Furanones and Diphenyl-Oxopyrrole Derivatives: From Analytical Reagents for Amino Groups to New Fluorochromes for Cytochemical Staining of Chromatin DNA and Chromosomes: Proposal for Intercalative Binding and Fluorescence Mechanism
Abstract
:1. Introduction
2. Materials and Methods
3. Results
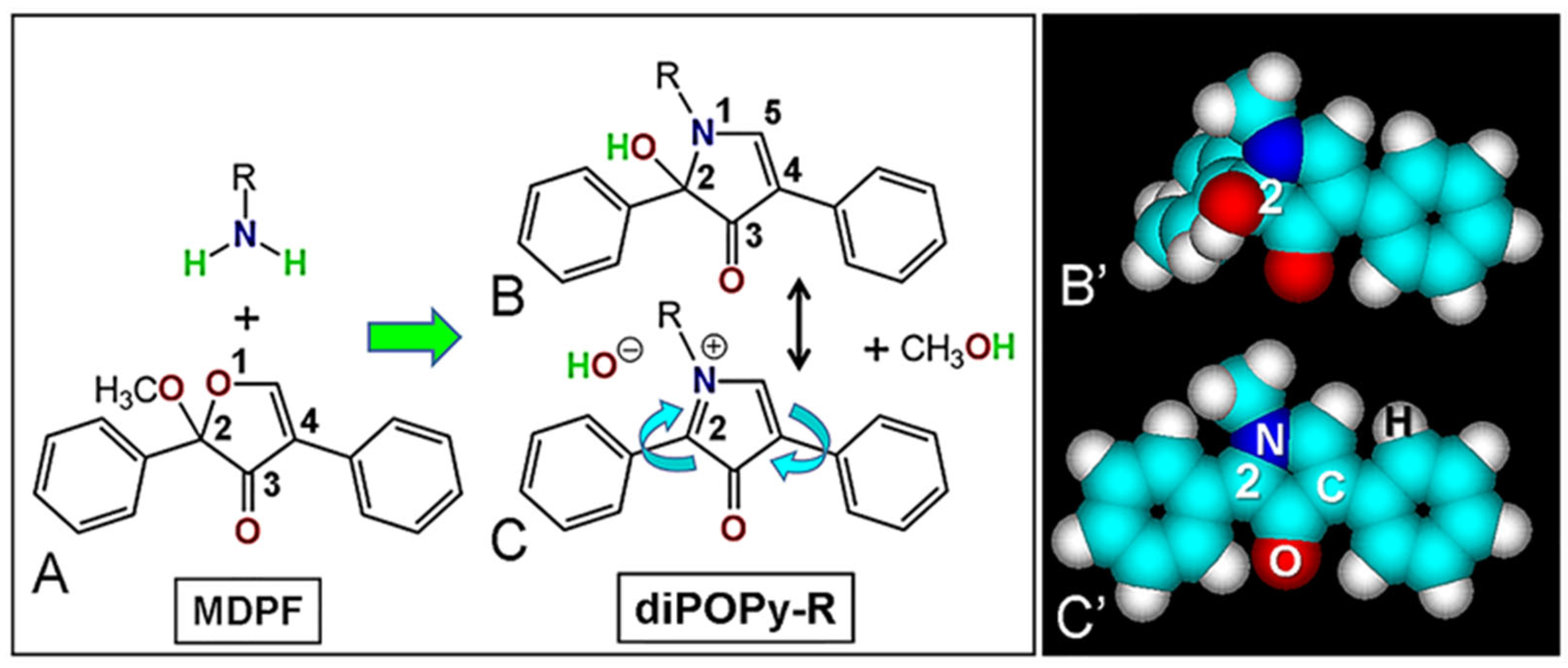
3.1. DiPOPy Fluorochromes from MDPF
3.2. BzPOPy Fluorochromes from Fluorescamine
4. Discussion
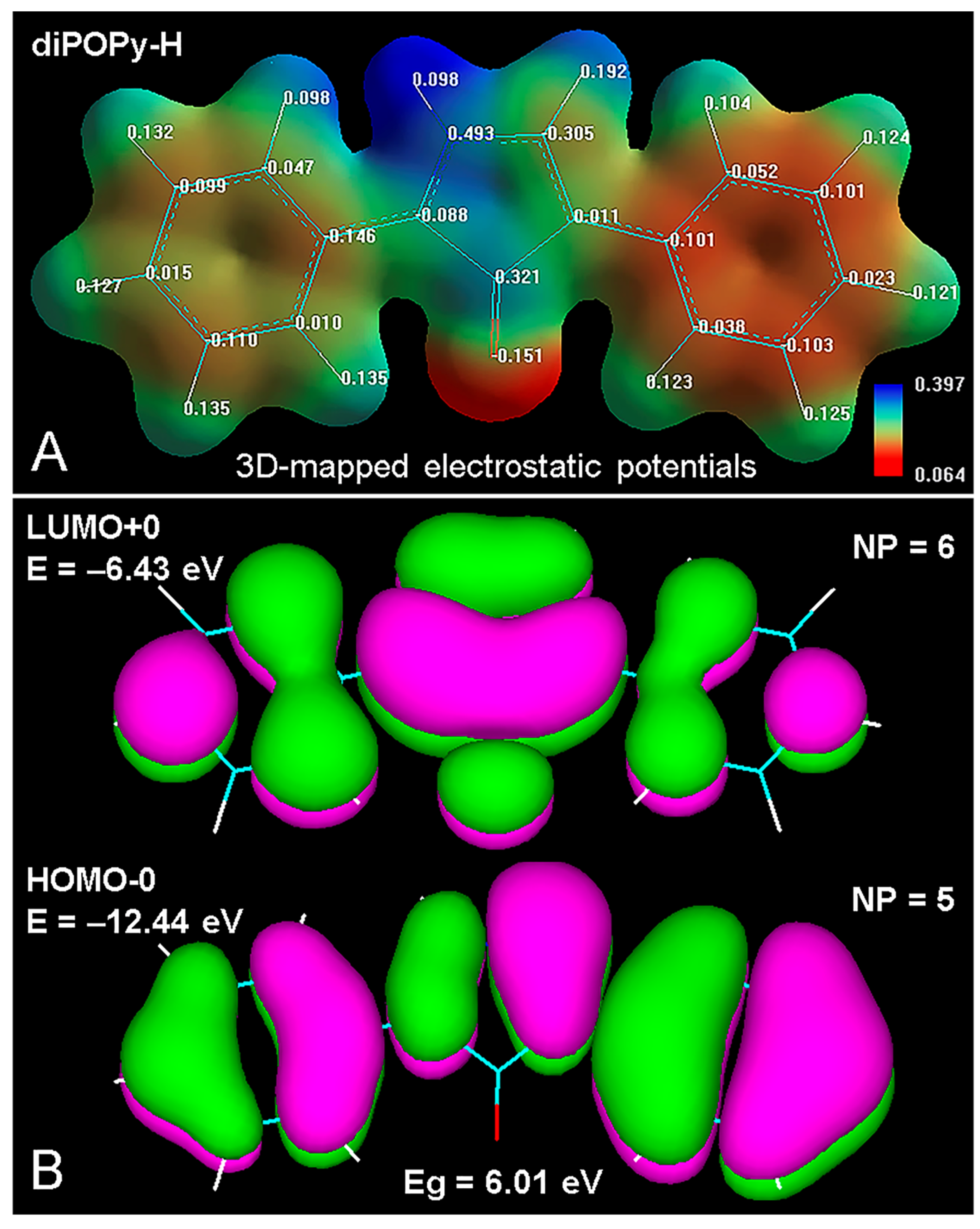
4.1. DiPOPy Derivatives as Fluorochromes
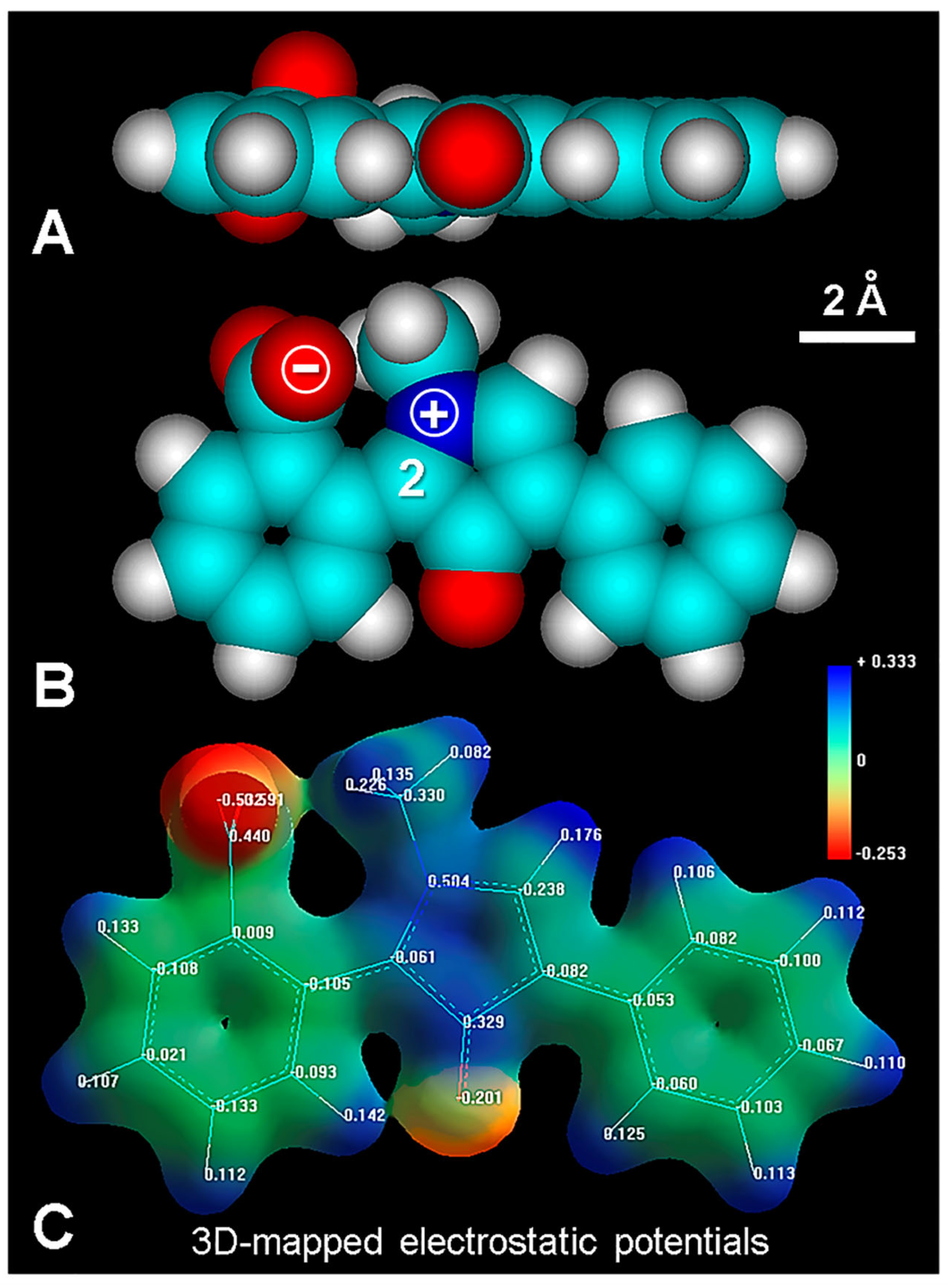
4.2. Bis-Intercalative Binding
4.3. Fluorescence Mechanisms of Fluorescamine Products
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Udenfriend, S.; Stein, S.; Böhlen, P.; Dairman, W.; Leimgruber, W.; Weigele, M. Fluorescamine: A reagent for assay of amino acids, peptides, proteins and primary amines in the picomole range. Science 1972, 178, 871–872. [Google Scholar] [CrossRef] [PubMed]
- Weigele, M.; De Bernardo, S.L.; Tengi, J.P.; Leimgruber, W. A novel reagent for the fluorometric assay of primary amines. J. Am. Chem. Soc. 1972, 94, 5927–5928. [Google Scholar] [CrossRef]
- Böhlen, P.; Stein, S.; Dairman, W.; Udenfriend, S. Fluorometric assay of proteins in the nanogram range. Arch. Biochem. Biophys. 1973, 155, 213–220. [Google Scholar] [CrossRef]
- Lai, C.Y. Detection of peptides by fluorescent methods. Meth. Enzymol. 1977, 47, 236–243. [Google Scholar]
- Toome, V.; Wegrzynski, B. Application of MDPF and fluorescamine. XV. Chiroptical properties of MDPF condensation compounds with dipeptides in situ. Biochem. Biophys. Res. Commun. 1983, 114, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Felix, A.M.; Jimenez, M.H. Usage of fluorescamine as a spray reagent for thin-layer chromatography. J. Chromatogr. 1974, 89, 361–364. [Google Scholar] [CrossRef]
- Samejima, K. Separation of fluorescamine derivatives of aliphatic diamines and polyamines by high-speed liquid chromatography. J. Chromatogr. 1974, 96, 250–254. [Google Scholar] [CrossRef]
- Tata, S.J.; Moir, G.F.J. Fluorescamine as a reagent for location of proteins after electrophoresis in starch gel or on paper. Anal. Biochem. 1976, 70, 495–498. [Google Scholar] [CrossRef]
- Nakamura, H.; Takagi, K.; Tamura, Z.; Yoda, R.; Yamamoto, Y. Stepwise fluorometric determination of primary and secondary amines by liquid chromatography after derivatization with 2 methoxy 2,4-diphenyl 3(2H)-furanone. Anal. Chem. 1984, 56, 919–922. [Google Scholar] [CrossRef]
- Bodhe, A.M.; Vartak, H.G.; Rele, M.V.; Dhamankar, V.S. Location and purification of enzymes on polyacrylamide gels using dialyzable fluorescent markers. Anal. Biochem. 1987, 164, 39–43. [Google Scholar] [CrossRef]
