Transcriptomic Profile of New Gene Markers Encoding Proteins Responsible for Structure of Porcine Ovarian Granulosa Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Collection of Porcine Ovarian Granulosa Cells
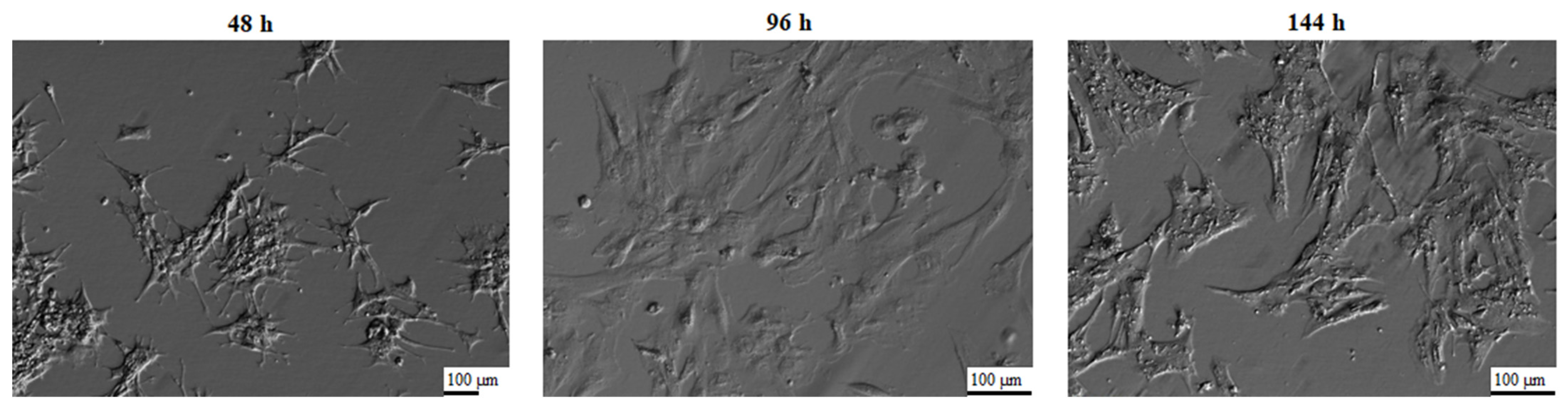
2.3. In Vitro Primary Culture of Porcine Granulosa Cells
2.4. Microarray Expression Analysis and Statistics
2.5. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Analysis
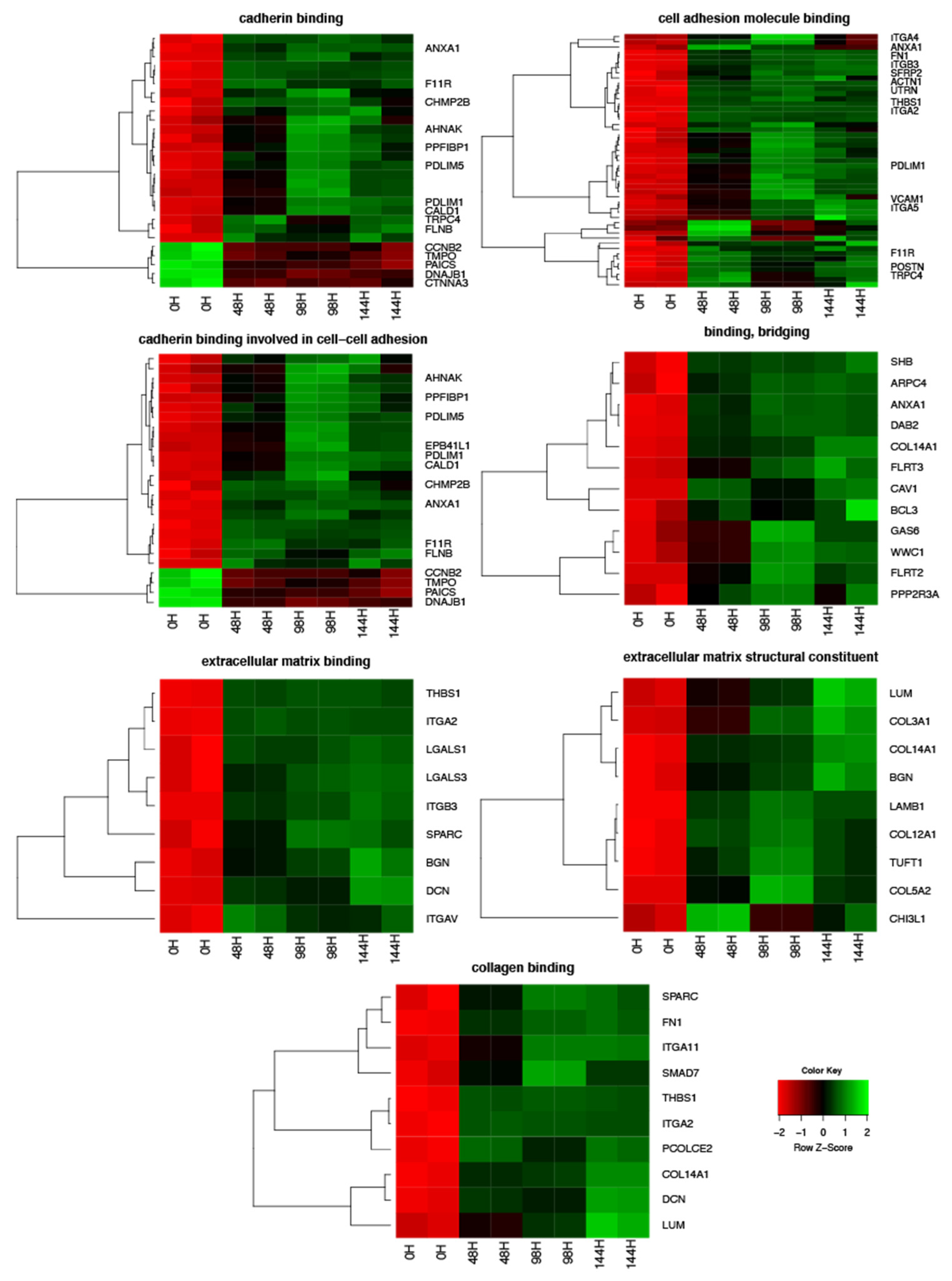
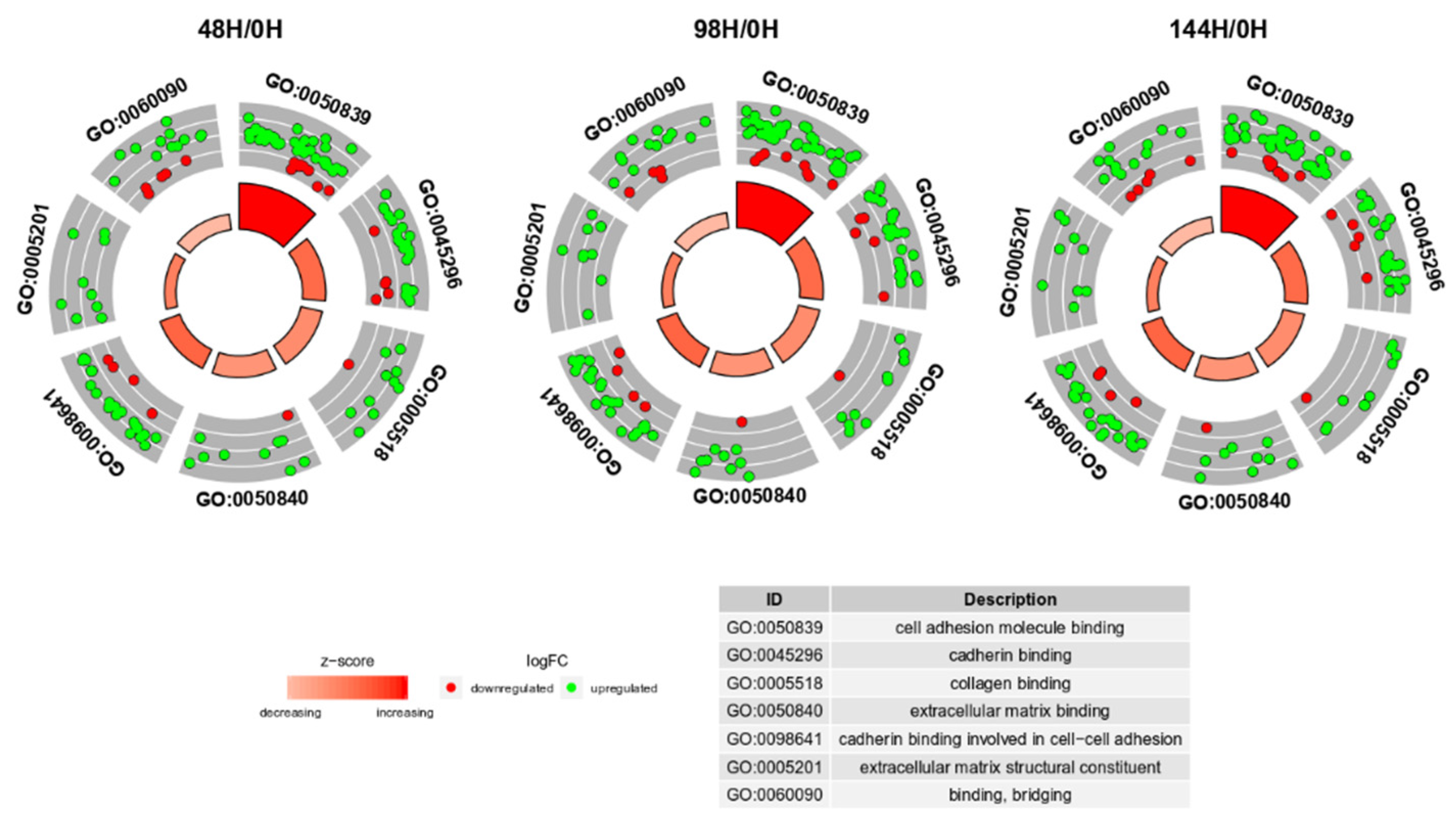
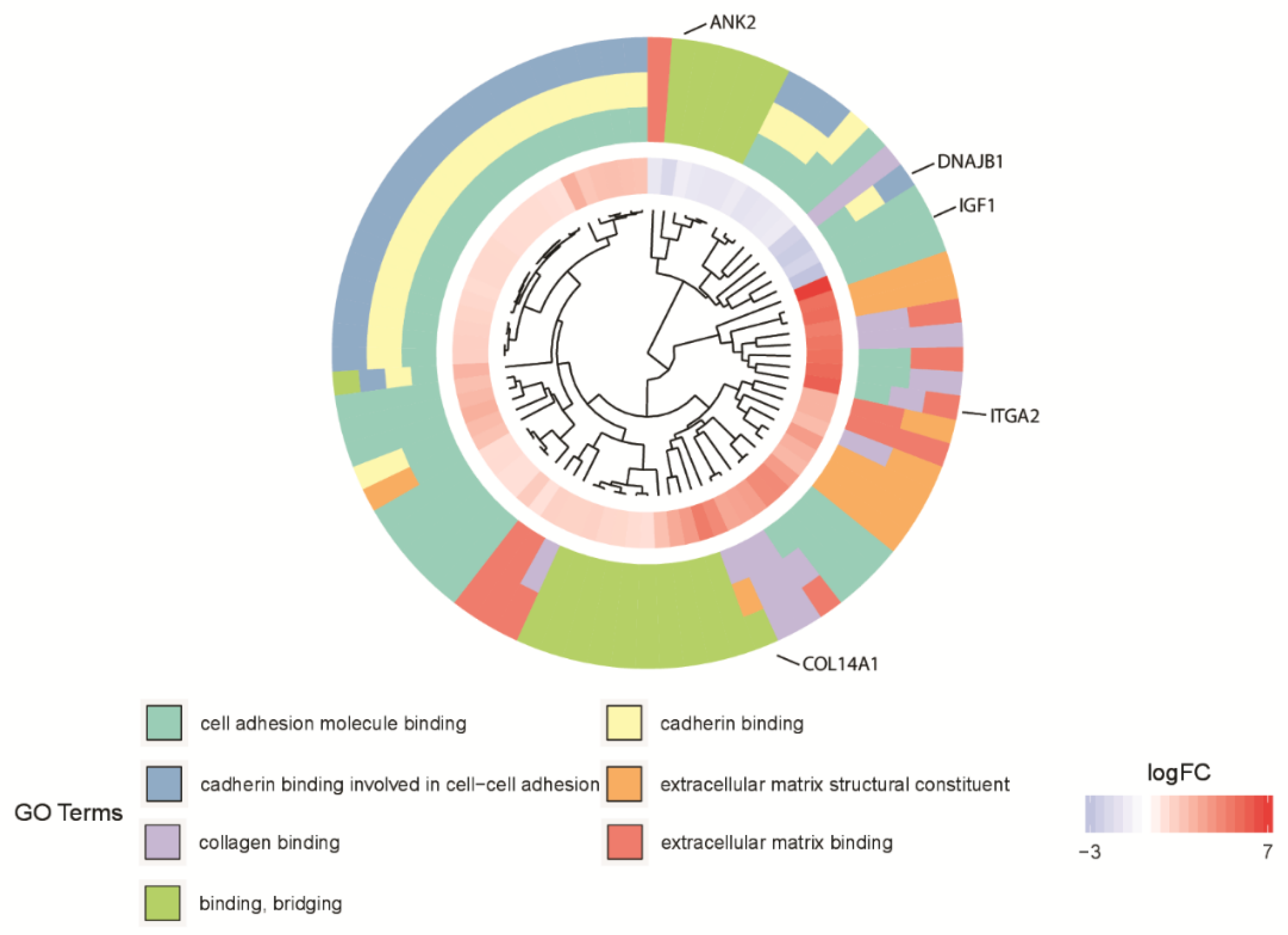
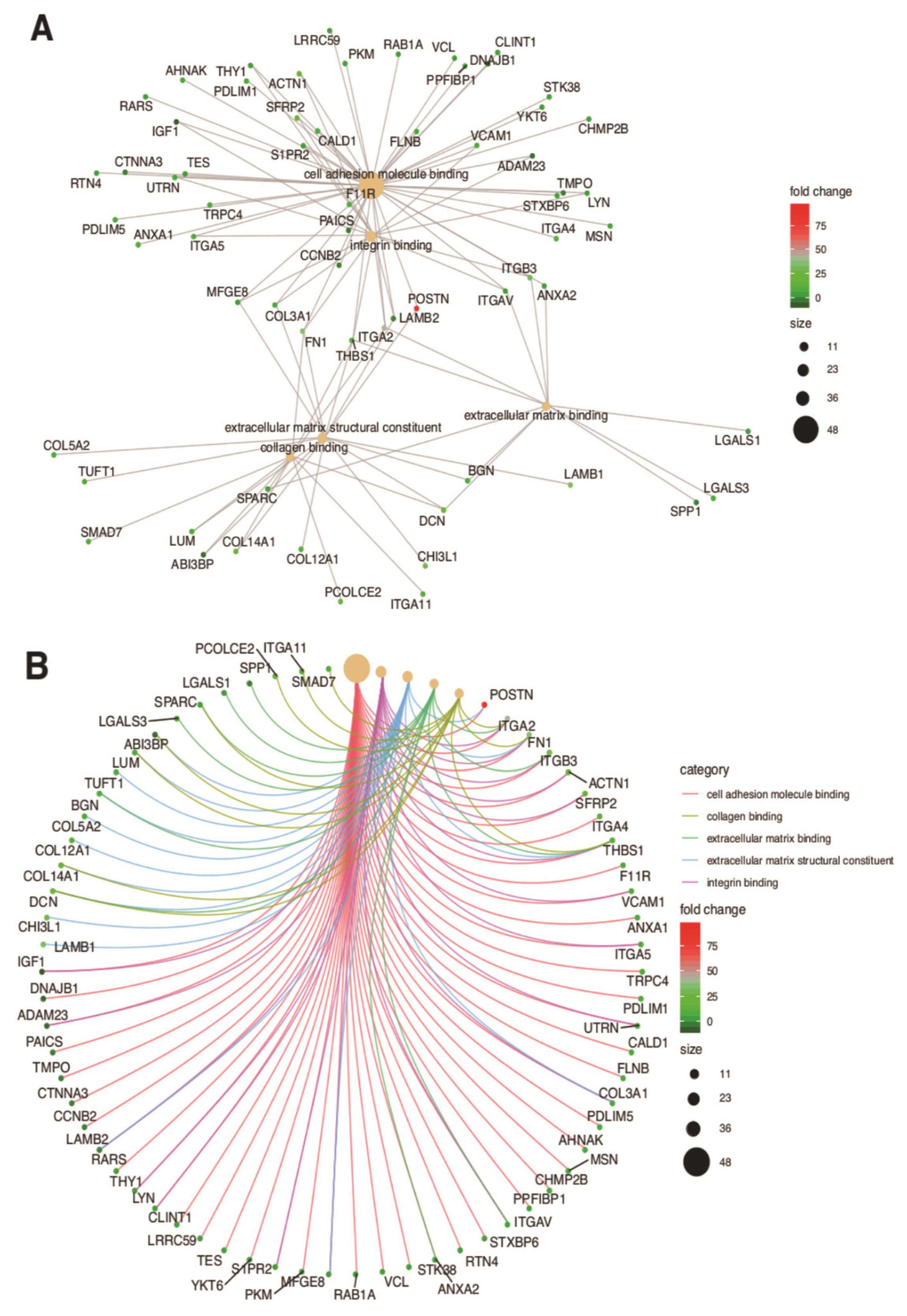
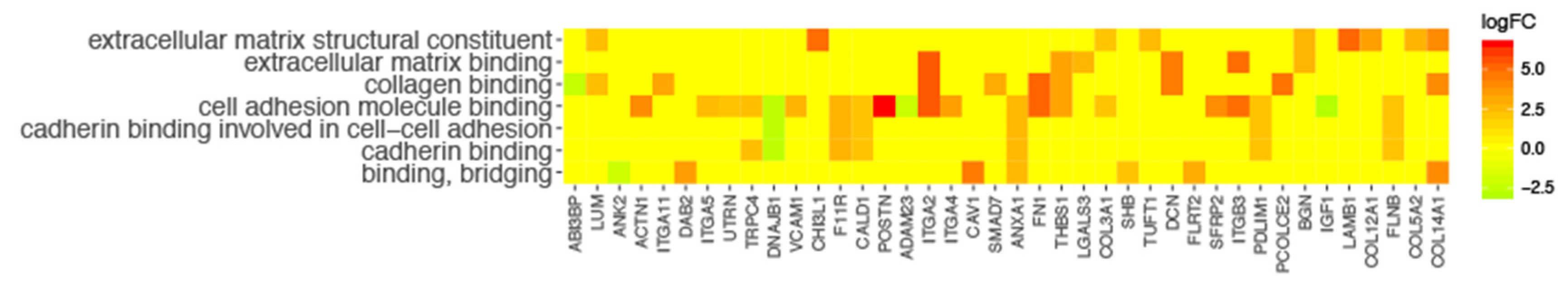
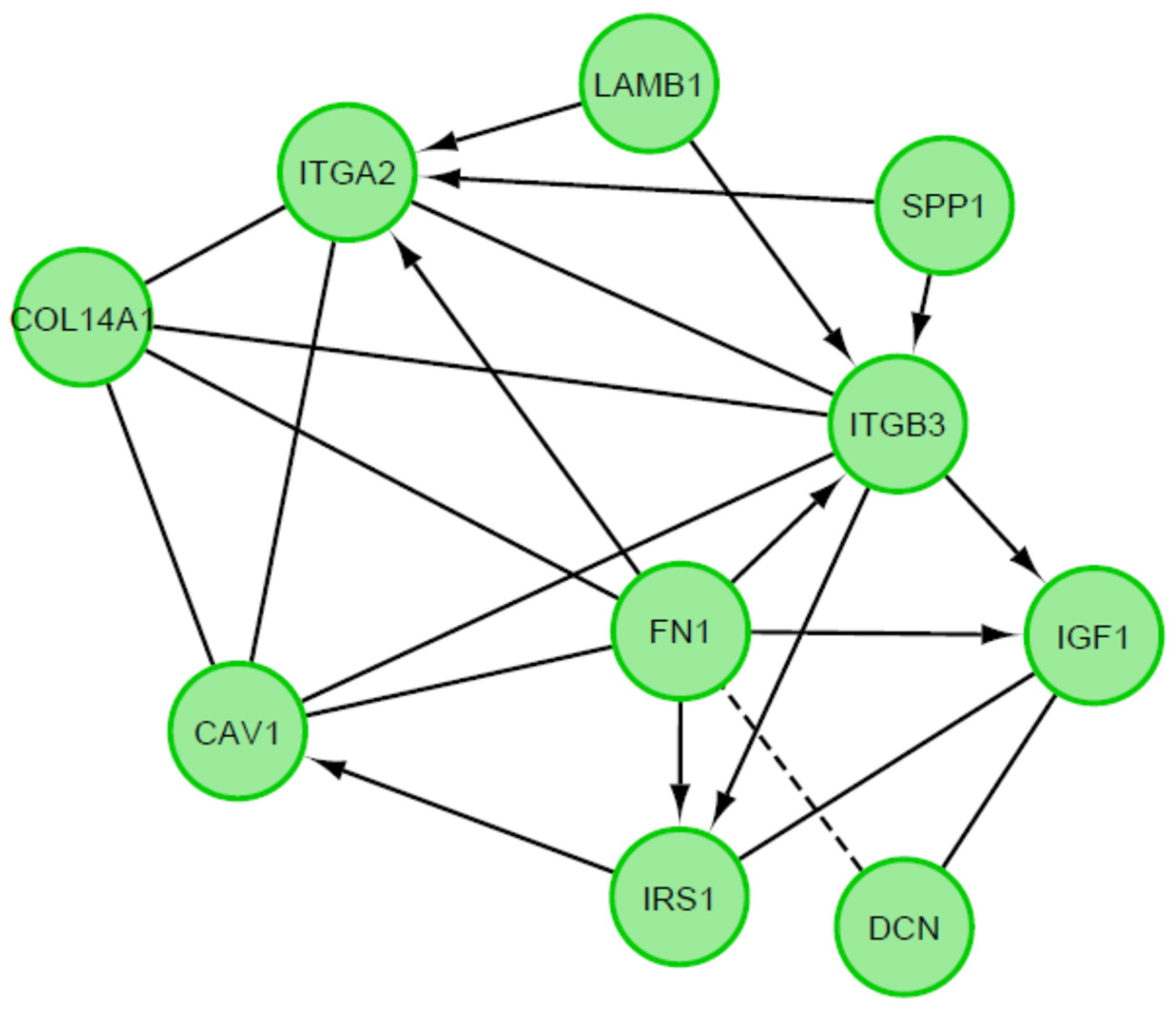
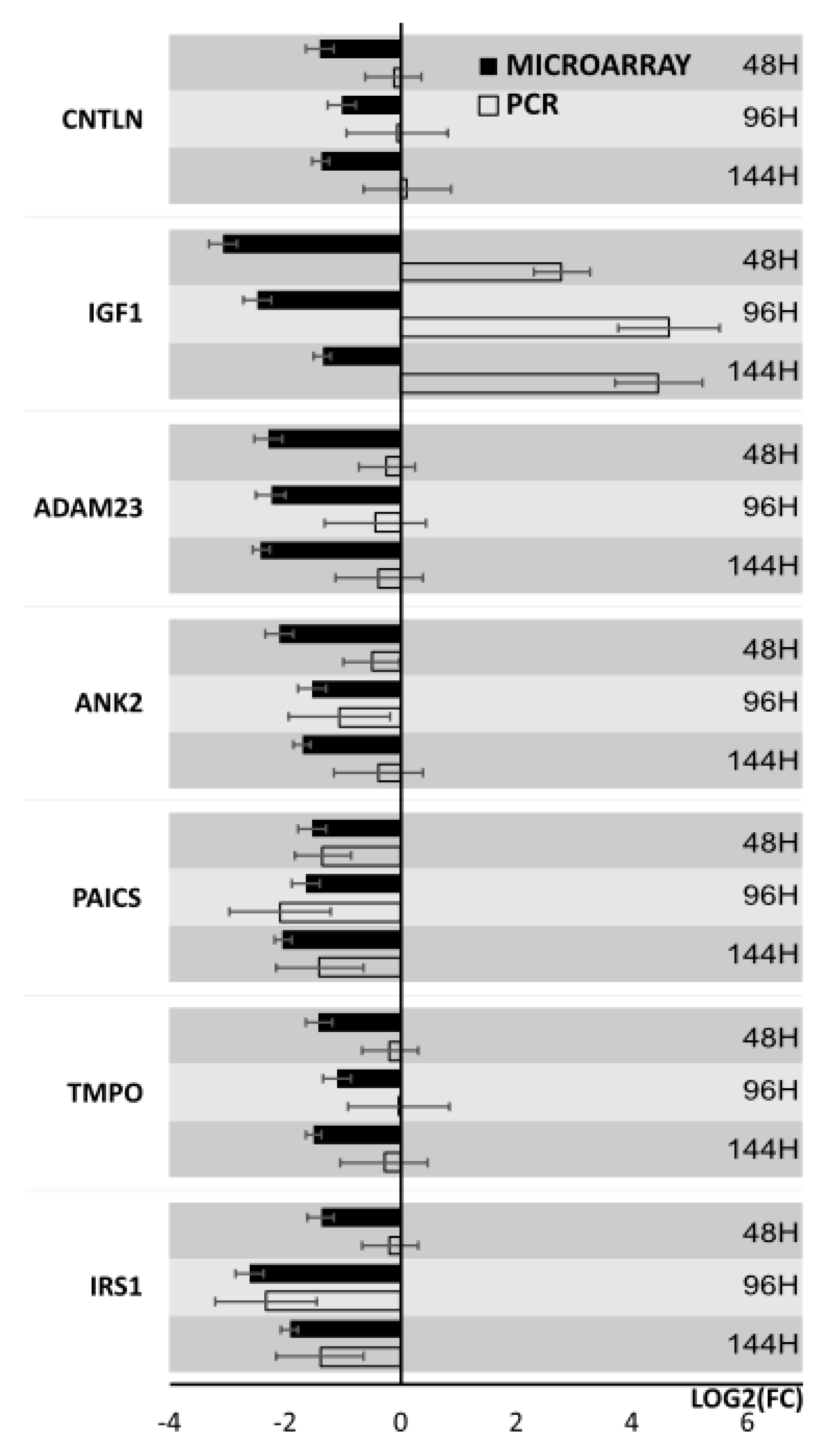
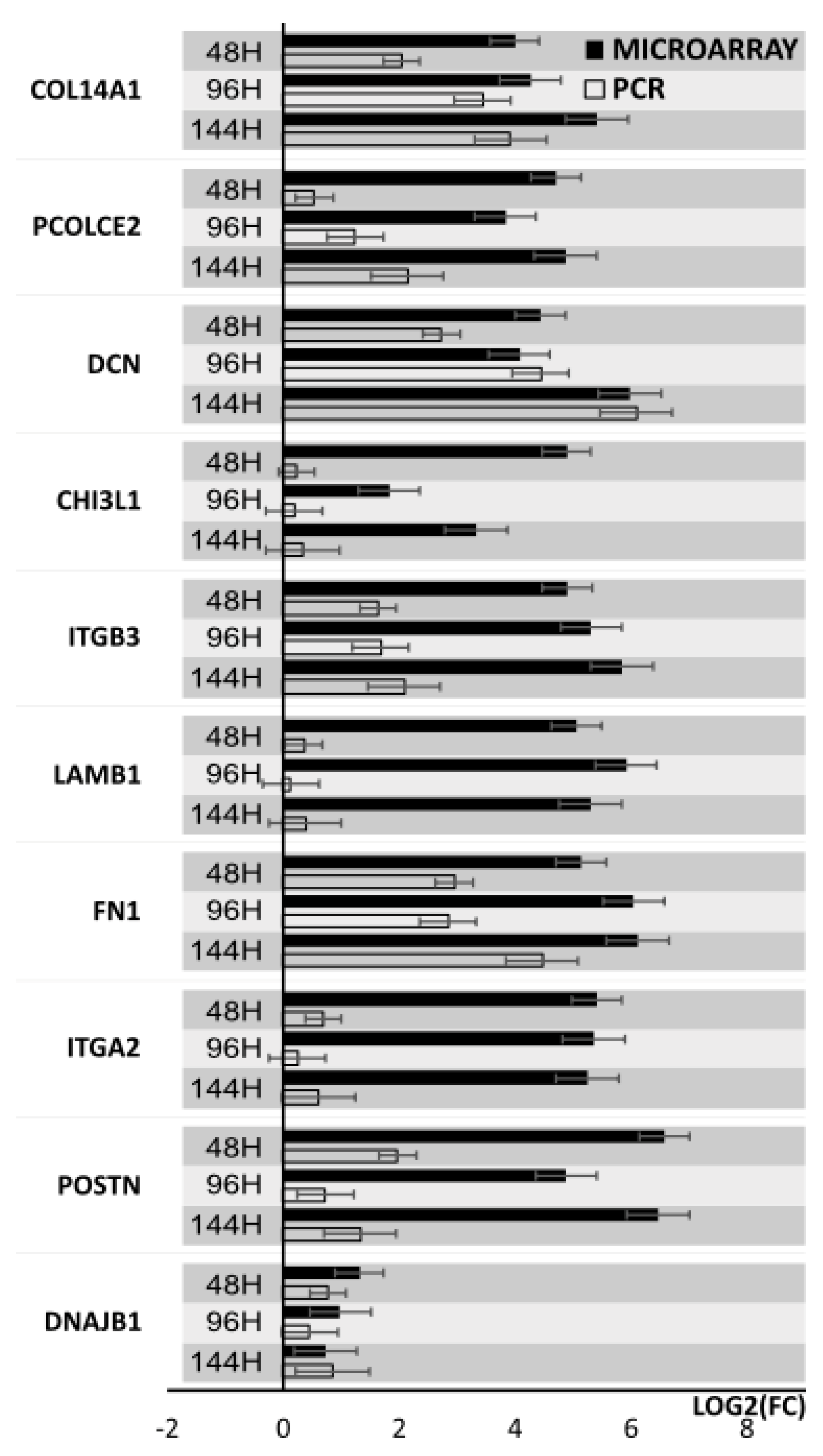
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A


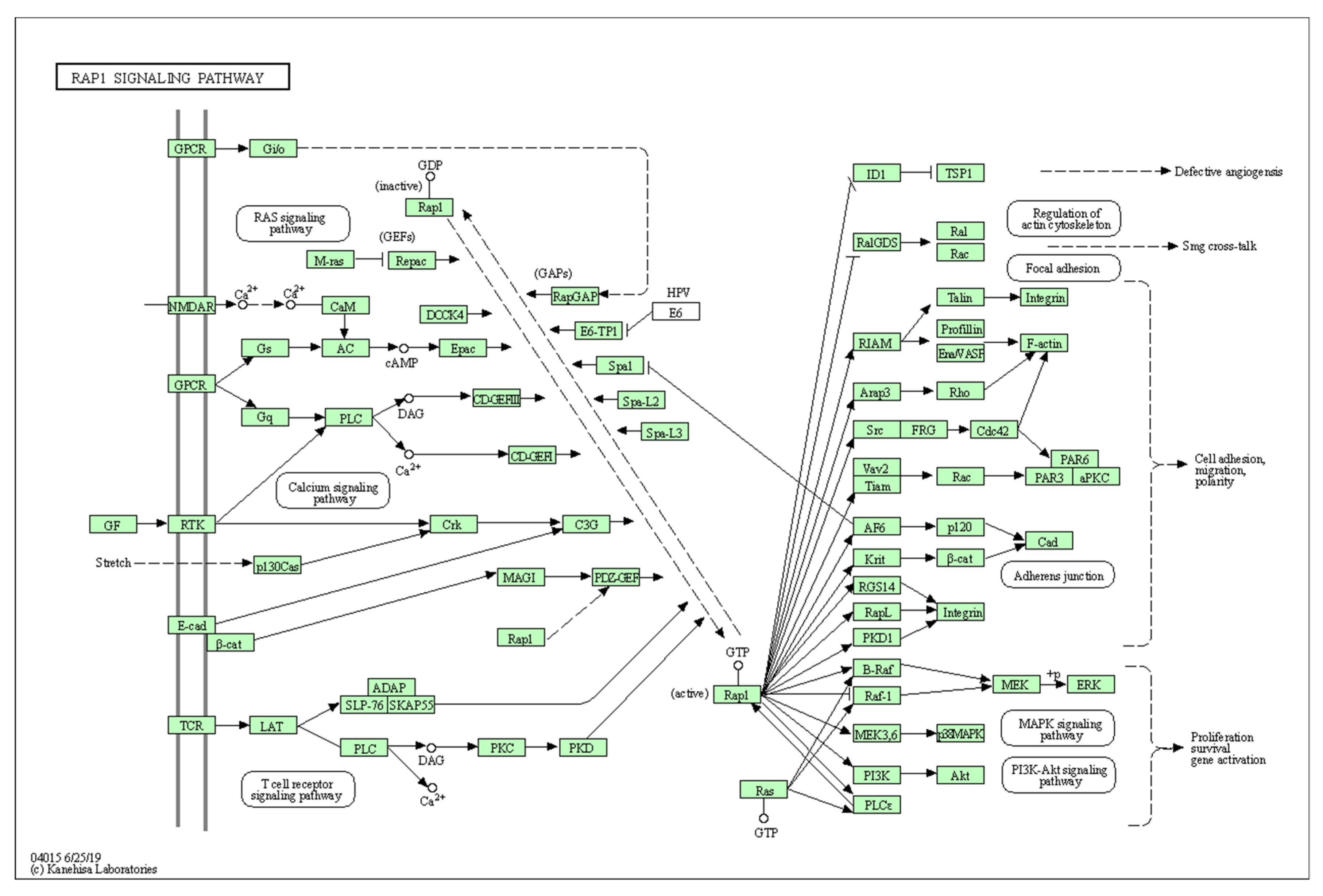
References
- Walma, D.A.C.; Yamada, K.M. The extracellular matrix in development. Development 2020, 147, dev175596. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O. The extracellular matrix: Not just pretty fibrils. Science 2009, 326, 1216–1219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouw, J.K.; Ou, G.; Weaver, V.M. Extracellular matrix assembly: A multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 2014, 15, 771–785. [Google Scholar] [CrossRef]
- Mecham, R.P. Overview of Extracellular Matrix. Curr. Protoc. Cell Biol. 2012, 57, 10.1.1–10.1.16. [Google Scholar] [CrossRef]
- Rodgers, R.J.; Irving-Rodgers, H.F.; Van Wezel, I.L. Extracellular matrix in ovarian follicles. Mol. Cell Endocrinol. 2000, 163, 73–79. [Google Scholar] [CrossRef]
- Cho, A.; Howell, V.M.; Colvin, E.K. The extracellular matrix in epithelial ovarian cancer—A piece of a puzzle. Front. Oncol. 2015, 5, 245. [Google Scholar] [CrossRef] [Green Version]
- Xiang, S.; Xia, M.-f.; Song, J.-y.; Liu, D.-q.; Lian, F. Effect of Electro-acupuncture on Expression of IRS-1/PI3K/GLUT4 Pathway in Ovarian Granulosa Cells of Infertile Patients with Polycystic Ovary Syndrome-Insulin Resistance of Phlegm-Dampness Syndrome. Chin. J. Integr. Med. 2020, 27, 330–335. [Google Scholar] [CrossRef]
- Chen, X.; Huo, L.; Ren, L.; Li, Y.; Sun, Y.; Li, Y.; Zhang, P.; Chen, S.; Song, G.Y. Polycystic Ovary Syndrome is Associated with Elevated Periostin Levels. Exp. Clin. Endocrinol. Diabetes 2019, 127, 571–577. [Google Scholar] [CrossRef]
- Patil, K.; Hinduja, I.; Mukherjee, S. Alteration in angiogenic potential of granulosa-lutein cells and follicular fluid contributes to luteal defects in polycystic ovary syndrome. Hum. Reprod. 2021, 36, 1052–1064. [Google Scholar] [CrossRef]
- Huang, K.; Dang, Y.; Zhang, P.; Shen, C.; Sui, X.; Xia, G.; Qin, Y.; Jiao, X.; Wang, C.; Huo, R.; et al. CAV1 regulates primordial follicle formation via the Notch2 signalling pathway and is associated with premature ovarian insufficiency in humans. Hum. Reprod. 2018, 33, 087–2095. [Google Scholar] [CrossRef]
- Henning, N.F.; LeDuc, R.D.; Even, K.A.; Laronda, M.M. Proteomic analyses of decellularized porcine ovaries identified new matrisome proteins and spatial differences across and within ovarian compartments. Sci. Rep. 2019, 9, 20001. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Feng, H.; Zhang, Y.; Zhang, J.V.; Wang, X.; Liu, D. Current Understandings of Core Pathways for the Activation of Mammalian Primordial Follicles. Cells 2021, 10, 1491. [Google Scholar] [CrossRef] [PubMed]
- Kinnear, H.M.; Tomaszewski, C.E.; Chang, F.L.; Moravek, M.B.; Xu, M.; Padmanabhan, V.; Shikanov, A. The ovarian stroma as a new frontier. Reproduction 2020, 160, R25–R39. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, T.K.; Shea, L.D. The Role of the Extracellular Matrix in Ovarian Follicle Development. Reprod. Sci. 2007, 14, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Van Wezel, I.L.; Rodgers, H.F.; Rodgers, R.J. Differential localization of laminin chains in bovine follicles. J. Reprod. Fertil. 1998, 112, 267–278. [Google Scholar] [CrossRef]
