Comparative Transcriptome Analysis Reveals the Molecular Immunopathogenesis of Chinese Soft-Shelled Turtle (Trionyx sinensis) Infected with Aeromonas hydrophila
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Bacteria Strain
2.2. Experimental Treatments, Pathological Observation, and Sampling
2.3. RNA Extraction, Library Preparation, and RNA Sequencing (RNA-Seq)
2.4. Identification of the Differentially Expressed Genes (DEGs)
2.5. KEGG Pathway Enrichment Analysis
2.6. Gene Expression Validation Using Quantitative PCR (qPCR)
3. Results
3.1. Symptom Description of the Turtles Challenged with A. hydrophila
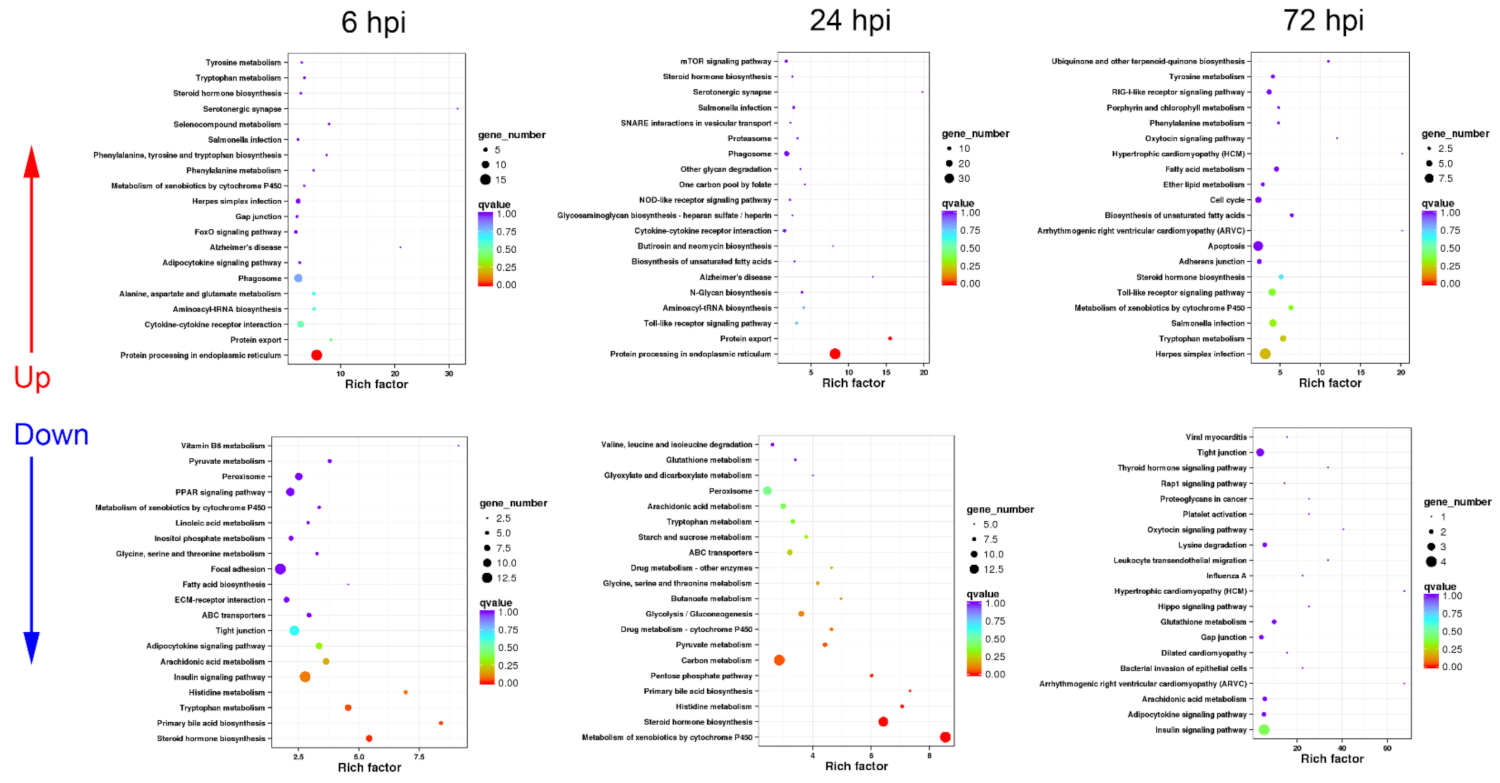
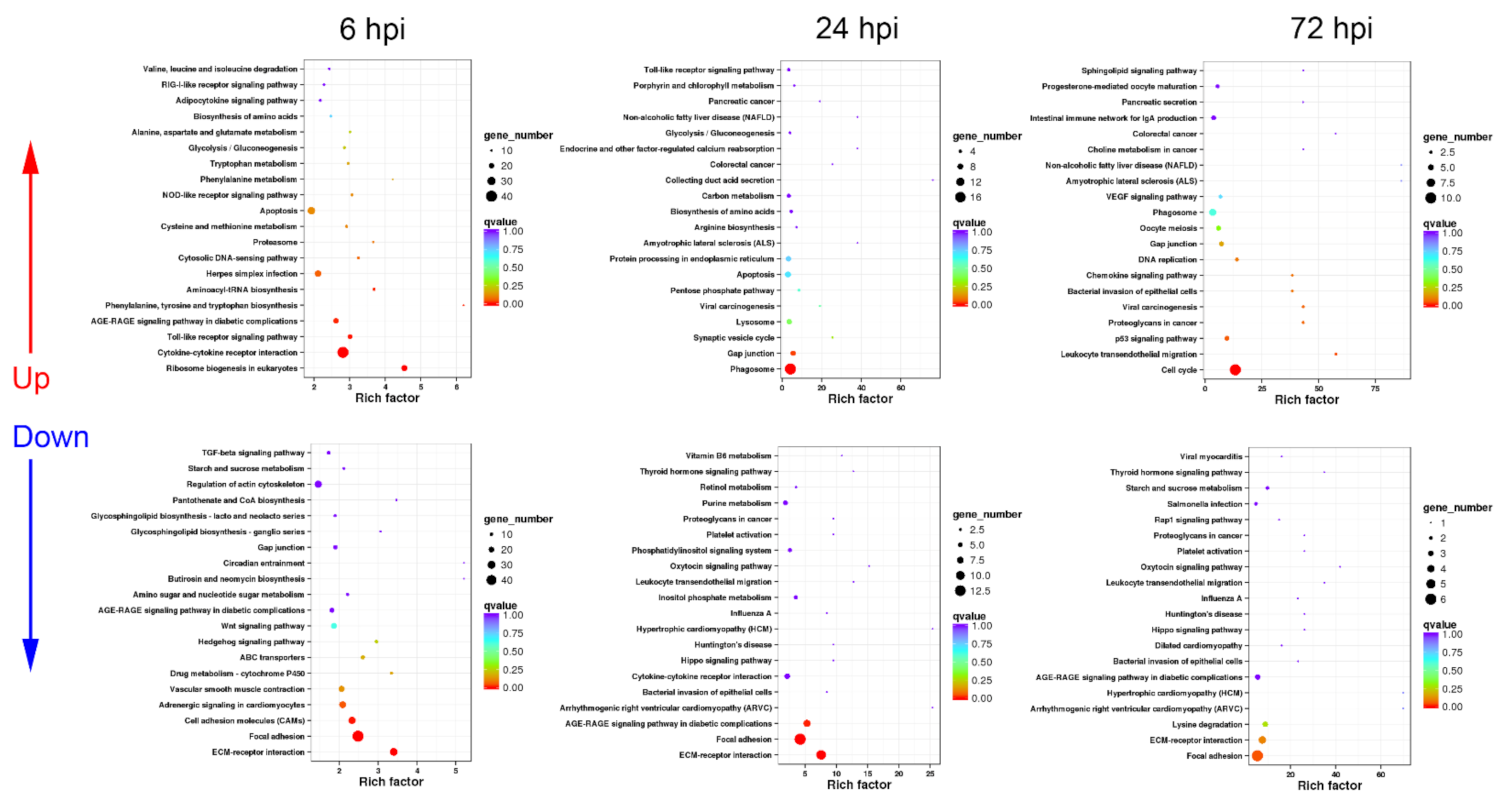
3.2. Functional Classification of DEGs in Turtle Liver Transcriptomes by KEGG
3.2.1. Sequential Changes of KEGG Enrichment in AL Group Turtles
3.2.2. Sequential Changes of KEGG Enrichment in IL Group Turtles
3.2.3. Expression Difference Analysis of Cytokine, Phagocytosis, and Apoptosis-Related Genes between AL and IL Group Turtles
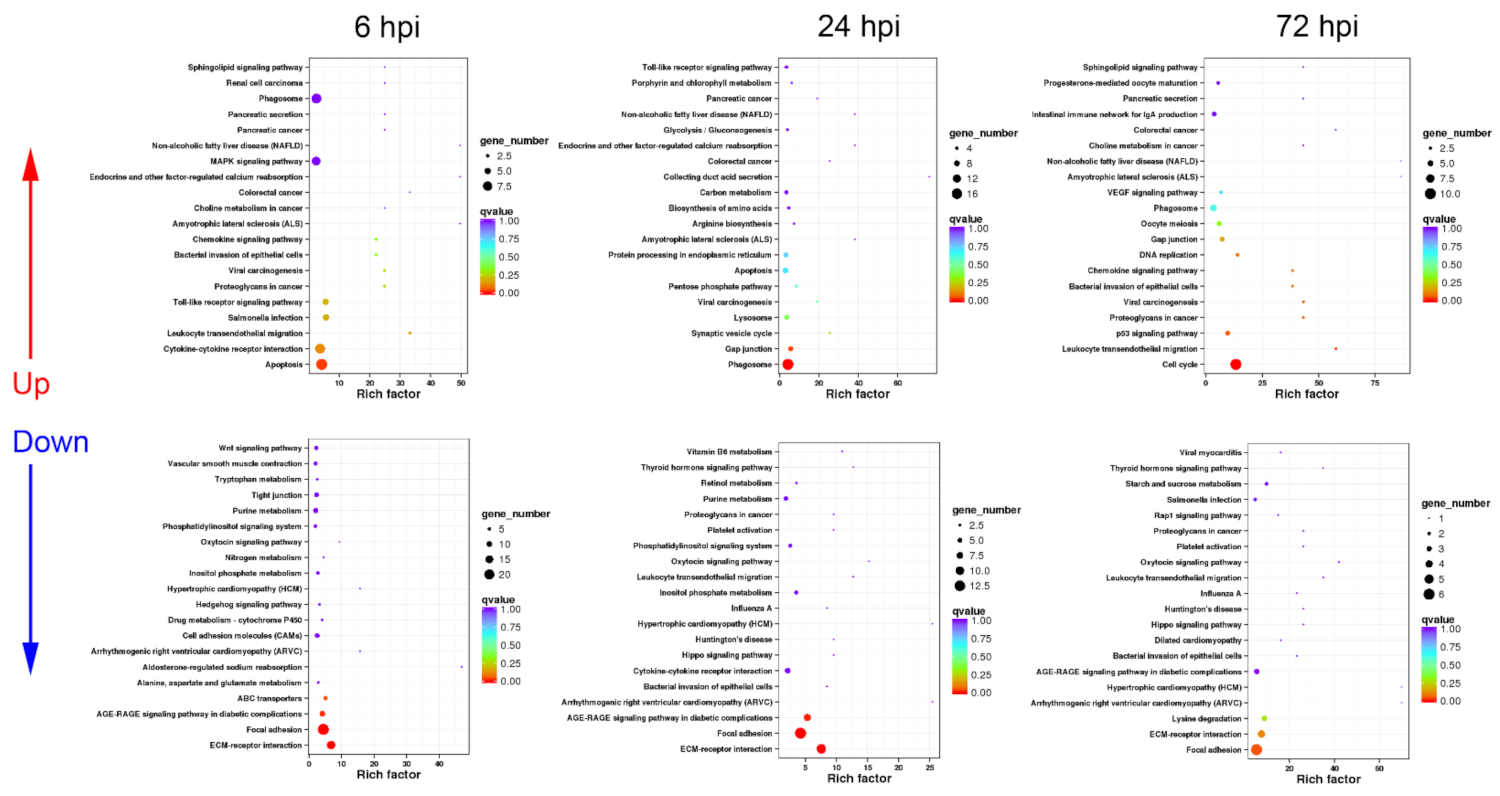
3.3. Functional Classification of DEGs in Turtle Spleen Transcriptomes by KEGG
3.3.1. Sequential Changes of KEGG Enrichment in AS Group Turtles
3.3.2. Sequential Changes of KEGG Enrichment in IS Group Turtles
3.3.3. Expression Difference Analysis of Cytokine, Phagocytosis, and Apoptosis-Related Genes between AS and IS Group Turtles
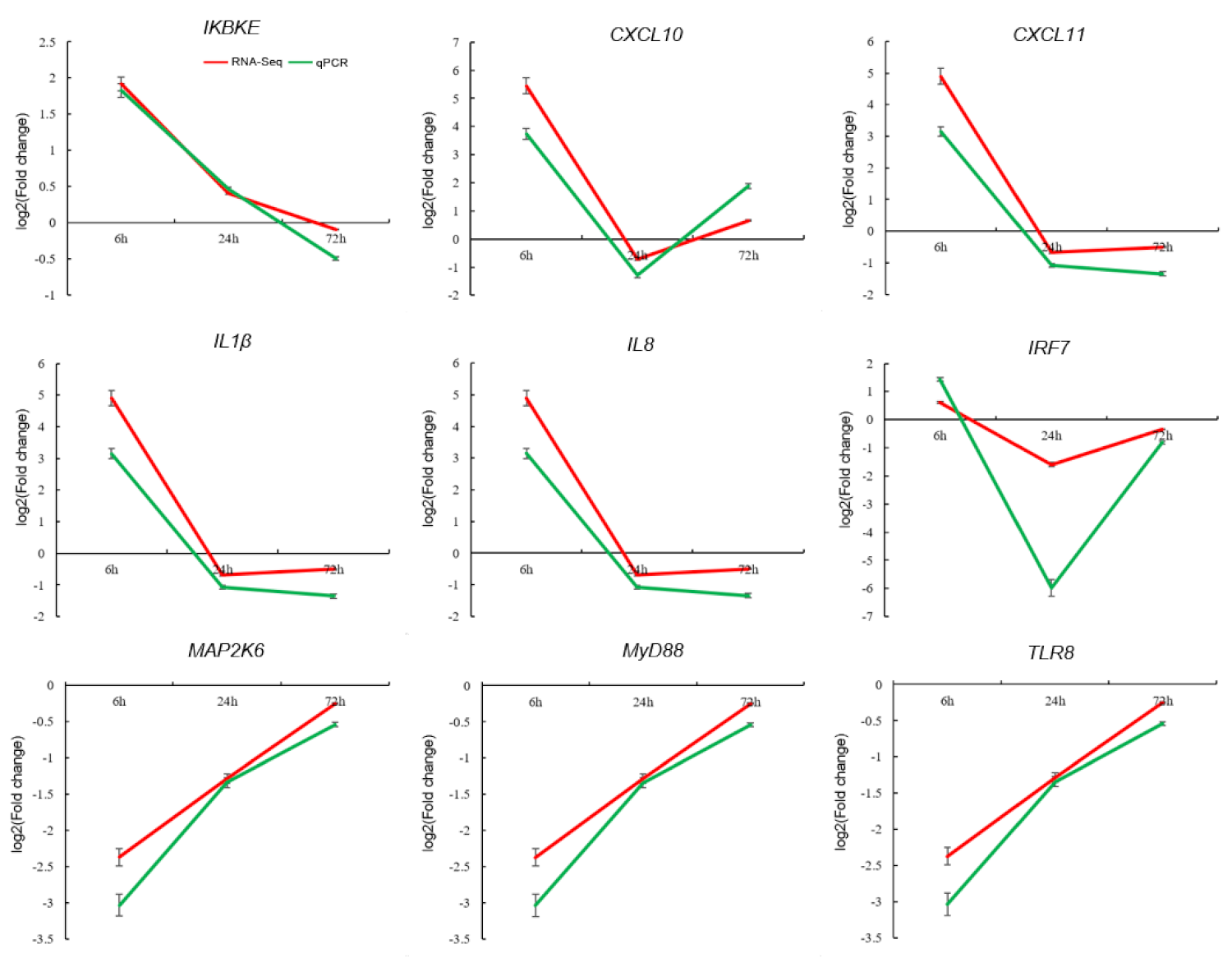
3.4. Validation of DEGs by qPCR
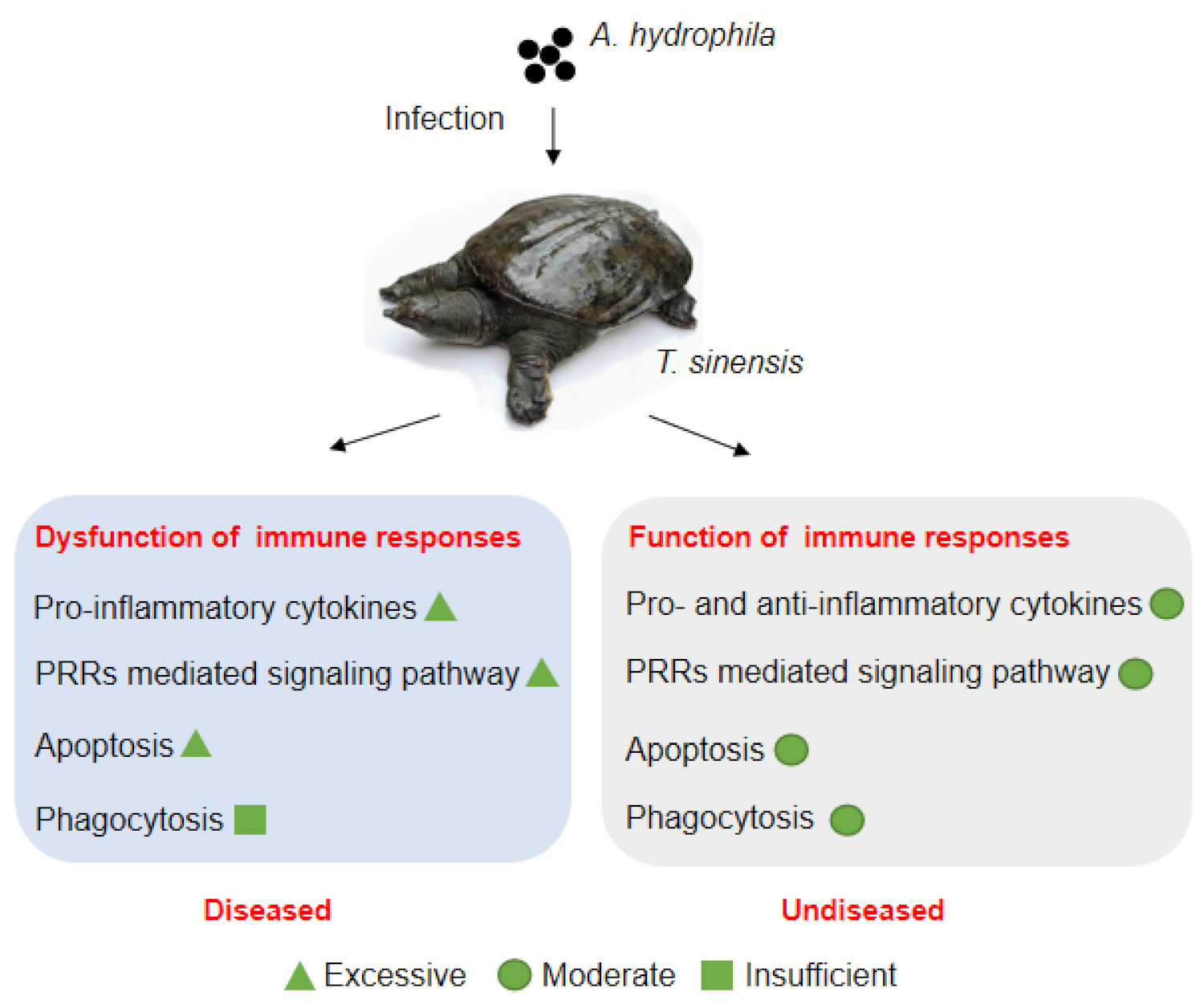
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Jia, Y.; Yang, Z.; Hao, Y.; Gao, Y. Effects of animal-plant protein ratio in extruded and expanded diets on nitrogen and energy budgets of juvenile Chinese soft-shelled turtle (Pelodiscus sinensis Wiegmann). Aquac. Res. 2005, 36, 61–68. [Google Scholar] [CrossRef]
- Chen, J.; Zhu, N.; Kong, L.; Bei, Y.; Zheng, T.; Ding, X.; He, Z. First reported fatal Bacillus thuringiensis infections in Chinese soft-shelled turtles (Trionyx sinensis). Aquaculture 2014, 428–429, 16–20. [Google Scholar] [CrossRef]
- Chen, Z.-X.; Zheng, J.-C.; Jiang, Y.-L. A new iridovirus isolated from soft-shelled turtle. Virus Res. 1999, 63, 147–151. [Google Scholar] [CrossRef]
- Chung, T.; Yi, S.; Kim, B.; Kim, W.; Shin, G. Identification and antibiotic resistance profiling of bacterial isolates from septicaemic soft-shelled turtles (Pelodiscus sinensis). Vet. Med. 2017, 62, 169–177. [Google Scholar] [CrossRef]
- Chen, J.; Zhu, N.; Kong, L.; Bei, Y.; Zheng, T.; Ding, X.; He, Z. First case of soft shell disease in Chinese soft-shelled turtle (Trionyx sinens) associated with Aeromonas sobria—A. veronii complex. Aquaculture 2013, 406, 62–67. [Google Scholar] [CrossRef]
- Cahill, M.M. Virulence factors in motile Aeromonas species. J. Appl. Bacteriol. 1990, 69, 1–16. [Google Scholar] [CrossRef]
- Pasquale, V.; Baloda, S.B.; Dumontet, S.; Krovacek, K. An Outbreak of Aeromonas hydrophila Infection in Turtles (Pseudemis scripta). Appl. Environ. Microbiol. 1994, 60, 1678–1680. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Sahoo, P.; Mohanty, B.; Jena, J. Pathophysiology of experimental Aeromonas hydrophila infection in Puntius sarana: Early changes in blood and aspects of the innate immune-related gene expression in survivors. Veter. Immunol. Immunopathol. 2011, 142, 207–218. [Google Scholar] [CrossRef]
- Mares, C.A.; Ojeda, S.S.; Morris, E.G.; Li, Q.; Teale, J.M. Initial delay in the immune response to Francisella tularensis is followed by Hypercytokinemia Characteristic of severe sepsis and correlating with upregulation and release of damage-associated molecular patterns. Infect. Immun. 2008, 76, 3001–3010. [Google Scholar] [CrossRef]
- Coperchini, F.; Chiovato, L.; Rotondi, M. Interleukin-6, CXCL10 and infiltrating macrophages in COVID-19-related cytokine storm: Not one for all but all for one! Front. Immunol. 2021, 12, 668507. [Google Scholar] [CrossRef]