- Miedel, M.C.; Hulmes, J.D.; Pan, Y.C. The use of fluorescamine as a detection reagent in protein microcharacterization. J. Biochem. Biophys. Meth. 1989, 18, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Kok, W.T. Postcolumn derivatization of peptides with fluorescamine in capillary electrophoresis. J. Chromatogr. A 1998, 814, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Weigele, M.; De Bernardo, S.L.; Leimgruber, W.; Cleeland, R.; Grunberg, E. Fluorescent labeling of proteins. A new methodology. Biochem. Biophys. Res. Commun. 1973, 54, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Tu, S.I.; Grosso, L. Fluorescent labeling of proteins in sodium dodecyl sulfate complexes with fluorescamine. Biochem. Biophys. Res. Commun. 1976, 72, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Barger, B.O.; White, F.C.; Pace, J.L.; Kemper, D.L.; Ragland, W.L. Estimation of molecular weight by polyacrylamide gel electrophoresis using heat stable fluorophors. Anal. Biochem. 1976, 70, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Fishman, M.A.; Hagen, S.; Trotter, J.L.; O’Connell, K.; Agrawal, H.C. Use of a stable fluorescent reagent, 2-methoxy-2,4-diphenyl-3(2H)-furanone, for the visualization and purification of myelin proteins. J. Neurochem. 1979, 32, 1077–1083. [Google Scholar] [CrossRef]
- Falk, B.W.; Elliott, C. Fluorescent monitoring of proteins during sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting. Anal. Biochem. 1985, 144, 537–541. [Google Scholar] [CrossRef]
- Jackson, P.; Urwin, V.E.; Mackay, C.D. Rapid imaging, using a cooled charge-coupled-device, of fluorescent two-dimensional polyacrylamide gels produced by labeling proteins in the first-dimensional isoelectric focusing gel with the fluorophore 2-methoxy-2,4-diphenyl-3(2H)furanone. Electrophoresis 1988, 9, 330–339. [Google Scholar] [CrossRef]
- Alba, F.J.; Daban, J.R. Nonenzymatic chemiluminescent detection and quantitation of total protein on Western and slot blots allowing subsequent immunodetection and sequencing. Electrophoresis 1997, 18, 1960–1966. [Google Scholar] [CrossRef]
- Sprinzl, M.; Faulhammer, H.G. Participation of X47-fluorescamine modified E. coli tRNAs in in vitro protein biosynthesis. Nucleic Acids Res. 1978, 5, 4837–4853. [Google Scholar] [CrossRef]
- Kasai, H.; Hayami, H.; Yamaizumi, Z.; Saito, H.; Nishimra, S. Detection and identification of mutagens and carcinogens as their adducts with guanosine derivatives. Nucleic Acids Res. 1984, 12, 2127–2136. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, S.P.; Meehan, T.D.; Bissell, M.J. The use of fluorescamine as a probe for labeling the outer surface of the plasma membrane. Biochem. Biophys Res. Commun. 1976, 68, 1226–1233. [Google Scholar] [CrossRef] [PubMed]
- Cross, J.W.; Briggs, W.R. Labeling of membranes from erythrocytes and corn with fluorescamine. Biochim. Biophys. Acta 1977, 471, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Håkanson, R.; Larsson, L.I.; Sundler, F. Fluorescamine: A novel reagent for the histochemical detection of amino groups. Histochemistry 1974, 39, 15–23. [Google Scholar] [CrossRef]
- Larsson, L.I.; Sundler, F.; Håkanson, R. Fluorescamine as a histochemical reagent: Demonstration of polypeptide hormone-secreting cells. Histochemistry 1975, 44, 245–251. [Google Scholar] [CrossRef]
- Bruni, A.; Fasulo, M.P.; Tosi, B.; Dall’Olio, G.; Vannini, G.L. Fluorogenic detection of primary amines in plant histochemistry with fluorescamine: A comparative study on the effects of coagulant and non-coagulant fixatives. Histochemistry 1976, 48, 269–281. [Google Scholar] [CrossRef]