- Hatzirodos, N.; Irving-Rodgers, H.F.; Hummitzsch, K.; Harland, M.L.; Morris, S.E.; Rodgers, R.J. Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes. BMC Genom. 2014, 15, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irving-Rodgers, H.F.; Hummitzsch, K.; Murdiyarso, L.S.; Bonner, W.M.; Sado, Y.; Ninomiya, Y.; Couchman, J.R.; Sorokin, L.M.; Rodgers, R.J. Dynamics of extracellular matrix in ovarian follicles and corpora lutea of mice. Cell Tissue Res. 2010, 339, 613–624. [Google Scholar] [CrossRef] [Green Version]
- Rodgers, R.J.; Irving-Rodgers, H.F.; Russell, D.L. Extracellular matrix of the developing ovarian follicle. Reproduction 2003, 126, 415–424. [Google Scholar] [CrossRef]
- Irving-Rodgers, H.F.; Rodgers, R.J. Extracellular matrix of the developing ovarian follicle. Semin. Reprod. Med. 2006, 24, 195–203. [Google Scholar] [CrossRef]
- Berkholtz, C.B.; Shea, L.D.; Woodruff, T.K. Extracellular matrix functions in follicle maturation. Semin. Reprod. Med. 2006, 24, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Piprek, R.P.; Kloc, M.; Mizia, P.; Kubiak, J.Z. The central role of cadherins in gonad development, reproduction, and fertility. Int. J. Mol. Sci. 2020, 21, 8264. [Google Scholar] [CrossRef] [PubMed]
- Sueoka, K.; Kuji, N.; Shiokawa, S.; Tanaka, M.; Miyazaki, T.; Yoshimura, Y. Integrins and reproductive physiology: Expression and modulation in fertilization, embryogenesis, and implantation. Fertil. Steril. 1997, 67, 799–811. [Google Scholar] [CrossRef]
- Bronson, R.A.; Fusi, F.M. Integrins and human reproduction. Mol. Hum. Reprod. 1996, 2, 153–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, G.F.; Bjerke, M.A.; DeSimone, D.W. Integrins and cadherins join forces to form adhesive networks. J. Cell Sci. 2011, 124, 1183–1193. [Google Scholar] [CrossRef] [Green Version]
- Mui, K.L.; Chen, C.S.; Assoian, R.K. The mechanical regulation of integrin-cadherin crosstalk organizes cells, signaling and forces. J. Cell Sci. 2016, 129, 1093–1100. [Google Scholar] [CrossRef] [Green Version]
- Ryan, P.L.; Valentine, A.F.; Bagnell, C.A. Expression of epithelial cadherin in the developing and adult pig ovary. Biol. Reprod. 1996, 55, 1091–1097. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Wen, J.; Zhang, T.; Zheng, W.; He, M.; Huang, K.; Guo, Q.; Chen, Q.; Yang, Y.; Deng, G.; et al. Oocyte-derived E-cadherin acts as a multiple functional factor maintaining the primordial follicle pool in mice. Cell Death Dis. 2019, 10, 160. [Google Scholar] [CrossRef]
- Caballero, J.N.; Gervasi, M.G.; Veiga, M.F.; Dalvit, G.C.; Perez-Martínez, S.; Cetica, P.D.; Vazquez-Levin, M.H. Epithelial cadherin is present in bovine oviduct epithelial cells and gametes, and is involved in fertilization-related events. Theriogenology 2014, 81, 1189–1206. [Google Scholar] [CrossRef]
- Kossowska-Tomaszczuk, K.; De Geyter, C. Cells with stem cell characteristics in somatic compartments of the ovary. Biomed. Res. Int. 2013, 2013, 310859. [Google Scholar] [CrossRef] [Green Version]
- Kossowska-Tomaszczuk, K.; De Geyter, C.; De Geyter, M.; Martin, I.; Holzgreve, W.; Scherberich, A.; Zhang, H. The Multipotency of Luteinizing Granulosa Cells Collected from Mature Ovarian Follicles. Stem Cells 2009, 27, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Brevini, T.A.L.; Pennarossa, G.; Rahman, M.M.; Paffoni, A.; Antonini, S.; Ragni, G.; DeEguileor, M.; Tettamanti, G.; Gandolfi, F. Morphological and Molecular Changes of Human Granulosa Cells Exposed to 5-Azacytidine and Addressed Toward Muscular Differentiation. Stem Cell Rev. Rep. 2014, 10, 633–642. [Google Scholar] [CrossRef]