- Sattler, S. The role of the immune system beyond the fight against infection. Adv. Exp. Med. Biol. 2017, 1003, 3–14. [Google Scholar] [CrossRef]
- Schultz, M.; van der Poll, M.J.S.A.T. Modulation of innate immune responses in the treatment of sepsis and pneumonia. Curr. Drug Target. 2004, 3, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Tu, G.-W.; Ju, M.-J.; Yang, C.; Luo, Z. The immune system regulation in Sepsis: From innate to adaptive. Curr. Protein Pept. Sci. 2019, 20, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.S.; Monneret, G.; Payen, D. Sepsis-induced immunosuppression: From cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 2013, 13, 862–874. [Google Scholar] [CrossRef]
- de Castro, I.F.; Guzmán-Fulgencio, M.; García-Álvarez, M.; Resino, S. First evidence of a pro-inflammatory response to severe infection with influenza virus H1N1. Crit. Care 2010, 14, 115. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yuan, S.; Jiang, S.-C.; Zhang, Z.-W.; Fu, Y.-F.; Hu, J.; Li, Z.-L. Quantification of cytokine storms during virus infections. Front. Immunol. 2021, 12, 659419. [Google Scholar] [CrossRef]
- Flajnik, M.F. A cold-blooded view of adaptive immunity. Nat. Rev. Immunol. 2018, 18, 438–453. [Google Scholar] [CrossRef]
- Liang, Q.; Li, W.; Guo, N.; Tong, C.; Zhou, Y.; Fang, W.; Li, X. Identification and functional analysis of Interleukin-1β in the Chinese soft-shelled turtle Pelodiscus sinensis. Genes 2016, 7, 18. [Google Scholar] [CrossRef]
- Zhang, Z.; Tian, M.; Song, R.; Xing, X.; Fan, Y.; Wang, L.; Niu, C.; Dalmo, R.A. A New IL6 Isoform in Chinese soft-shelled turtle (Pelodiscus sinesis) discovered: Its regulation during cold stress and infection. Biology 2020, 9, 111. [Google Scholar] [CrossRef]
- Zhou, X.; Feng, H.; Guo, Q.; Dai, H. Identification and characterization of the first reptilian CD9, and its expression analysis in response to bacterial infection. Dev. Comp. Immunol. 2010, 34, 150–157. [Google Scholar] [CrossRef]
- Liu, T.; Han, Y.; Chen, S.; Zhao, H. Genome-wide identification of Toll-like receptors in the Chinese soft-shelled turtle Pelodiscus sinensis and expression analysis responding to Aeromonas hydrophila infection. Fish. Shellfish. Immunol. 2019, 87, 478–489. [Google Scholar] [CrossRef]
- Zhou, X.; Guo, Q.; Dai, H. Molecular characterization and expression profiles in response to bacterial infection of Chinese soft-shelled turtle interleukin-8 (IL-8), the first reptilian chemokine gene. Dev. Comp. Immunol. 2009, 33, 838–847. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, B.; Yuan, L.; Niu, C. Acute cold stress improved the transcription of pro-inflammatory cytokines of Chinese soft-shelled turtle against Aeromonas hydrophila. Dev. Comp. Immunol. 2015, 49, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Elbahnaswy, S.; Elshopakey, G.E. Differential gene expression and immune response of Nile tilapia (Oreochromis niloticus) challenged intraperitoneally with Photobacterium damselae and Aeromonas hydrophila demonstrating immunosuppression. Aquaculture 2020, 526, 735364. [Google Scholar] [CrossRef]
- Li, C.; Jiang, J.; Xie, J.; Yang, W.; Wang, Y. Transcriptome profiling and differential expression analysis of the immune-related genes during the acute phase of infection with Mycobacterium marinum in the goldfish (Carassius auratus L.). Aquaculture 2020, 533, 736198. [Google Scholar] [CrossRef]
- Zhu, C.; Pan, Z.; Chang, G.; Wu, N.; Wang, H.; Ding, H.; Qiang, X.; Zhang, L.; Qiang, J.; Zhang, J.; et al. Transcriptomic insights into immune responses to ulcerative syndrome in Pseudobagrus ussuriensis. Aquaculture 2021, 537, 736504. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, G.-R.; Ji, W.; Shi, Z.-C.; Ma, X.-F.; Luo, Z.-L.; Wei, K.-J. The dynamic immune response of yellow catfish (Pelteobagrus fulvidraco) infected with Edwardsiella ictaluri presenting the inflammation process. Front. Immunol. 2021, 12, 625928. [Google Scholar] [CrossRef] [PubMed]