- Sciorra, L.J.; Lee, M.L.; Wynnyckyj, H. Study of human chromosomes. IV. Labeling of chromosomal proteins with the amino group specific fluorescent reagent fluorescamine. J. Histochem. Cytochem. 1985, 33, 1252–1255. [Google Scholar] [CrossRef]
- Cuéllar, T.; Gosálvez, J.; Del Castillo, P.; Stockert, J.C. Fluram induces species-dependent C and G bands in mammalian chromosomes, revealing heterogeneous distribution of chromosomal proteins. Genome 1991, 34, 772–776. [Google Scholar] [CrossRef]
- Stockert, J.C.; Trigoso, C.I. Fluorescence of eosinophil leukocyte granules induced by the fluorogenic reagent 2 methoxy 2,4-diphenyl 3(2H)-furanone. Blood Cells 1993, 19, 423–430. [Google Scholar]
- Sundler, F.; Uddman, R.; Larsson, L.I.; Telenius-Berg, M.; Berg, B.; Håkanson, R. Fluorescamine in diagnosis of thyroid carcinomas. Acta Cytol. 1978, 22, 54–56. [Google Scholar]
- Uddman, R.; Larsson, L.I.; Sundler, F. Fluorescamine as a cytochemical detection reagent for mammary carcinoma cells. Acta Cytol. 1978, 22, 273–275. [Google Scholar]
- Lorch-Jorgensen, L.; Ingemansson, S.; Larsson, L.I. Formaldehyde-fluorescamine-induced fluorescence as a property of carcinoma cells. Virchows Arch. B Cell Pathol. 1979, 30, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Parry, G.; Blenis, J.; Hawkes, S.P. Detection of transformed cells using a fluorescent probe: The molecular basis for the differential reaction of fluorescamine with normal and transformed cells. Cytometry 1982, 3, 97–103. [Google Scholar] [CrossRef] [PubMed]
- De Bernardo, S.; Weigele, M.; Toome, V.; Manhart, K.; Leimgruber, W.; Böhlen, P.; Stein, S.; Udenfriend, S. Studies on the reaction of fluorescamine with primary amines. Arch. Biochem. Biophys. 1974, 163, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Weigele, M.; Tengi, J.P.; De Bernardo, S.; Czajkowski, R.; Leimgruber, W. Fluorometric reagents for primary amines. Synthesis of 2-alkoxy- and 2-acyloxy-3(2H)-furanones. J. Org. Chem. 1976, 41, 388–389. [Google Scholar] [CrossRef]
- Stockert, J.C.; Blázquez-Castro, A. Fluorescence Microscopy in Life Sciences; Bentham Science Publishers: Sharjah, United Arab Emirates, 2017. [Google Scholar] [CrossRef]
- Stockert, J.C. Reactive staining reagents and fluorescent labels. In Conn’s Biological Stains. A Handbook of Dyes, Stains and Fluorochromes for Use in Biology and Medicine, 10th ed.; Horobin, R.W., Kiernan, J.A., Eds.; Bios Scientific Publishers: Oxford, UK, 2002; pp. 77–88. ISBN 859960995. [Google Scholar]
- Stockert, J.C.; Blázquez, A.; Galaz, S.; Juarranz, A. A mechanism for the fluorogenic reaction of amino groups with fluorescamine and 2 methoxy 2,4-diphenyl 3(2H)-furanone. Acta Histochem. 2008, 110, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Stockert, J.C.; Abasolo, M.I. Inaccurate chemical structure of dyes and fluorochromes found in the literature can be problematic for teaching and research. Biotech. Histochem. 2011, 86, 52–60. [Google Scholar] [CrossRef]
- Stockert, J.C.; Romero, S.A.; Felix-Pozzi, M.N.; Blázquez-Castro, A. In Vitro polymerization of the dopamine-borate melanin precursor: A proof-of-concept regarding 10boron neutron-capture therapy for melanoma. Biocell 2023, 47, 919–928. [Google Scholar] [CrossRef]