- Józkowiak, M.; Hutchings, G.; Jankowski, M.; Kulcenty, K.; Mozdziak, P.; Kempisty, B.; Spaczyński, R.Z.; Piotrowska-Kempisty, H. The Stemness of Human Ovarian Granulosa Cells and the Role of Resveratrol in the Differentiation of MSCs-A Review Based on Cellular and Molecular Knowledge. Cells 2020, 9, 1418. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, M.; Gloria, A.; Turriani, M.; Berardinelli, P.; Russo, V.; Nardinocchi, D.; Curini, V.; Baratta, M.; Martignani, E.; Barboni, B. Osteo-regenerative potential of ovarian granulosa cells: An in vitro and in vivo study. Theriogenology 2012, 77, 1425–1437. [Google Scholar] [CrossRef] [Green Version]
- Oki, Y.; Ono, H.; Motohashi, T.; Sugiura, N.; Nobusue, H.; Kano, K. Dedifferentiated follicular granulosa cells derived from pig ovary can transdifferentiate into osteoblasts. Biochem. J. 2012, 447, 239–248. [Google Scholar] [CrossRef] [Green Version]
- Rybska, M.; Knap, S.; Jankowski, M.; Jeseta, M.; Bukowska, D.; Antosik, P.; Nowicki, M.; Zabel, M.; Kempisty, B.; Jaśkowski, J.M. Characteristic of factors influencing the proper course of folliculogenesis in mammals. Med. J. Cell Biol. 2018, 6, 33–38. [Google Scholar] [CrossRef] [Green Version]
- Chamier-Gliszczyńska, A.; Kałuzna, S.; Stefańska, K.; Celichowski, P.; Antosik, P.; Bukowska, D.; Bruska, M.; Zakova, J.; Machatkova, M.; Jeseta, M.; et al. Analysis of expression of genes responsible for regulation of cellular proliferation and migration-Microarray approach based on porcine oocyte model. Med. J. Cell Biol. 2019, 7, 48–57. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.; Lee, S.; Hatzirodos, N.; Hummitzsch, K.; Sullivan, T.R.; Rodgers, R.J.; Irving-Rodgers, H.F. Spatial differences within the membrana granulosa in the expression of focimatrix and steroidogenic capacity. Mol. Cell Endocrinol. 2012, 363, 62–73. [Google Scholar] [CrossRef]
- Mushahary, D.; Spittler, A.; Kasper, C.; Weber, V.; Charwat, V. Isolation, cultivation, and characterization of human mesenchymal stem cells. Cytom. Part A 2018, 93, 19–31. [Google Scholar] [CrossRef] [Green Version]
- Hendijani, F. Explant culture: An advantageous method for isolation of mesenchymal stem cells from human tissues. Cell Prolif. 2017, 50, e12334. [Google Scholar] [CrossRef] [Green Version]
- Stefańska, K.; Sibiak, R.; Hutchings, G.; Dompe, C.; Moncrieff, L.; Janowicz, K.; Jeseta, M.; Kempisty, B.; Machatkova, M.; Mozdziak, P. Evidence for existence of molecular stemness markers in porcine ovarian follicular granulosa cells. Med. J. Cell Biol. 2019, 7, 183–188. [Google Scholar] [CrossRef] [Green Version]
- Moncrieff, L.; Mozdziak, P.; Jeseta, M.; Machatkova, M.; Kranc, W.; Kempisty, B. Ovarian follicular cells—Living in the shadow of stemness cellular competence. Med. J. Cell Biol. 2019, 7, 134–140. [Google Scholar] [CrossRef] [Green Version]
- Trejter, M.; Hochol, A.; Tyczewska, M.; Ziolkowska, A.; Jopek, K.; Szyszka, M.; Malendowicz, L.K.; Rucinski, M. Sex-related gene expression profiles in the adrenal cortex in the mature rat: Microarray analysis with emphasis on genes involved in steroidogenesis. Int. J. Mol. Med. 2015, 35, 702–714. [Google Scholar] [CrossRef] [PubMed]
- Chamier-Gliszczyńska, A.; Brązert, M.; Sujka-Kordowska, P.; Popis, M.; Ożegowska, K.; Stefańska, K.; Kocherova, I.; Celichowski, P.; Kulus, M.; Bukowska, D.; et al. Genes involved in angiogenesis and circulatory system development are differentially expressed in porcine epithelial oviductal cells during long-term primary in vitro culture—A transcriptomic study. Med. J. Cell Biol. 2018, 6, 163–173. [Google Scholar] [CrossRef] [Green Version]
- Nawrocki, M.J.; Celichowski, P.; Jankowski, M.; Kranc, W.; Bryja, A.; Borys-Wójcik, S.; Jeseta, M.; Antosik, P.; Bukowska, D.; Bruska, M.; et al. Ontology groups representing angiogenesis and blood vessels development are highly up-regulated during porcine oviductal epithelial cells long-term real-time proliferation—A primary cell culture approach. Med. J. Cell Biol. 2018, 6, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Stefańska, K.; Chamier-Gliszczyńska, A.; Jankowski, M.; Celichowski, P.; Kulus, M.; Rojewska, M.; Antosik, P.; Bukowska, D.; Bruska, M.; Nowicki, M.; et al. Epithelium morphogenesis and oviduct development are regulated by significant increase of expression of genes after long-term in vitro primary culture—A microarray assays. Med. J. Cell Biol. 2018, 6, 195–204. [Google Scholar] [CrossRef] [Green Version]