- Eslamloo, K.; Caballero-Solares, A.; Inkpen, S.M.; Emam, M.; Kumar, S.; Bouniot, C.; Avendaño-Herrera, R.; Jakob, E.; Rise, M.L. Transcriptomic profiling of the adaptive and innate immune responses of atlantic salmon to Renibacterium salmoninarum infection. Front. Immunol. 2020, 11, 567838. [Google Scholar] [CrossRef] [PubMed]
- Moraleda, C.P.; Robledo, D.; Gutiérrez, A.P.; Del-Pozo, J.; Yáñez, J.M.; Houston, R.D. Investigating mechanisms underlying genetic resistance to Salmon Rickettsial Syndrome in Atlantic salmon using RNA sequencing. BMC Genom. 2021, 22, 156. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Tao, C.; Olyarchuk, J.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Xin, L. An improvement of the 2(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath. 2013, 3, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Pan, H.; Sun, H. The immune response and protective efficacy of oral alginate microparticle Aeromonas sobria vaccine in soft-shelled turtles (Trionyx sinensis). Veter. Immunol. Immunopathol. 2007, 119, 299–302. [Google Scholar] [CrossRef]
- Musgrave, K.E.; Diehl, K.; Mans, C. Aeromonas Hydrophila Keratitis in Freshwater Turtles. J. Exot. Pet. Med. 2016, 25, 26–29. [Google Scholar] [CrossRef]
- Zhang, B.; Li, C.; Wang, X.; Liu, C.; Zhou, H.; Mai, K.; He, G. Administration of commensal Shewanella sp. MR-7 ameliorates lipopolysaccharide-induced intestine dysfunction in turbot (Scophthalmus maximus L.). Fish. Shellfish. Immunol. 2020, 102, 460–468. [Google Scholar] [CrossRef]
- Pradel, B.; Robert-Hebmann, V.; Espert, L. Regulation of Innate Immune Responses by Autophagy: A Goldmine for Viruses. Front. Immunol. 2020, 11, 578038. [Google Scholar] [CrossRef]
- Yang, M.; Wang, Q.; Wang, S.; Wang, Y.; Zeng, Q.; Qin, Q. Transcriptomics analysis reveals candidate genes and pathways for susceptibility or resistance to Singapore grouper iridovirus in orange-spotted grouper (Epinephelus coioides). Dev. Comp. Immunol. 2019, 90, 70–79. [Google Scholar] [CrossRef]
- Zhu, M.; Nan, Y.; Zhai, M.; Wang, M.; Shao, Y.; Blair, H.T.; Morris, S.T.; Kenyon, P.R.; Zhao, Z.; Zhang, H. Comparative profiling of the resistance of different genotypes of mannose-binding lectin to Mycoplasma pneumoniae infection in Chinese Merino sheep based on high-throughput sequencing technology. Veter. Immunol. Immunopathol. 2021, 233, 110183. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.G.; Song, C.Y.; Wu, X.S.; Zhao, X.T.; Wang, X.Y.; Li, B.C.; Gan, Y. RNA-seq transcriptome profiling of porcine lung from two pig breeds in response to Mycoplasma hyopneumoniae infection. PeerJ 2019, 7, e7900. [Google Scholar] [CrossRef] [PubMed]
- Truong, A.D.; Hong, Y.H.; Lillehoj, H.S. High-throughput sequencing reveals differing immune responses in the intestinal mucosa of two inbred lines afflicted with necrotic enteritis. Veter. Immunol. Immunopathol. 2015, 166, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Peatman, E.; Li, C.; Peterson, B.C.; Straus, D.L.; Farmer, B.D.; Beck, B.H. Basal polarization of the mucosal compartment in Flavobacterium columnare susceptible and resistant channel catfish (Ictalurus punctatus). Mol. Immunol. 2013, 56, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Feghali, C.; Wright, T. Cytokines in acute and chronic inflammation. Front. Biosci. 1997, 2, d12–d26. [Google Scholar] [CrossRef]
- Shachar, I.; Karin, N. The dual roles of inflammatory cytokines and chemokines in the regulation of autoimmune diseases and their clinical implications. J. Leukoc. Biol. 2013, 93, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Murtaugh, M.P.; Baarsch, M.J.; Zhou, Y.; Scamurra, R.W.; Lin, G. Inflammatory cytokines in animal health and disease. Veter. Immunol. Immunopathol. 1996, 54, 45–55. [Google Scholar] [CrossRef]