- Molero, M.L.; Hazen, M.J.; Pérez Gorroño, A.I.; Stockert, J.C. Simple ß-carboline alkaloids as nucleic acids fluorochromes. Acta Histochem. 1995, 97, 165–173. [Google Scholar] [CrossRef]
- Stockert, J.C.; Cañete, M.; Villanueva, A.; Juarranz, A.; Trigoso, C.I.; Braña, M.F. Fluorescence of chromatin DNA induced by antitumoral naphthalimides. Z. Nat. 1997, 52, 408–411. [Google Scholar] [CrossRef]
- Juarranz, A.; Villanueva, A.; Cañete, M.; Polo, S.; Domínguez, V.; Stockert, J.C. Microscopical and spectroscopic studies on the fluorescence of a daunomycin-aluminum complex. Histochem. J. 1999, 31, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Stockert, J.C.; Del Castillo, P.; Llorente, A.R.; Rasskin, D.M.; Romero, J.B.; Gómez, A. New fluorescence reactions in DNA cytochemistry. 1. Microscopic and spectroscopic studies on nonrigid fluorochromes. Anal. Quant. Cytol. Histol. 1990, 12, 1–10. [Google Scholar] [PubMed]
- Stockert, J.C.; Trigoso, C.I.; Cuéllar, T.; Bella, J.L.; Lisanti, J.L. A new fluorescence reaction in DNA cytochemistry: Microscopic and spectroscopic studies on the aromatic diamidino compound M&B 938. J. Histochem. Cytochem. 1997, 45, 97–105. [Google Scholar] [PubMed]
- Stockert, J.C.; Del Castillo, P.; Romero, J.B.; Tato, A.; Llorente, A.R.; Ferrer, J.M. Orientation of nucleosomes in the 30 nm chromatin fiber as revealed by microscopic studies of linear dichroism and polarized fluorescence. Biol. Zentralbl. 1990, 109, 471–480. [Google Scholar]
- Mello, M.L.S.; Vidal, B.C. Polarization microscopy of extended chromatin fibers. Meth. Mol. Biol. 2014, 1094, 71–78. [Google Scholar] [CrossRef]
- Stockert, J.C. Monomerizing effect of caffeine, o-phenanthroline and tannin on cationic dyes: A model system to analyze spectral characteristics of the intercalative binding to nucleic acids. Acta Histochem. 1989, 87, 33–42. [Google Scholar] [CrossRef]
- Trigoso, C.I.; Del Castillo, P.; Stockert, J.C. Influence of inorganic salts on the staining reaction of eosinophil lecocyte granules by anionic dyes. Acta Histochem. 1992, 93, 313–318. [Google Scholar] [CrossRef]
- Stockert, J.C. The horse eosinophil as a model leucocyte for morphological and cytochemical studies. Braz. J. Morphol. Sci. 2005, 22, 73–84. [Google Scholar]
- Del Castillo, P.; Horobin, R.W.; Blázquez-Castro, A.; Stockert, J.C. Binding of cationic dyes to DNA: Distinguishing intercalation and groove binding mechanisms using simple experimental and numerical models. Biotech. Histochem. 2010, 85, 247–256. [Google Scholar] [CrossRef]
- Stockert, J.C.; Espada, J.; Blázquez-Castro, A. Melanin-binding colorants: Updating molecular modeling, staining and labeling mechanisms, and biomedical perspectives. Colorants 2022, 1, 91–120. [Google Scholar] [CrossRef]
- Stockert, J.C. Molecular modeling of phenyl-boronic acids and catechol-borate esters: A mechanistic rationale for boron binding to melanin catechols, regarding the 10boron neutron-capture therapy of melanoma. Ann. Rev. Res. 2022, 8, 555726. [Google Scholar] [CrossRef]