- Kranc, W.; Brązert, M.; Ożegowska, K.; Budna-Tukan, J.; Celichowski, P.; Jankowski, M.; Bryja, A.; Nawrocki, M.J.; Popis, M.; Jeseta, M.; et al. Response to abiotic and organic substances stimulation belongs to ontologic groups significantly up-regulated in porcine immature oocytes. Med. J. Cell Biol. 2018, 6, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Borys-Wójcik, S.; Kocherova, I.; Celichowski, P.; Popis, M.; Jeseta, M.; Bukowska, D.; Antosik, P.; Nowicki, M.; Kempisty, B. Protein oligomerization is the biochemical process highly up-regulated in porcine oocytes before in vitro maturation (IVM). Med. J. Cell Biol. 2018, 6, 155–162. [Google Scholar] [CrossRef] [Green Version]
- Budna, J.; Celichowski, P.; Bryja, A.; Jeseta, M.; Jankowski, M.; Bukowska, D.; Antosik, P.; Nowicki, A.; Brüssow, K.P.; Bruska, M.; et al. Expression Changes in Fatty acid Metabolic Processrelated Genes in Porcine Oocytes During in Vitro Maturation. Med. J. Cell Biol. 2018, 6, 48–54. [Google Scholar] [CrossRef] [Green Version]
- Walter, W.; Sánchez-Cabo, F.; Ricote, M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Onodera, T.; Sakai, T.; Hsu, J.C.F.; Matsumoto, K.; Chiorini, J.A.; Yamada, K.M. Btbd7 regulates epithelial cell dynamics and branching morphogenesis. Science 2010, 329, 562–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kedem, A.; Ulanenko-Shenkar, K.; Yung, Y.; Yerushalmi, G.M.; Maman, E.; Hourvitz, A. Elucidating Decorin’s role in the preovulatory follicle. J. Ovarian Res. 2020, 13, 15. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.Y.; Gao, K.X.; Xin, H.Y.; Han, P.; Zhu, G.Q.; Cao, B.Y. Molecular cloning, expression analysis, and function of decorin in goat ovarian granulosa cells. Domest. Anim. Endocrinol. 2016, 57, 108–116. [Google Scholar] [CrossRef]
- Chermuła, B.; Brazert, M.; Izycki, D.; Ciesiółka, S.; Kranc, W.; Celichowski, P.; Ozegowska, K.; Nawrocki, M.J.; Jankowski, M.; Jeseta, M.; et al. New Gene Markers of Angiogenesis and Blood Vessels Development in Porcine Ovarian Granulosa Cells during Short-Term Primary Culture in Vitro. Biomed. Res. Int. 2019, 2019, 6545210. [Google Scholar] [CrossRef]
- Kulus, M.; Kranc, W.; Sujka-Kordowska, P.; Mozdziak, P.; Jankowski, M.; Konwerska, A.; Kulus, J.; Bukowska, D.; Skowroński, M.; Piotrowska-Kempisty, H.; et al. The processes of cellular growth, aging, and programmed cell death are involved in lifespan of ovarian granulosa cells during short-term IVC—Study based on animal model. Theriogenology 2020, 148, 76–88. [Google Scholar] [CrossRef]
- Sawada, Y.; Sato, T.; Saito, C.; Ozawa, F.; Ozaki, Y.; Sugiura-Ogasawara, M. Clinical utility of decorin in follicular fluid as a biomarker of oocyte potential. Reprod. Biol. 2018, 18, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Xin, H.; Han, P.; Gao, K.; Gao, T.; Lei, Y.; Ji, S.; An, X.; Cao, B. Expression and regulative function of tissue inhibitor of metalloproteinase 3 in the goat ovary and its role in cultured granulosa cells. Mol. Cell Endocrinol. 2015, 412, 104–115. [Google Scholar] [CrossRef]
- Messini, C.I.; Vasilaki, A.; Korona, E.; Anifandis, G.; Georgoulias, P.; Dafopoulos, K.; Garas, A.; Daponte, A.; Messinis, I.E. Effect of resistin on estradiol and progesterone secretion from human luteinized granulosa cells in culture. Syst. Biol. Reprod. Med. 2019, 65, 350–356. [Google Scholar] [CrossRef]