- D’Elia, R.V.; Harrison, K.; Oyston, P.C.; Lukaszewski, R.A.; Clark, G.C. Targeting the “Cytokine Storm” for therapeutic benefit. Clin. Vaccine Immunol. 2013, 20, 319–327. [Google Scholar] [CrossRef]
- Chen, N.; Jiang, J.; Gao, X.; Li, X.; Zhang, Y.; Liu, X.; Yang, H.; Bing, X.; Zhang, X. Histopathological analysis and the immune related gene expression profiles of mandarin fish (Siniperca chuatsi) infected with Aeromonas hydrophila. Fish. Shellfish. Immunol. 2018, 83, 410–415. [Google Scholar] [CrossRef]
- Liu, X.; Sun, W.; Zhang, Y.; Zhou, Y.; Xu, J.; Gao, X.; Zhang, S.; Zhang, X. Impact of Aeromonas hydrophila and infectious spleen and kidney necrosis virus infections on susceptibility and host immune response in Chinese perch (Siniperca chuatsi). Fish. Shellfish. Immunol. 2020, 105, 117–125. [Google Scholar] [CrossRef]
- Gallani, S.U.; Valladão, G.M.R.; Alves, L.D.O.; Kotzent, S.; Hashimoto, D.T.; Wiegertjes, G.; Kirsten, K.; Kreutz, L.C.; Pilarski, F. Patterns of the innate immune response in tambaqui Colossoma macropomum: Modulation of gene expression in haemorrhagic septicaemia caused by Aeromonas hydrophila. Microb. Pathog. 2020, 150, 104638. [Google Scholar] [CrossRef]
- Zhou, J.; Zhao, H.; Zhang, L.; Ye, X.; Wang, Z.; Li, Q.; Ke, H.; Zhao, G.; Du, J.; Yang, S.; et al. Effects of bacterial haemorrhagic septicemia on the immune response of Leiocassis longirostris by RNA-Seq and microRNA-Seq. Comp. Biochem. Physiol. Part. D Genom. Proteom. 2020, 34, 100659. [Google Scholar] [CrossRef]
- Couper, K.N.; Blount, D.G.; Riley, E.M. IL-10: The master regulator of immunity to infection. J. Immunol. 2008, 180, 5771–5777. [Google Scholar] [CrossRef] [PubMed]
- González, L.A.; Melo-González, F.; Sebastián, V.P.; Vallejos, O.P.; Noguera, L.P.; Suazo, I.D.; Schultz, B.M.; Manosalva, A.H.; Peñaloza, H.F.; Soto, J.A.; et al. Characterization of the anti-inflammatory capacity of IL-10-producing neutrophils in response to Streptococcus pneumoniae infection. Front. Immunol. 2021, 12, 578038. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, M.; O’Garra, A. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 2010, 10, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Cao, X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 2016, 16, 35–50. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like receptors and the control of immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef]
- Stok, J.E.; Quiroz, M.E.V.; Van Der Veen, A.G. Self RNA Sensing by RIG-I–like Receptors in viral infection and sterile inflammation. J. Immunol. 2020, 205, 883–891. [Google Scholar] [CrossRef]
- Platnich, J.M.; Muruve, D.A. NOD-like receptors and inflammasomes: A review of their canonical and non-canonical signaling pathways. Arch. Biochem. Biophys. 2019, 670, 4–14. [Google Scholar] [CrossRef]
- Lu, Y.; Li, X.; Liu, S.; Zhang, Y.; Zhang, D. Toll-like receptors and inflammatory bowel disease. Front. Immunol. 2018, 9, 72. [Google Scholar] [CrossRef]
- Place, D.E.; Lee, S.; Kanneganti, T.-D. PANoptosis in microbial infection. Curr. Opin. Microbiol. 2021, 59, 42–49. [Google Scholar] [CrossRef]
- Jorgensen, I.; Rayamajhi, M.; Miao, E.A. Programmed cell death as a defence against infection. Nat. Rev. Immunol. 2017, 17, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Lacey, C.; Miao, E. Programmed cell death in the evolutionary race against bacterial virulence factors. Cold Spring Harb. Perspect. Biol. 2019, 12, a036459. [Google Scholar] [CrossRef] [PubMed]
- Amarante-Mendes, G.P.; Adjemian, S.; Branco, L.M.; Zanetti, L.; Weinlich, R.; Bortoluci, K.R. Pattern recognition receptors and the host cell death molecular machinery. Front. Immunol. 2018, 9, 2379. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kong, X.; Liu, X.; Wang, X.; Wang, Z.; Liu, J.; Zhang, Q.; Yu, H. Edwardsiella tarda triggers the pyroptosis of the macrophage of Japanese flounder (Paralichthys olivaceus). Aquaculture 2020, 533, 736153. [Google Scholar] [CrossRef]