- Stockert, J.C.; Blázquez-Castro, A.; Horobin, R.W. Identifying different types of chromatin using Giemsa staining. Meth. Mol. Biol. 2014, 1094, 25–38. [Google Scholar]
- Stockert, J.C. Cytochemistry of nucleic acids: Binding mechanism of dyes and fluorochromes. Biocell 1985, 9, 89–131. [Google Scholar]
- Blackburn, G.M.; Gait, M.J. (Eds.) Nucleic Acids in Chemistry and Biology; IRL Press: Oxford, UK; New York, NY, USA, 1990; pp. 313–326. [Google Scholar]
- Mason, S.F. Color and the Electronic States of Organic Molecules. Chemistry of the Synthetic Dyes; J. Wiley and Sons: New York, NY, USA, 1970; pp. 169–221. [Google Scholar]
- Stockert, J.C. Lipid peroxidation assay using BODIPY-phenylbutadiene probes: A methodological overview. Meth. Mol. Biol. 2021, 2202, 199–214. [Google Scholar] [CrossRef]
- Stockert, J.C.; Pinna-Senn, E.; Bella, J.L.; Lisanti, J.A. DNA-binding fluorochromes: Correlation between C-banding of mouse metaphase chromosomes and hydrogen bonding to adenine-thymine base pairs. Acta Histochem. 2005, 106, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Horobin, R.W.; Kiernan, J.A. Conn’s Biological Stains. A Handbook of Dyes, Stains and Fluorochromes for Use in Biology and Medicine, 10th ed.; Bios Scientific Publishers: Oxford, UK, 2002; ISBN 859960995. [Google Scholar]
- Moutschen, J.; Degraeve, N.; Moutschen-Dahmen, M. Chromosome fluorescence with berberine. Cytobiologie 1973, 8, 112–117. [Google Scholar]
- Cañete, M.; Villanueva, A.; Juarranz, A.; Stockert, J.C. A study of interaction of thioflavine T with DNA: Evidence for intercalation. Cell. Mol. Biol. 1987, 33, 191–199. [Google Scholar]
- Stockert, J.C.; Trigoso, C.I.; Llorente, A.R.; Del Castillo, P. DNA fluorescence induced by polymethine cation pyrvinium binding. Histochem. J. 1991, 23, 548–552. [Google Scholar] [CrossRef]
- Glazer, A.N.; Rye, H.S. Stable dye-DNA intercalation complexes as reagents for high-sensitivity fluorescence detection. Nature 1992, 359, 859–861. [Google Scholar] [CrossRef]
- Stockert, J.C. 2,5-bis(4-aminophenyl)-1,3,4-oxadiazol: Fluorescence reaction of chromatin and basophilic cytoplasm. Acta Histochem. Cytochem. 1983, 16, 66–69. [Google Scholar] [CrossRef]
- Stockert, J.C.; Pelling, C.; Espada, J. New cationic fluorochromes from diaryloxazole scintillators: Fluorescence of chromatin DNA induced by N-quaternary POPOP derivatives. Acta Histochem. 1997, 99, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Pinna-Senn, E.; Lisanti, J.A.; Ortiz, M.I.; Dalmasso, G.; Bella, J.L.; Gosálvez, J.; Stockert, J.C. Specific heterochromatic banding of metaphase chromosomes using Nuclear Yellow. Biotech. Histochem. 2000, 75, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Stockert, J.C.; Del Castillo, P. Linear dichroism and polarized fluorescence of dye- complexed DNA fibers. Histochemistry 1989, 91, 263–264. [Google Scholar] [CrossRef]
- Saitoh, Y.; Laemmli, U.K. Metaphase chromosome structure: Bands arise from a differential folding path of the highly AT-rich scaffold. Cell 1994, 76, 609–622. [Google Scholar] [CrossRef]