- Balzac, F.; Avolio, M.; Degani, S.; Kaverina, I.; Torti, M.; Silengo, L.; Small, J.V.; Retta, S.F. E-cadherin endocytosis regulates the activity of Rap1: A traffic light GTPase at the crossroads between cadherin and integrin function. J. Cell Sci. 2005, 118, 4765–4783. [Google Scholar] [CrossRef] [Green Version]
- Asem, E.K.; Carnegie, J.A.; Tsang, B.K. Fibronectin production by chicken granulosa cells in vitro: Effect of follicular development. Eur. J. Endocrinol. 1992, 127, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Kitasaka, H.; Kawai, T.; Hoque, S.A.M.; Umehara, T.; Fujita, Y.; Shimada, M. Inductions of granulosa cell luteinization and cumulus expansion are dependent on the fibronectin-integrin pathway during ovulation process in mice. PLoS ONE 2018, 13, e0192458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huet, C.; Pisselet, C.; Mandon-Pépin, B.; Monget, P.; Monniaux, D. Extracellular matrix regulates ovine granulosa cell survival, proliferation and steroidogenesis: Relationships between cell shape and function. J. Endocrinol. 2001, 169, 347–360. [Google Scholar] [CrossRef] [Green Version]
- Dias, F.C.F.; Khan, M.I.R.; Sirard, M.A.; Adams, G.P.; Singh, J. Differential gene expression of granulosa cells after ovarian superstimulation in beef cattle. Reproduction 2013, 146, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Lin, J.; Han, B.; Wang, L.; Chen, Y.; Liu, M.; Huang, J. Proteomic profiling of follicle fluids after superstimulation in one-month-old lambs. Reprod. Domest. Anim. 2018, 53, 186–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lussier, J.G.; Diouf, M.N.; Lévesque, V.; Sirois, J.; Ndiaye, K. Gene expression profiling of upregulated mRNAs in granulosa cells of bovine ovulatory follicles following stimulation with hCG. Reprod. Biol. Endocrinol. 2017, 15, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudo, A. Periostin in fibrillogenesis for tissue regeneration: Periostin actions inside and outside the cell. Cell Mol. Life Sci. 2011, 68, 3201–3207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.-W.; Chiang, Y.-C.; Sun, N.-Y.; Chen, Y.-L.; Chang, C.-F.; Tai, Y.-J.; Chen, C.-A.; Cheng, W.-F. CHI3L1 results in poor outcome of ovarian cancer by promoting properties of stem-like cells. Endocr.-Relat. Cancer 2019, 26, 73–88. [Google Scholar] [CrossRef]
- Zhao, T.; Su, Z.; Li, Y.; Zhang, X.; You, Q. Chitinase-3 like-protein-1 function and its role in diseases. Signal Transduct. Target. Ther. 2020, 5, 201. [Google Scholar] [CrossRef]
- Albacete-Albacete, L.; Navarro-Lérida, I.; López, J.A.; Martín-Padura, I.; Astudillo, A.M.; Ferrarini, A.; Van-Der-Heyden, M.; Balsinde, J.; Orend, G.; Vázquez, J.; et al. ECM deposition is driven by caveolin-1–dependent regulation of exosomal biogenesis and cargo sorting. J. Cell Biol. 2020, 219, e202006178. [Google Scholar] [CrossRef]
- Diouf, M.N.; Lefebvre, R.; Silversides, D.W.; Sirois, J.; Lussier, J.G. Induction of alpha-caveolin-1 (αCAV1) expression in bovine granulosa cells in response to an ovulatory dose of human chorionic gonadotropin. Mol. Reprod. Dev. 2006, 73, 1353–1360. [Google Scholar] [CrossRef]
- Sá, N.A.R.; Araújo, V.R.; Correia, H.H.V.; Ferreira, A.C.A.; Guerreiro, D.D.; Sampaio, A.M.; Escobar, E.; Santos, F.W.; Moura, A.A.; Lôbo, C.H.; et al. Anethole improves the in vitro development of isolated caprine secondary follicles. Theriogenology 2017, 89, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Lin, W.; Wen, L.; Li, G. Lgr5 in cancer biology: Functional identification of Lgr5 in cancer progression and potential opportunities for novel therapy. Stem Cell Res. Ther. 2019, 10, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kayampilly, P.P.; Menon, K.M.J. Dihydrotestosterone inhibits insulin-stimulated cyclin D2 messenger ribonucleic acid expression in rat ovarian granulosa cells by reducing the phosphorylation of insulin receptor substrate-1. Endocrinology 2006, 147, 464–471. [Google Scholar] [CrossRef] [Green Version]