- Lu, Z.; Yang, M.; Zhang, K.; Zhan, F.; Li, F.; Shi, F.; Li, Y.; Zhao, L.; Li, J.; Lin, L.; et al. Aeromonas hydrophila infection activates death receptor apoptosis pathway in the red blood cells of grass carp (Ctenopharyngodon idellus). Aquaculture 2021, 532, 735956. [Google Scholar] [CrossRef]
- Wang, Z.; Gu, Z.; Hou, Q.; Chen, W.; Mu, D.; Zhang, Y.; Liu, Q.; Liu, Z.; Yang, D. Zebrafish GSDMEb cleavage-gated pyroptosis drives septic acute kidney injury in vivo. J. Immunol. 2020, 204, 1929–1942. [Google Scholar] [CrossRef]
- Greenberg, S.; Grinstein, S. Phagocytosis and innate immunity. Curr. Opin. Immunol. 2002, 14, 136–145. [Google Scholar] [CrossRef]
- Stuart, L.M.; Ezekowitz, R.A. Phagocytosis and comparative innate immunity: Learning on the fly. Nat. Rev. Immunol. 2008, 8, 131–141. [Google Scholar] [CrossRef]
- Browne, N.; Heelan, M.; Kavanagh, K. An analysis of the structural and functional similarities of insect hemocytes and mammalian phagocytes. Virulence 2013, 4, 597–603. [Google Scholar] [CrossRef]
- Flannagan, R.S.; Cosío, G.; Grinstein, S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 2009, 7, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Underhill, D.M.; Goodridge, H.S. Information processing during phagocytosis. Nat. Rev. Immunol. 2012, 12, 492–502. [Google Scholar] [CrossRef] [PubMed]
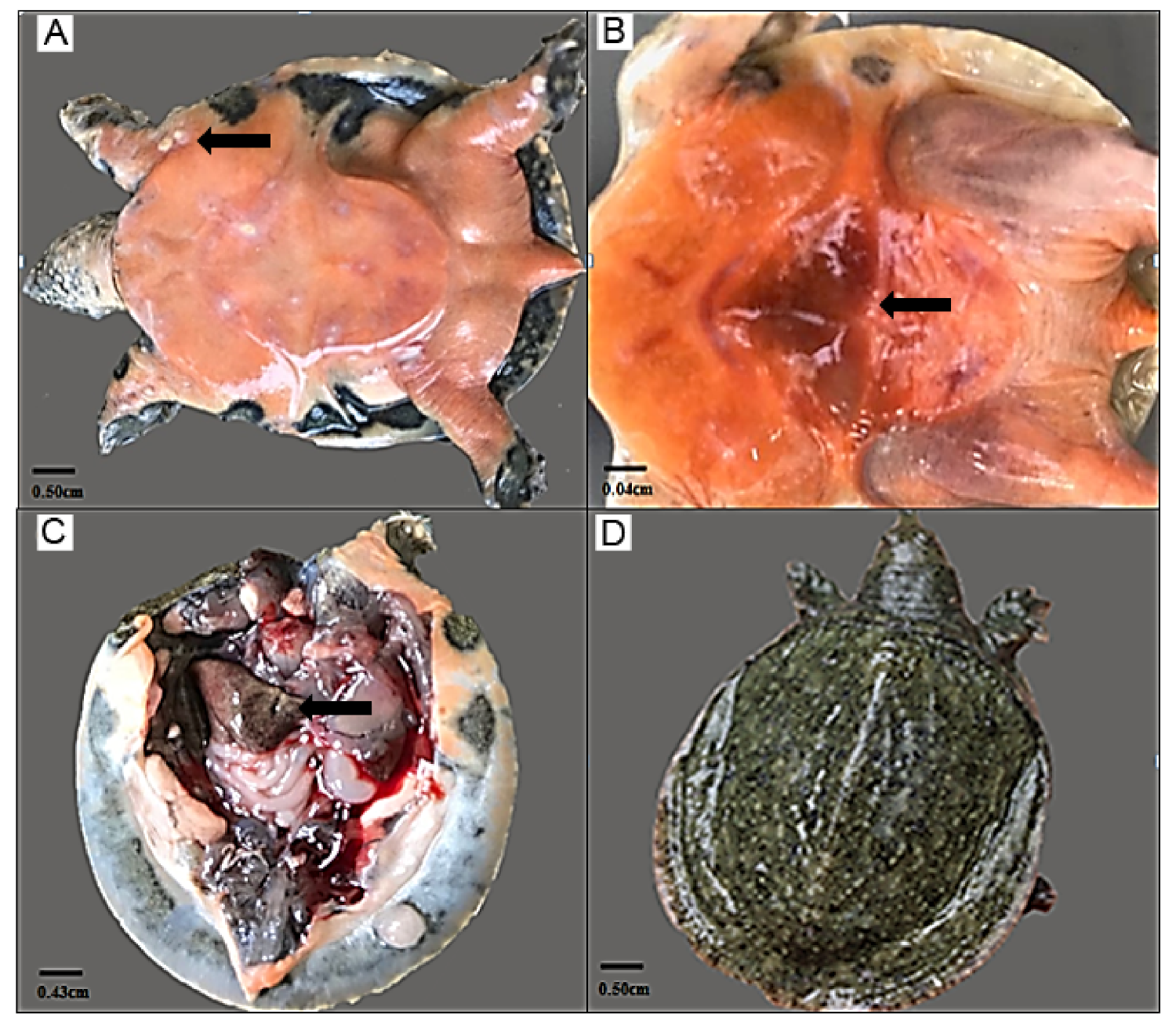
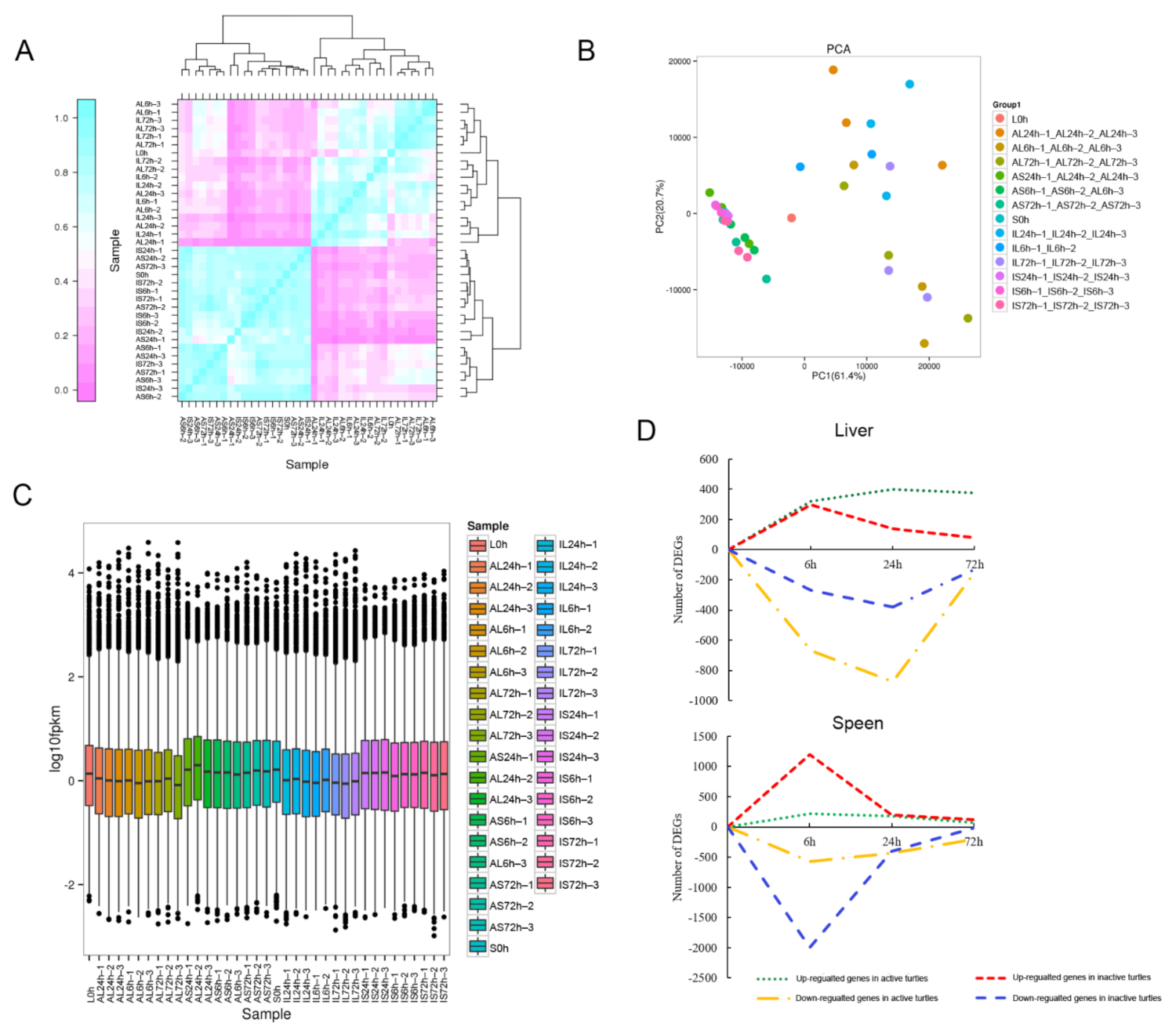

| Categories/ Gene Name | Description | Log2 (Fold Changes) | |||||
|---|---|---|---|---|---|---|---|
| AL Group | IL Group | ||||||
| 6 Hpi | 24 Hpi | 72 Hpi | 6 Hpi | 24 Hpi | 72 Hpi | ||
| Interleukins and interleukin receptors | |||||||
| IL1β | interleukin 1 beta | 2.18 | 4.82 | ||||
| IL8 | interleukin 8 | 3.55 | |||||
| IL10 | interleukin 10 | 6.08 | 4.41 | 2.31 | |||
| IL1R1 | interleukin 1 receptor type I | 1.66 | |||||
| IL1R2 | interleukin 1 receptor type II | 5.88 | 3.16 | 7.06 | |||
| IL5RA | interleukin 5 receptor alpha | −2.57 | −2.40 | −3.01 | |||
| IL18R1 | interleukin 18 receptor 1 | −5.30 | |||||
| Chemokines and chemokine receptors | |||||||
| CCL5 | C-C motif chemokine 5 | 4.71 | |||||
| CCL20 | C-C motif chemokine 20 | 9.34 | 7.63 | 12.18 | |||
| CX3CL1 | C-X3-C motif chemokine 1 | −4.91 | 3.28 | ||||
| CCR5 | C-C chemokine receptor type 5 | 3.02 | |||||
| TNF family members and TNF receptors | |||||||
| TNFSF10 | tumor necrosis factor ligand superfamily member 10 | −2.56 | −2.08 | ||||
| TNFSF15 | tumor necrosis factor ligand superfamily member 15 | −2.40 | |||||
| SF6B | tumor necrosis factor receptor superfamily member 6B | −2.57 | |||||
| SF12A | tumor necrosis factor receptor superfamily member 12A | 4.49 | 4.60 | ||||
| Toll-like receptor (TLR) signaling pathway | |||||||
| TLR2 | toll-like receptor 2 | 4.56 | 2.80 | ||||
| TLR5 | toll-like receptor 5 | 3.61 | 1.51 | 2.89 | |||
| TRIF | toll-like receptor adapter molecule 1 | −3.15 | −2.87 | −1.99 | −3.40 | −3.42 | |
| TAB1 | TAK1-binding protein 1 | 1.77 | |||||
| PI3K | phosphoinositide-3-kinase | 2.48 | 2.96 | ||||
| MAP2K1 | mitogen-activated protein kinase kinase 1 | 2.13 | |||||
| MAP2K6 | mitogen-activated protein kinase kinase 6 | −2.04 | -2.63 | −2.59 | |||
| AP-1 | proto-oncogene protein c-fos | 1.43 | |||||
| STAT1 | signal transducer and activator of transcription 1 | 1.49 | |||||
| RIG-I-like receptor (RLR) signaling pathway | |||||||
| RIG-I | retinoic acid inducible gene I | 2.38 | |||||
| LGP2 | laboratory of genetics and physiology 2 | 2.21 | |||||
| MDA5 | melanoma differentiation-associated gene 5 | 2.43 | |||||
| TRAF3 | TNF receptor-associated factor 3 | 5.07 | 3.54 | ||||
| IRF7 | interferon regulatory factor 7 | −1.83 | 2.86 | ||||
| DDX3X | ATP-dependent RNA helicase DDX3X | −1.86 | −1.69 | −2.08 | −2.02 | ||