- Weisblum, B.; De Haseth, P.L. Quinacrine, a chromosome stain specific for deoxyadenylate-deoxythymidylate rich regions in DNA. Proc. Natl. Acad. Sci. USA 1972, 69, 629–632. [Google Scholar] [CrossRef] [PubMed]
- Korenberg, J.R.; Engels, W.R. Base ratio DNA content and quinacrine brightness of human chromosomes. Proc. Nat. Acad. Sci. USA 1978, 75, 3382–3386. [Google Scholar] [CrossRef]
- Ferrucci, L.; Mezzanotte, R. A cytological approach to the role of guanine in determining quinacrine fluorescence response in eukaryotic chromosomes. J. Histochem. Cytochem. 1982, 30, 1289–1292. [Google Scholar] [CrossRef] [PubMed]
- Bickmore, W.; Craig, J. Chromosome Bands: Patterns in the Genome; Springer: New York, NY, USA, 1997. [Google Scholar]
- Diekmann, S.; Zarling, D.A. Unique poly(dA) poly(dT) B′-conformation in cellular and synthetic DNAs. Nucleic Acids Res. 1987, 15, 6063–6074. [Google Scholar] [CrossRef]
- Diekmann, S. DNA curvature. Nucleic Acids Mol. Biol. 1987, 1, 138–156. [Google Scholar]
- Hörz, W.; Altenburger, W. Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res. 1981, 9, 683–696. [Google Scholar] [CrossRef]
- Redi, C.A.; Garagna, S.; Della Valle, G.; Bottiroli, G.; Dell’Orto, P.; Viale, G.; Peverali, F.A.; Raimondi, E.; Forejt, J. Differences in the organization and chromosomal allocation of satellite DNA between the European long tailed house mice Mus domesticus and Mus musculus. Chromosoma 1990, 99, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Saenger, W. Principles of Nucleic Acid Structure; Springer-Verlag: New York, NY, USA; Berlin, Germany, 1984; pp. 385–404. [Google Scholar]
- Assa-Munt, N.; Denny, W.A.; Leupin, W.; Kearns, R.D. Proton NMR study of the binding of bis(acridines) to d(AT)5·d(AT)5. 1. Mode of binding. Biochemistry 1985, 24, 1441–1449. [Google Scholar] [CrossRef] [PubMed]
- Tekola, P.; Baak, J.P.; Beliën, J.A.; Brugghe, J. Highly sensitive, specific, and stable new fluorescent DNA stains for confocal laser microscopy and image processing of normal paraffin sections. Cytometry 1994, 17, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Ploeger, L.; Dullens, H.F.J.; Huisman, A.; van Diest, P.J. Fluorescent stains for quantification of DNA by confocal laser scanning microscopy in 3-D. Biotech. Histochem. 2008, 83, 63–69. [Google Scholar] [CrossRef]
- Nygren, J.; Svanvik, N.; Kubista, M. The interactions between the fluorescent dye thiazole orange and DNA. Biopolymers 1998, 46, 39–51. [Google Scholar] [CrossRef]
- Prodhomme, S.; Demaret, J.P.; Vinogradov, S.; Asseline, U.; Morin-Allory, L.; Vigny, P. A theoretical and experimental study of two thiazole orange derivatives with single- and double-stranded oligonucleotides, polydeoxyribonucleotides and DNA. J. Photochem. Photobiol. B Biol. 1999, 53, 60–69. [Google Scholar] [CrossRef]
- Hendry, L.B.; Mahesh, V.B.; Edwin, D.; Bransome, E.D.; Ewing, D.E. Small molecule intercalation with double stranded DNA: Implications for normal gene regulation and for predicting the biological efficacy and genotoxicity of drugs and other chemicals. Mutat. Res. 2007, 623, 53–71. [Google Scholar] [CrossRef]
