Metabolomics and Transcriptomics Analyses of Two Contrasting Cherry Rootstocks in Response to Drought Stress
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, Stress Treatment, and Sampling
2.2. RNA Sequencing and Functional Annotation
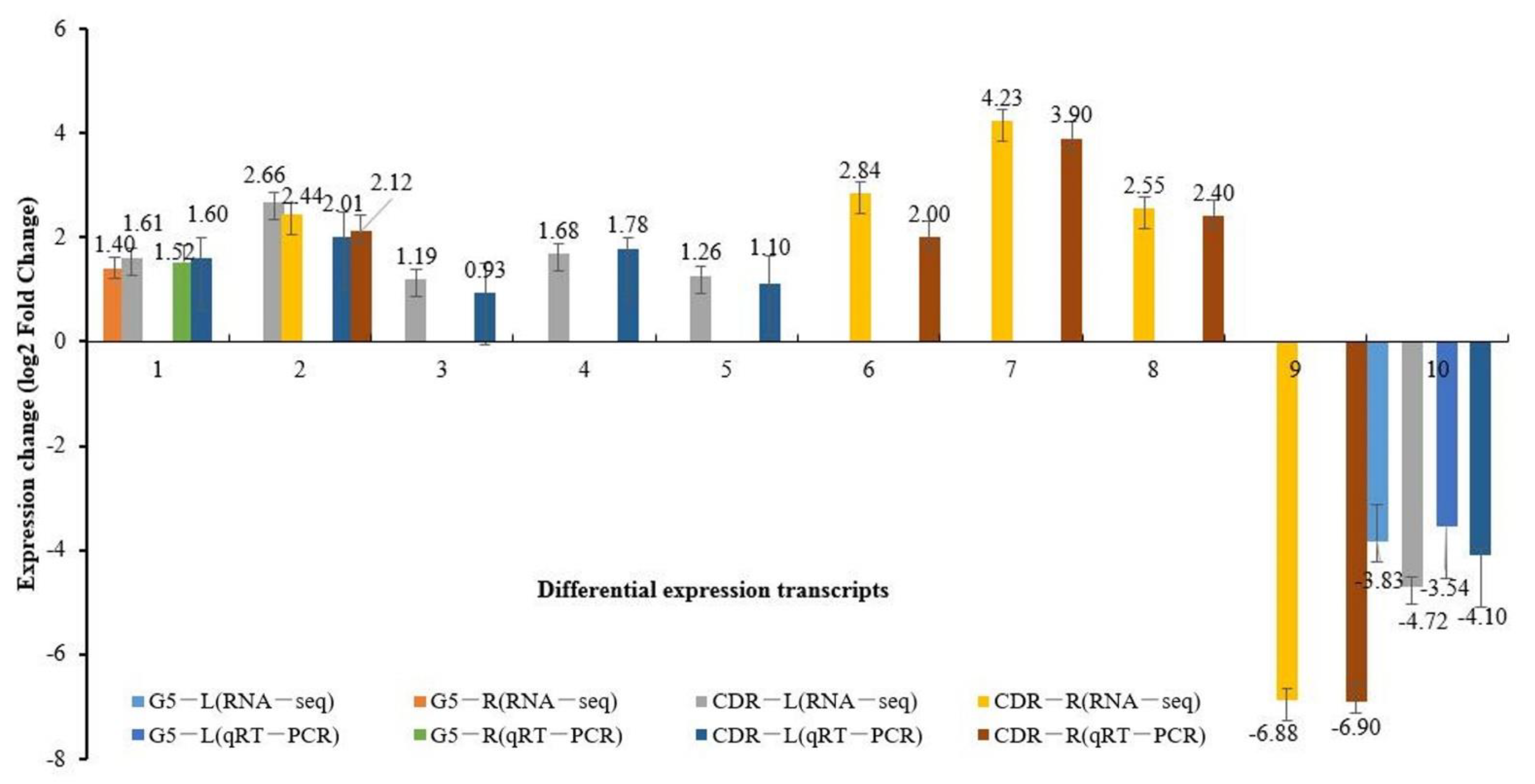
2.3. Quantitative Real-Time PCR (qRT-PCR)