| NOD-like receptor (NLR) signaling pathway | |||||||
| ASC | apoptosis-associated speck-like protein containing a CARD | 1.99 | |||||
| RIPK2 | receptor-interacting serine/threonine-protein kinase 2 | 2.47 | |||||
| cIAP | baculoviral IAP repeat-containing protein 2/3 | 2.50 | |||||
| TNFAIP3 | tumor necrosis factor alpha-induced protein 3 | 1.56 | 3.75 | ||||
| Categories/ Gene Name | Description | Log2 (Fold Changes) | |||||
|---|---|---|---|---|---|---|---|
| AL Group | IL Group | ||||||
| 6 Hpi | 24 Hpi | 72 Hpi | 6 Hpi | 24 Hpi | 72 Hpi | ||
| Internalization and formation of the phagosomes | |||||||
| TLR2 | toll-like receptor 2 | 4.56 | 2.80 | ||||
| MR | mannose receptor | 1.87 | 1.07 | −2.18 | |||
| iC3b | the fragment of complement component 3 | 4.38 | 2.79 | ||||
| Collectin | C-type lectin | 4.81 | 3.69 | 1.47 | −2.63 | ||
| F-actin | actin beta/gamma 1 | 5.57 | 5.31 | 4.39 | 5.09 | 5.68 | 4.64 |
| Early phagosome | |||||||
| Rab5 | ras-related protein Rab-5B | 2.28 | |||||
| vATPase | V-type H+-transporting ATPase | 2.33 | 2.05 | 3.85 | −2.08 | ||
| CALR | calreticulin | 4.05 | |||||
| Mature phagosome | |||||||
| TUBA | tubulin alpha | 2.26 | |||||
| TUBB | tubulin beta | 4.63 | 3.25 | 1.42 | 2.72 | ||
| vATPase | V-type H+-transporting ATPase | 2.33 | 2.05 | 3.85 | −2.08 | ||
| Phagolysosome | |||||||
| sec61 | protein transport protein SEC61 subunit beta | 2.72 | 3.26 | 2.95 | |||
| vATPase | V-type H+-transporting ATPase | 2.33 | 2.05 | 3.85 | −2.08 | ||
| Activation of NADPH oxidase | |||||||
| p40phox | neutrophil cytosolic factor 4 | 3.50 | |||||
| p47phox | neutrophil cytosolic factor 1 | 2.79 | 2.42 | ||||
| gp91 | NADPH oxidase 1 | 1.59 | |||||
| Antigen presentation | |||||||
| MHC II | MHC class II antigen | −2.21 | |||||
| sec22 | vesicle transport protein SEC22 | 4.21 | 3.54 | ||||
| Gene Name | Description | Log2 (Fold Changes) | |||||
|---|---|---|---|---|---|---|---|
| AL Group | IL Group | ||||||
| 6 Hpi | 24 Hpi | 72 Hpi | 6 Hpi | 24 Hpi | 72 Hpi | ||
| p53 | tumor protein p53 | 1.94 | |||||
| IP3R | inositol 1,4,5-triphosphate receptor type 3 | 1.35 | 1.56 | 1.92 | |||
| Perforin | perforin 1 | 3.12 | 2.57 | ||||
| PI3K | phosphoinositide-3-kinase | 2.48 | 2.96 | ||||
| MERK2 | mitogen-activated protein kinase kinase 2 | 2.13 | |||||
| PERK | protein kinase RNA (PKR)-like ER kinase | 1.85 | 2.30 | ||||
| Cathepsin | cathepsin B | 5.11 | 4.27 | ||||
| NOXA | phorbol-12-myristate-13-acetate-induced protein 1 | 1.62 | |||||
| AP1 | proto-oncogene protein c-fos | 1.43 | |||||
| GZMB | granzyme B | 3.87 | |||||
| IL3R | cytokine receptor common subunit beta | 3.43 | |||||
| A1 | hematopoietic Bcl-2-related protein A1 | 4.59 | |||||
| Categories/ Gene Name | Description | Log2 (Fold Changes) | |||||
|---|---|---|---|---|---|---|---|
| AS Group | IS Group | ||||||
| 6 Hpi | 24 Hpi | 72 Hpi | 6 Hpi | 24 Hpi | 72 Hpi | ||
| Interleukins and interleukin receptors | |||||||
| IL1β | interleukin 1 beta | 2.46 | |||||
| IL6 | interleukin 6 | 5.20 | |||||
| IL7 | interleukin 7 | −2.79 | |||||
| IL8 | interleukin 8 | 3.27 | 2.43 | 2.41 | |||
| IL10 | interleukin 10 | 6.82 | 5.69 | 1.21 | 7.30 | ||
| IL1R2 | interleukin 1 receptor type II | 4.13 | 3.81 | ||||
| IL1RAP | interleukin 1 receptor accessory protein | 2.24 | |||||
| IL3RB | cytokine receptor common subunit beta | 3.56 | |||||
| IL4R | interleukin 4 receptor | 1.18 | |||||
| IL5RA | interleukin 5 receptor alpha | −1.95 | −3.81 | −2.85 | |||
| IL5RB | interleukin 5 receptor alpha | 3.56 | |||||
| IL8RB | interleukin 8 receptor | 3.07 | 2.32 | ||||
| IL12RB1 | interleukin 12 receptor beta-1 | 2.60 | |||||
| IL12RB2 | interleukin 12 receptor beta-1 | 3.20 | |||||
| IL15RA | interleukin 15 receptor alpha | 3.68 | |||||
| IL21R | interleukin 21 receptor | 1.98 | |||||
| IL22RA2 | interleukin 22 receptor alpha 2 | 5.21 | 4.77 | 4.39 | |||