- Corbould, A.; Kim, Y.B.; Youngren, J.F.; Pender, C.; Kahn, B.B.; Lee, A.; Dunaif, A. Insulin resistance in the skeletal muscle of women with PCOS involves intrinsic and acquired defects in insulin signaling. Am. J. Physiol. Endocrinol. Metab. 2005, 288, E1047–E1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, P.X.; Huang, Z.; Li, C.C.; Fan, B.; Li, K.; Liu, B.; Yu, M.; Zhao, S.H. Cloning, chromosomal localization, SNP detection and association analysis of the porcine IRS-1 gene. Mol. Biol. Rep. 2009, 36, 2087–2092. [Google Scholar] [CrossRef]
- Cai, G.; Ma, X.; Chen, B.; Huang, Y.; Liu, S.; Yang, H.; Zou, W. MicroRNA-145 Negatively Regulates Cell Proliferation Through Targeting IRS1 in Isolated Ovarian Granulosa Cells from Patients with Polycystic Ovary Syndrome. Reprod. Sci. 2017, 24, 902–910. [Google Scholar] [CrossRef]
- Fan, H.Y.; Liu, Z.; Shimada, M.; Sterneck, E.; Johnson, P.F.; Hedrick, S.M.; Richards, J.S. MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science 2009, 324, 938–941. [Google Scholar] [CrossRef] [Green Version]
- Buck, C.A.; Horwitz, A.F. Integrin, a transmembrane glycoprotein complex mediating cell-substratum adhesion. J. Cell Sci. Suppl. 1987, 1987, 231–250. [Google Scholar] [CrossRef] [Green Version]
- Parise, L.V.; Lee, J.W.; Juliano, R.L. New aspects of integrin signaling in cancer. Semin. Cancer Biol. 2000, 10, 407–414. [Google Scholar] [CrossRef]
- Hood, J.D.; Cheresh, D.A. Role of integrins in cell invasion and migration. Nat. Rev. Cancer 2002, 2, 91–100. [Google Scholar] [CrossRef]
- Shield, K.; Riley, C.; Quinn, M.A.; Rice, G.E.; Ackland, M.L.; Ahmed, N. α2β1 integrin affects metastatic potential of ovarian carcinoma spheroids by supporting disaggregation and proteolysis. J. Carcinog. 2007, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Sun, Y.; Li, D.; Li, H.; Jin, X.; Ren, D. Overexpressed ITGA2 contributes to paclitaxel resistance by ovarian cancer cells through the activation of the AKT/FoxO1 pathway. Aging 2020, 12, 5336–5351. [Google Scholar] [CrossRef] [PubMed]
- Clark, E.A.; Brugge, J.S. Integrins and signal transduction pathways: The road taken. Science 1995, 268, 233–239. [Google Scholar] [CrossRef]
- Giancotti, F.G.; Ruoslahti, E. Integrin signaling. Science 1999, 285, 1028–1032. [Google Scholar] [CrossRef]
- Retta, S.F.; Balzac, F.; Avolio, M. Rap1: A turnabout for the crosstalk between cadherins and integrins. Eur. J. Cell Biol. 2006, 85, 283–293. [Google Scholar] [CrossRef]
- Watanabe, T.; Sato, K.; Kaibuchi, K. Cadherin-mediated intercellular adhesion and signaling cascades involving small GTPases. Cold Spring Harb. Perspect. Biol. 2009, 1, a003020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulus, M.; Sujka-Kordowska, P.; Konwerska, A.; Celichowski, P.; Kranc, W.; Kulus, J.; Piotrowska-Kempisty, H.; Antosik, P.; Bukowska, D.; Iżycki, D.; et al. New molecular markers involved in regulation of ovarian granulosa cell morphogenesis, development and differentiation during short-term primary in vitro culture-transcriptomic and histochemical study based on ovaries and individual separated follicles. Int. J. Mol. Sci. 2019, 20, 3966. [Google Scholar] [CrossRef] [Green Version]
- Ozegowska, K.; Brazert, M.; Ciesiółka, S.; Nawrocki, M.J.; Kranc, W.; Celichowski, P.; Jankowski, M.; Bryja, A.; Jeseta, M.; Antosik, P.; et al. Genes Involved in the Processes of Cell Proliferation, Migration, Adhesion, and Tissue Development as New Potential Markers of Porcine Granulosa Cellular Processes In Vitro: A Microarray Approach. DNA Cell Biol. 2019, 38, 549–560. [Google Scholar] [CrossRef] [Green Version]
- Antosik, P.; Jeseta, M.; Kranc, W.; Chachuła, A.; Bryja, A.; Budna, J.; Ciesiołka, S.; Wojtanowicz-Markiewicz, K.; Bukowska, D.; Brussow, K.P.; et al. Expression of integrins and GDF9 mRNAs is associated with Ovarian follicle size and donor puberty status in pigs. Med. Weter. 2016, 72, 750–754. [Google Scholar] [CrossRef] [Green Version]