2.4. Metabolomics Data Processing and Analysis
2.5. DEGs and Conjoint Analysis of Central Metabolites
2.6. Screening of Core Candidate Genes and Bioinformatics Analysis
3. Results
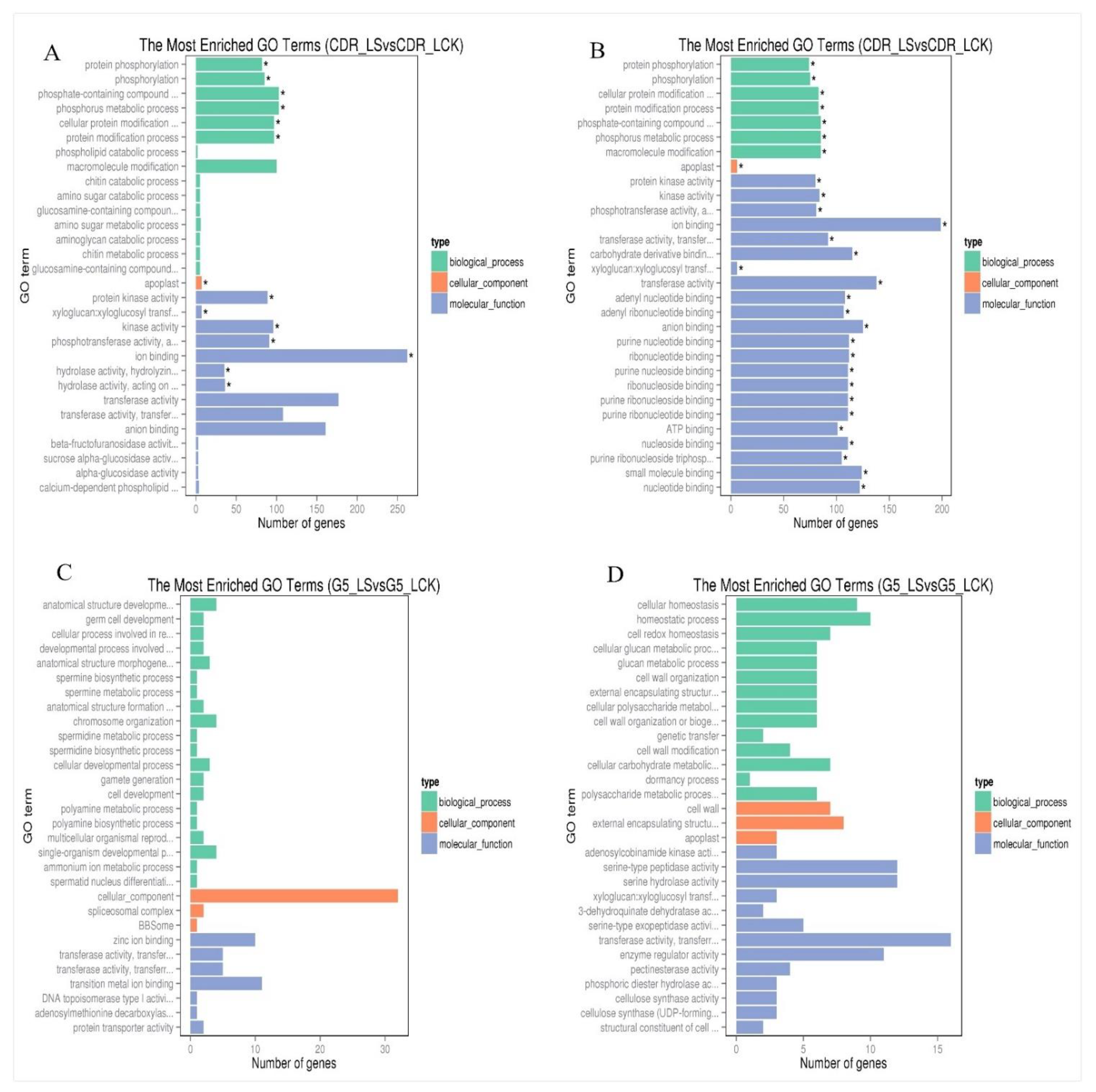
3.1. Transcriptional Characteristics of the Responses of Cherry Rootstock to Drought Stress