| Chemokines and chemokine receptors | |||||||
| CCL20 | C-C motif chemokine 20 | 6.33 | 6.72 | 2.56 | |||
| CXCL10 | C-X-C motif chemokine 10 | 5.36 | |||||
| CXCL11 | C-X-C motif chemokine 11 | 4.32 | |||||
| CXCL12 | C-X-C motif chemokine 12 | −2.00 | |||||
| CXCL13 | C-X-C motif chemokine 13 | 3.47 | 2.41 | ||||
| CXCL14 | C-X-C motif chemokine 14 | 1.81 | 2.07 | ||||
| CX3CL1 | C-X3-C motif chemokine 1 | −3.70 | 3.90 | ||||
| CCR5 | C-C chemokine receptor type 5 | 1.26 | |||||
| CXCR4 | C-X-C chemokine receptor type 4 | −1.75 | |||||
| XCR1 | XC chemokine receptor 1 | −4.81 | |||||
| TNF family members and TNF receptors | |||||||
| TNFSF8 | tumor necrosis factor ligand superfamily member 8 | 3.06 | |||||
| TNFSF10 | tumor necrosis factor ligand superfamily member 10 | −2.86 | −4.23 | −3.17 | |||
| TNFSF12 | tumor necrosis factor ligand superfamily member 12 | −1.99 | |||||
| TNFSF15 | tumor necrosis factor ligand superfamily member 15 | 3.09 | |||||
| TNFSF18 | tumor necrosis factor ligand superfamily member 18 | 4.56 | |||||
| EDA | ectodysplasin-A | −2.93 | |||||
| SF6B | tumor necrosis factor receptor superfamily member 6B | 7.29 | |||||
| SF9 | tumor necrosis factor receptor superfamily member 9 | 3.26 | |||||
| SF12A | tumor necrosis factor receptor superfamily member 12A | 6.32 | 7.32 | 7.37 | |||
| SF13B | tumor necrosis factor receptor superfamily member 13B | 2.33 | 3.08 | ||||
| SF19L | tumor necrosis factor receptor superfamily member 19-like | 5.98 | |||||
| FAS | tumor necrosis factor receptor superfamily member 6 | 2.34 | |||||
| NGFR | tumor necrosis factor receptor superfamily member 16 | −3.24 | |||||
| EDAR | ectodysplasin-A receptor | −4.72 | |||||
| Toll-like receptor (TLR) signaling pathway | |||||||
| TLR4 | toll-like receptor 4 | 2.30 | 1.71 | ||||
| TLR5 | toll-like receptor 5 | 2.77 | 1.82 | 3.00 | |||
| MyD88 | myeloid differentiation factor 88 | 1.08 | |||||
| TRIF | toll-like receptor adapter molecule 1 | 1.96 | |||||
| Rac | Ras-related C3 botulinum toxin substrate 1 | 2.37 | 2.52 | 1.97 | 1.84 | 2.51 | 2.04 |
| PI3K | phosphoinositide-3-kinase | −4.06 | |||||
| AKT | RAC serine/threonine-protein kinase | 1.26 | |||||
| MAP2K1 | mitogen-activated protein kinase kinase 1 | 2.88 | 2.82 | 2.50 | 2.85 | 2.94 | 2.57 |
| MAP2K6 | mitogen-activated protein kinase kinase 6 | −2.37 | |||||
| AP-1 | proto-oncogene protein c-fos | 1.60 | 1.33 | ||||
| STAT1 | signal transducer and activator of transcription 1 | 2.03 | |||||
| IKBKE | inhibitor of nuclear factor kappa-B kinase subunit epsilon | 1.92 | |||||
| RIG-I-like receptor (RLR) signaling pathway | |||||||
| RIG-I | retinoic acid inducible gene I | 2.32 | |||||
| LGP2 | laboratory of genetics and physiology 2 | 1.90 | |||||
| MDA5 | melanoma differentiation-associated gene 5 | 1.54 | |||||
| TRAF2 | TNF receptor-associated factor 2 | 2.19 | |||||
| MITA | Mediator of IFN regulatory transcription factor 3 activation | 1.29 | |||||
| NOD-like receptor (NLR) signaling pathway | |||||||
| NLRP12 | NACHT, LRR and PYD domains-containing protein 12 | −2.65 | −2.05 | −3.11 | |||
| ASC | apoptosis-associated speck-like protein containing a CARD | 2.27 | |||||
| RIPK2 | receptor-interacting serine/threonine-protein kinase 2 | 1.46 | |||||
| CARD8 | caspase recruitment domain-containing protein 8 | 1.49 | |||||
| Categories/ Gene Name | Description | Log2 (Fold Changes) | |||||
|---|---|---|---|---|---|---|---|
| AS Group | IS Group | ||||||
| 6 Hpi | 24 Hpi | 72 Hpi | 6 Hpi | 24 Hpi | 72 Hpi | ||
| Internalization and formation of the phagosome | |||||||
| TLR4 | toll-like receptor 4 | 2.30 | 1.71 | ||||
| MR | mannose receptor | −2.71 | −1.43 | −3.02 | |||
| CR1 | complement receptor 1 | 1.69 | 2.15 | ||||
| αvβ5 | integrin αvβ5 | −2.45 | |||||
| iC3b | the fragment of complement component 3 | −3.36 | |||||
| Collectin | C-type lectin | −4.00 | −1.66 | −4.85 | |||