- Hassani, F.; Oryan, S.; Eftekhari-Yazdi, P.; Bazrgar, M.; Moini, A.; Nasiri, N.; Sharifi-Zarchi, A. Downregulation of extracellular matrix and cell adhesion molecules in cumulus cells of infertile polycystic ovary syndrome women with and without insulin resistance. Cell J. 2019, 21, 35–42. [Google Scholar] [CrossRef]
- Hodgkinson, C.P.; Naidoo, V.; Patti, K.G.; Gomez, J.A.; Schmeckpeper, J.; Zhang, Z.; Davis, B.; Pratt, R.E.; Mirotsou, M.; Dzau, V.J. Abi3bp is a multifunctional autocrine/paracrine factor that regulates mesenchymal stem cell biology. Stem Cells 2013, 31, 1669–1682. [Google Scholar] [CrossRef] [Green Version]
- Manabe, R.; Tsutsui, K.; Yamada, T.; Kimura, M.; Nakano, I.; Shimono, C.; Sanzen, N.; Furutani, Y.; Fukuda, T.; Oguri, Y.; et al. Transcriptome-based systematic identification of extracellular matrix proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 12849–12854. [Google Scholar] [CrossRef] [Green Version]
- Hao, J.; Tuck, A.R.; Prakash, C.R.; Damdimopoulos, A.; Sjödin, M.O.D.; Lindberg, J.; Niklasson, B.; Pettersson, K.; Hovatta, O.; Damdimopoulou, P. Culture of human ovarian tissue in xeno-free conditions using laminin components of the human ovarian extracellular matrix. J. Assist. Reprod. Genet. 2020, 37, 2137–2150. [Google Scholar] [CrossRef] [PubMed]
- Hatzirodos, N.; Hummitzsch, K.; Irving-Rodgers, H.F.; Rodgers, R.J. Transcriptome comparisons identify new cell markers for theca interna and granulosa cells from small and large antral ovarian follicles. PLoS ONE 2015, 10, e0119800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallelli, M.F.; Bianchi, C.; Lombardo, D.; Rey, F.; Rodriguez, F.M.; Castillo, V.A.; Miragaya, M. Leptin and IGF1 receptors in alpaca (Vicugna pacos) ovaries. Anim. Reprod. Sci. 2019, 200, 96–104. [Google Scholar] [CrossRef]
- Velazquez, M.A.; Zaraza, J.; Oropeza, A.; Webb, R.; Niemann, H. The role of IGF1 in the in vivo production of bovine embryos from superovulated donors. Reproduction 2009, 137, 161–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mani, A.M.; Fenwick, M.A.; Cheng, Z.; Sharma, M.K.; Singh, D.; Wathes, D.C. IGF1 induces up-regulation of steroidogenic and apoptotic regulatory genes via activation of phosphatidylinositol-dependent kinase/AKT in bovine granulosa cells. Reproduction 2010, 139, 139–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, K.G.; Ushizawa, K.; Hosoe, M.; Takahashi, T. Differential genome-wide gene expression profiling of bovine largest and second-largest follicles: Identification of genes associated with growth of dominant follicles. Reprod. Biol. Endocrinol. 2010, 8, 11. [Google Scholar] [CrossRef] [Green Version]
- Skinner, M.K.; Schmidt, M.; Savenkova, M.I.; Sadler-Riggleman, I.; Nilsson, E.E. Regulation of granulosa and theca cell transcriptomes during ovarian antral follicle development. Mol. Reprod. Dev. 2008, 75, 1457–1472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamort, A.S.; Giopanou, I.; Psallidas, I.; Stathopoulos, G.T. Osteopontin as a Link between Inflammation and Cancer: The Thorax in the Spotlight. Cells 2019, 8, 815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zalkin, H.; Dixon, J.E. De novo purine nucleotide biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 1992, 42, 259–287. [Google Scholar] [CrossRef] [PubMed]
- Bønsdorff, T.; Gautier, M.; Farstad, W.; Rønningen, K.; Lingaas, F.; Olsaker, I. Mapping of the bovine genes of the de novo AMP synthesis pathway. Anim. Genet. 2004, 35, 438–444. [Google Scholar] [CrossRef] [PubMed]

| Gene | Primer Sequence (5′-3′) | Product Size (bp) | |
|---|---|---|---|
| IRS1 | F | CCTAGCACCAACAGGACTCA | 239 |
| R | GAAGAGATGAAACCGCCGTC | ||
| TMPO | F | GCTCAGTGGAAAGTCAGCAG | 241 |
| R | CCTGTCAATTTGCTGCCACT | ||
| PAICS | F | AGTCATGCTACACAGGCCAT | 235 |
| R | TTACCATCTGCAGCCCTTCA | ||