- Shang, X.F.; Morris-Natschke, S.L.; Liu, Y.Q.; Guo, X.; Xu, X.S.; Goto, M.; Li, J.C.; Yang, G.Z.; Lee, K.H. Biologically active quinoline and quinazoline alkaloids. Part I. Med. Res. Rev. 2018, 38, 775–828. [Google Scholar] [CrossRef]
- Ughetto, G.; Wang, A.H.; Quigley, G.J.; van der Marel, G.A.; van Boom, J.H.; Rich, A. A comparison of the structure of echinomycin and triostin A complexed to a DNA fragment. Nucleic Acids Res. 1985, 13, 2305–2323. [Google Scholar] [CrossRef]
- Lown, J.W.; Hanstock, C.C. Structure and function of the antitumor antibiotic carzinophilin A: The first natural intercalative bisalkylator. J. Am. Chem. Soc. 1982, 104, 3213–3214. [Google Scholar] [CrossRef]
- Huang, C.H.; Mong, S.; Crooke, S.T. Interactions of a new antitumor antibiotic BBM-928A with deoxyribonucleic acid. Bifunctional intercalative binding studied by fluorometry and viscometry. Biochemistry 1980, 19, 5537–5542. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Mirabelli, C.K.; Mong, S.; Crooke, S.T. Intermolecular cross-linking of DNA through bifunctional intercalation of an antitumor antibiotic, luzopeptin A (BBM-928A). Cancer Res. 1983, 43, 2718–2724. [Google Scholar] [PubMed]
- Dai, J.; Punchihewa, C.; Mistry, P.; Ooi, A.T.; Yang, D. Novel DNA bis-intercalation by MLN944, a potent clinical bisphenazine anticancer drug. J. Biol. Chem. 2004, 279, 46096–46103. [Google Scholar] [CrossRef] [PubMed]
- Jobson, A.G.; Willmore, E.; Tilby, M.J.; Mistry, P.; Charlton, P.; Austin, C.A. Effect of phenazine compounds XR11576 and XR5944 on DNA topoisomerases. Cancer Chemother. Pharmacol. 2009, 63, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Mathad, R.L.; Shang, Z.; Sidell, N.; Yang, D. Solution structure of a 2:1 complex of anticancer drug XR5944 with TFF1 estrogen response element: Insights into DNA recognition by a bis-intercalator. Nucleic Acids Res. 2014, 42, 6012–6024. [Google Scholar] [CrossRef]
- Moorthy, N.S.H.N.; Pratheepa, V.; Ramos, M.J.; Vasconcelos, V.; Fernandes, P.A. Fused aryl-phenazines: Scaffold for the development of bioactive molecules. Curr. Drug Targets 2014, 15, 681–688. [Google Scholar] [CrossRef]
- Antony, S.; Agama, Z.; Miao, K.K.; Hollingshead, M.; Holbeck, S.L.; Wright, M.H.; Varticovski, L.; Nagarajan, M.; Morrell, A.; Cushman, M.; et al. Bisindenoisoquinoline bis-1,3-{(5,6-dihydro-5,11-diketo-11H-indeno[1,2-c]isoquinoline)-6-propylamino}propane bis(trifluoroacetate) (NSC 727357), a DNA intercalator and topoisomerase inhibitor with antitumor activity. Mol. Pharmacol. 2006, 70, 1109–1120. [Google Scholar] [CrossRef]
- Gao, Q.; Williams, L.D.; Egli, M.; Rabinovich, D.; Chen, L.; Quigley, G.J.; Rich, A. Drug-induced DNA repair: X-ray structure of a DNA ditercalinium complex. Proc. Natl. Acad. Sci. USA 1991, 88, 2422–2426. [Google Scholar] [CrossRef]
- Braña, M.F.; Castellano, J.-M.; Morán, M.; Pérez de Vega, M.J.; Romerdahl, C.R.; Qian, X.D.; Bousquet, P.; Emling, F.; Schlick, E.; Keilhauer, G. Bis-naphthalimides: A new class of antitumor agents. Anticancer Drug Des. 1993, 8, 257–268. [Google Scholar]
- Wainwright, M. The use of dyes in modern biomedicine. Biotech. Histochem. 2003, 78, 147–155. [Google Scholar] [CrossRef]