| F-actin | actin beta/gamma 1 | 3.60 | −3.11 | −3.66 | −3.71 | −2.46 | |
| Early phagosome | |||||||
| Rab5 | ras-related protein Rab-5B | 2.47 | 1.84 | 1.82 | |||
| vATPase | V-type H+-transporting ATPase | 2.21 | −1.01 | 1.59 | |||
| CALR | calreticulin | 2.10 | |||||
| Mature phagosome | |||||||
| TUBA | tubulin alpha | 1.14 | 1.58 | 1.63 | |||
| TUBB | tubulin beta | 3.63 | 1.60 | −1.21 | 2.28 | 2.34 | |
| vATPase | V-type H+-transporting ATPase | 2.21 | −1.01 | 1.59 | |||
| Dynein | dynein heavy chain 2 | −1.82 | |||||
| Phagolysosome | |||||||
| sec61 | protein transport protein SEC61 subunit beta | 1.36 | |||||
| vATPase | V-type H+-transporting ATPase | 2.21 | −1.01 | 1.59 | |||
| NOS | nitric-oxide synthase | −3.07 | |||||
| TAP | ATP-binding cassette subfamily B member 3 | 2.17 | |||||
| Activation of NADPH oxidase | |||||||
| gp91 | NADPH oxidase 1 | 1.62 | 1.57 | 1.40 | |||
| p40phox | neutrophil cytosolic factor 4 | 1.27 | |||||
| p47phox | neutrophil cytosolic factor 1 | 2.75 | 2.32 | 2.53 | 2.04 | 1.52 | |
| P67phox | neutrophil cytosolic factor 2 | 1.40 | |||||
| Rac | Ras-related C3 botulinum toxin substrate 1 | 2.37 | 2.52 | 1.97 | 1.84 | 2.51 | 2.04 |
| Antigen presentation | |||||||
| MHC II | MHC class II antigen | 3.14 | −1.87 | −2.54 | |||
| Gene Name | Description | Log2 (Fold Changes) | |||||
|---|---|---|---|---|---|---|---|
| AS Group | IS Group | ||||||
| 6 Hpi | 24 Hpi | 72 Hpi | 6 Hpi | 24 Hpi | 72 Hpi | ||
| p53 | tumor protein p53 | 1.66 | |||||
| IP3R | inositol 1,4,5-triphosphate receptor type 3 | 1.17 | |||||
| Perforin | perforin 1 | 3.24 | 5.58 | ||||
| MERK2 | mitogen-activated protein kinase kinase 2 | 2.88 | 2.82 | 2.50 | 2.85 | 2.95 | 2.57 |
| Cathepsin | cathepsin B | 6.19 | 3.24 | 1.57 | 2.81 | ||
| AP1 | proto-oncogene protein c-fos | 1.60 | 1.33 | ||||
| GZMB | granzyme B | 1.22 | 1.57 | 5.59 | 5.34 | ||
| IL-3R | cytokine receptor common subunit beta | 2.56 | |||||
| TRAF12 | TNF receptor-associated factor 2 | 2.91 | 2.19 | ||||
| Fas | tumor necrosis factor receptor superfamily member 6 | 2.34 | |||||
| TrkA | neurotrophic tyrosine kinase receptor type 1 | 6.49 | |||||
| NIK | mitogen-activated protein kinase kinase kinase 14 | 1.24 | |||||
| FLIP | CASP8 and FADD-like apoptosis regulator | 1.26 | |||||
| eiF2α | translation initiation factor 2 subunit 1 | 1.25 | |||||
| Calpain | calpain-1 | 1.49 | |||||
| ARTS | septin 4 | 2.45 | |||||
| AIF | apoptosis-inducing factor 1 | 1.40 | |||||
| Gadd45 | growth arrest and DNA-damage-inducible protein | 2.32 | |||||
| ASK1 | mitogen-activated protein kinase kinase kinase 5 | 5.27 | |||||
| CytC | cytochrome c | 1.90 | 1.80 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, Z.; Hu, Y.; Tan, J.; Wang, X.; Liu, X.; Zeng, C. Comparative Transcriptome Analysis Reveals the Molecular Immunopathogenesis of Chinese Soft-Shelled Turtle (Trionyx sinensis) Infected with Aeromonas hydrophila. Biology 2021, 10, 1218. https://doi.org/10.3390/biology10111218
Lv Z, Hu Y, Tan J, Wang X, Liu X, Zeng C. Comparative Transcriptome Analysis Reveals the Molecular Immunopathogenesis of Chinese Soft-Shelled Turtle (Trionyx sinensis) Infected with Aeromonas hydrophila. Biology. 2021; 10(11):1218. https://doi.org/10.3390/biology10111218
Chicago/Turabian StyleLv, Zhao, Yazhou Hu, Jin Tan, Xiaoqing Wang, Xiaoyan Liu, and Cong Zeng. 2021. "Comparative Transcriptome Analysis Reveals the Molecular Immunopathogenesis of Chinese Soft-Shelled Turtle (Trionyx sinensis) Infected with Aeromonas hydrophila" Biology 10, no. 11: 1218. https://doi.org/10.3390/biology10111218
APA StyleLv, Z., Hu, Y., Tan, J., Wang, X., Liu, X., & Zeng, C. (2021). Comparative Transcriptome Analysis Reveals the Molecular Immunopathogenesis of Chinese Soft-Shelled Turtle (Trionyx sinensis) Infected with Aeromonas hydrophila. Biology, 10(11), 1218. https://doi.org/10.3390/biology10111218