| ANK2 | F | TTGTAACGGAGGAGGTCACC | 222 |
| R | AACGCAGGTAGTTCATCCCA | ||
| ADAM23 | F | AGCAGCTCAATACCAGGGTT | 235 |
| R | TTCACACCAACTCCCCTTGT | ||
| IGF1 | F | TTCTACTTGGCCCTGTGCTT | 222 |
| R | CTCCAGCCTCCTCAGATCAC | ||
| CNTLN | F | ACCTCAACCCATAAAGCCCA | 186 |
| R | TGTGGCAAAAGGAAGCTGTC | ||
| DNAJB1 | F | AGGACCATACCCGTTGTGTT | 167 |
| R | AGGACGGTTCTTGAGGTCTG | ||
| POSTN | F | ATTGACCGTGTCCTCACACA | 212 |
| R | GCCACTTTGTCTCCCATGAT | ||
| ITGA2 | F | CATGCCAGATCCCTTCATCT | 153 |
| R | CGCTTAAGGCTTGGAAACTG | ||
| FN1 | F | TGAGCCTGAAGAGACCTGCT | 113 |
| R | CAGCTCCAATGCAGGTACAG | ||
| LAMB1 | F | CTTCACCACCTTGGACCACT | 216 |
| R | AGCTGTGGCTCATAGCGAAT | ||
| ITGB3 | F | GGCTTCAAAGACAGCCTCAC | 175 |
| R | AGTCCTTTTCCGAGCACTCA | ||
| CHI3L1 | F | GGATGCAAGTTCCGACAGAT | 202 |
| R | GAGGATCCCTTTCTCCTTGG | ||
| DCN | F | CTCTCTGGCCAACACTCCTC | 155 |
| R | GCGGGCAGAAGTCATTAGAG | ||
| PCOLCE2 | F | TGTAAACGGACTGGGACTCC | 184 |
| R | CGATGACCTTGGCACTCATG | ||
| COL14A1 | F | AGTTCCAGCCCAGCAATACT | 229 |
| R | ATCGTCCAGTACAGCCAACA | ||
| Gene Symbol | Gene Name | Fold Change | Adj. p. val. |
|---|---|---|---|
| POSTN | periostin, osteoblast specific factor | 95.2 | 3.5 × 107 |
| ITGA2 | Integrin, alpha 2 (CD49B, alpha 2 subunit of VLA -2 receptor) | 42.7 | 3.5 × 107 |
| FN1 | Fibronectin 1 | 35.4 | 3.6 × 107 |
| LAMB1 | Laminin, beta 1 | 33.3 | 3.5 × 107 |
| ITGB3 | Integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | 29.9 | 3.5 × 107 |
| CHI3L1 | Chitinase 3-like 1 (cartilage glycoprotein-39) | 29.6 | 1.4 × 105 |
| PCOLCE2 | Procollagen C-endopeptidase enhancer 2 | 26.2 | 4.6 × 107 |
| CAV1 | Caveolin 1, caveolae protein, 22kDa | 22.3 | 4.3 × 107 |
| DCN | decorin | 21.6 | 4.4 × 107 |
| COL14A1 | Collagen, type XIV, alpha 1 | 16.0 | 5.3 × 107 |
| SPP1 | Secreted phosphoprotein 1 | −2.5 | 1.4 × 102 |
| IRS1 | Insulin receptor substrate 1 | −2.6 | 1.9 × 104 |
| CNTLN | Centlein, centrosmal protein | −2.6 | 5.1 × 104 |
| TMPO | thymopoietin | −2.7 | 1.3 × 104 |
| PAICS | Phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | −2.9 | 8.0 × 105 |
| ANK2 | Ankyrin 2 | −4.3 | 2.1 × 105 |
| ADAM23 | ADAM metalloptidase domain 23 | −4.9 | 1.0 × 105 |
| ABI3BP | ABI family, member 3 (NESH) binding protein | −5.3 | 2.1 × 105 |
| DNAJB1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | −6.4 | 2.3 × 106 |
| IGF1 | Insulin-like growth factor 1 (somatomedin C) | −8.3 | 1.0 × 104 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kulus, J.; Kulus, M.; Kranc, W.; Jopek, K.; Zdun, M.; Józkowiak, M.; Jaśkowski, J.M.; Piotrowska-Kempisty, H.; Bukowska, D.; Antosik, P.; et al. Transcriptomic Profile of New Gene Markers Encoding Proteins Responsible for Structure of Porcine Ovarian Granulosa Cells. Biology 2021, 10, 1214. https://doi.org/10.3390/biology10111214
Kulus J, Kulus M, Kranc W, Jopek K, Zdun M, Józkowiak M, Jaśkowski JM, Piotrowska-Kempisty H, Bukowska D, Antosik P, et al. Transcriptomic Profile of New Gene Markers Encoding Proteins Responsible for Structure of Porcine Ovarian Granulosa Cells. Biology. 2021; 10(11):1214. https://doi.org/10.3390/biology10111214
Chicago/Turabian StyleKulus, Jakub, Magdalena Kulus, Wiesława Kranc, Karol Jopek, Maciej Zdun, Małgorzata Józkowiak, Jędrzej M. Jaśkowski, Hanna Piotrowska-Kempisty, Dorota Bukowska, Paweł Antosik, and et al. 2021. "Transcriptomic Profile of New Gene Markers Encoding Proteins Responsible for Structure of Porcine Ovarian Granulosa Cells" Biology 10, no. 11: 1214. https://doi.org/10.3390/biology10111214
APA StyleKulus, J., Kulus, M., Kranc, W., Jopek, K., Zdun, M., Józkowiak, M., Jaśkowski, J. M., Piotrowska-Kempisty, H., Bukowska, D., Antosik, P., Mozdziak, P., & Kempisty, B. (2021). Transcriptomic Profile of New Gene Markers Encoding Proteins Responsible for Structure of Porcine Ovarian Granulosa Cells. Biology, 10(11), 1214. https://doi.org/10.3390/biology10111214