- Feughelman, M.; Langridge, R.; Seeds, W.E.; Stokes, A.R.; Wilson, H.R.; Hooper, C.W.; Wilkins, M.H.F.; Barclay, R.K.; Hamilton, L.D. Molecular structure of deoxyribose nucleic acid and nucleoprotein. Nature 1955, 175, 834–838. [Google Scholar] [CrossRef] [PubMed]
- De Santis, P.; Forni, E.; Rizzo, R. Conformational analysis of DNA-basic polypeptide complexes: Possible models of nucleoprotamines and nucleohistones. Biopolymers 1974, 13, 3113–3326. [Google Scholar] [CrossRef] [PubMed]
- Suau, P.; Subirana, J.A. X-ray diffraction studies of nucleoprotamine structure. J. Mol. Biol. 1977, 117, 909–926. [Google Scholar] [CrossRef] [PubMed]
- Warrant, R.W.; Kim, S.H. α-Helix-double helix interaction shown in the structure of a protamine-transfer RNA complex and a nucleoprotamine model. Nature 1978, 271, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Fita, I.; Campos, J.L.; Puigjaner, L.C.; Subirana, J.A. X-ray diffraction study of DNA complexes with arginine peptides and their relation to nucleoprotamine structure. J. Mol. Biol. 1983, 167, 157–177. [Google Scholar] [CrossRef]
- Rohs, R.; West, S.M.; Sosinsky, A.; Liu, P.; Mann, R.S.; Honig, B. The role of DNA shape in protein–DNA recognition. Nature 2009, 461, 1248–1254. [Google Scholar] [CrossRef]
- Pullman, B. Electrostatics of polymorphic DNA. J. Biomol. Struct. Dyn. 1983, 1, 773–794. [Google Scholar] [CrossRef]
- Aymami, J.; Coll, M.; Frederick, C.A.; Wang, A.H.J.; Rich, A. The propeller DNA conformation of poly(dA)·poly(dT). Nucleic Acids Res. 1989, 17, 3229–3245. [Google Scholar] [CrossRef]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stockert, J.C.; Romero, S.A.; Felix-Pozzi, M.N.; Blázquez-Castro, A. Diphenyl-Furanones and Diphenyl-Oxopyrrole Derivatives: From Analytical Reagents for Amino Groups to New Fluorochromes for Cytochemical Staining of Chromatin DNA and Chromosomes: Proposal for Intercalative Binding and Fluorescence Mechanism. Colorants 2023, 2, 245-263. https://doi.org/10.3390/colorants2020016
Stockert JC, Romero SA, Felix-Pozzi MN, Blázquez-Castro A. Diphenyl-Furanones and Diphenyl-Oxopyrrole Derivatives: From Analytical Reagents for Amino Groups to New Fluorochromes for Cytochemical Staining of Chromatin DNA and Chromosomes: Proposal for Intercalative Binding and Fluorescence Mechanism. Colorants. 2023; 2(2):245-263. https://doi.org/10.3390/colorants2020016
Chicago/Turabian StyleStockert, Juan C., Silvina A. Romero, Marcelo N. Felix-Pozzi, and Alfonso Blázquez-Castro. 2023. "Diphenyl-Furanones and Diphenyl-Oxopyrrole Derivatives: From Analytical Reagents for Amino Groups to New Fluorochromes for Cytochemical Staining of Chromatin DNA and Chromosomes: Proposal for Intercalative Binding and Fluorescence Mechanism" Colorants 2, no. 2: 245-263. https://doi.org/10.3390/colorants2020016
APA StyleStockert, J. C., Romero, S. A., Felix-Pozzi, M. N., & Blázquez-Castro, A. (2023). Diphenyl-Furanones and Diphenyl-Oxopyrrole Derivatives: From Analytical Reagents for Amino Groups to New Fluorochromes for Cytochemical Staining of Chromatin DNA and Chromosomes: Proposal for Intercalative Binding and Fluorescence Mechanism. Colorants, 2(2), 245-263. https://doi.org/10.3390/colorants2020016