3.2. Interaction Networks of DEGs between DT and DS
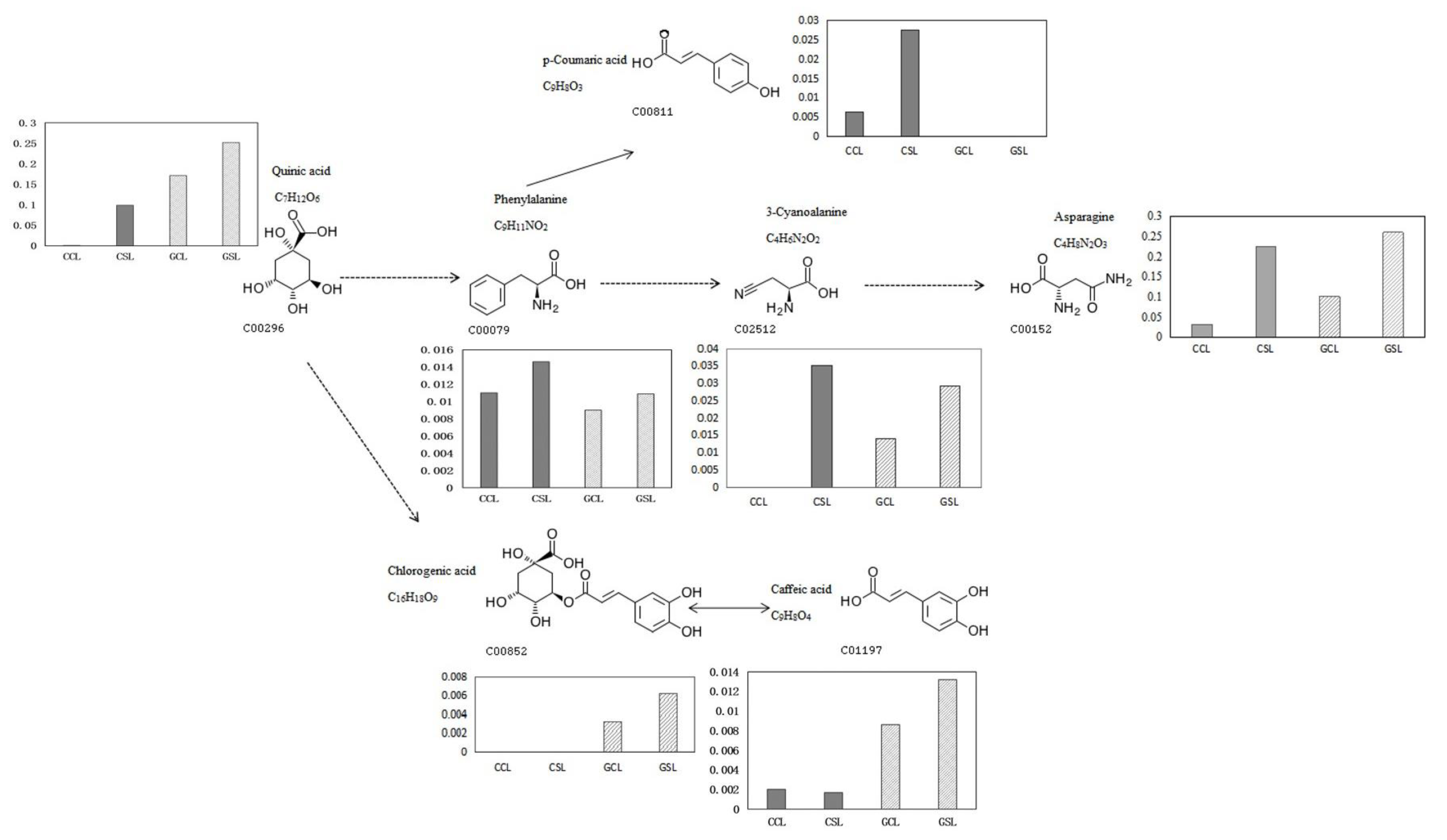
3.3. Metabolome Analysis and Screening of Central Metabolites
3.4. Identification and Cluster Analyses of Cherry Metabolites in Response to Drought
3.5. Analysis of Metabolic Pathways and DEGs
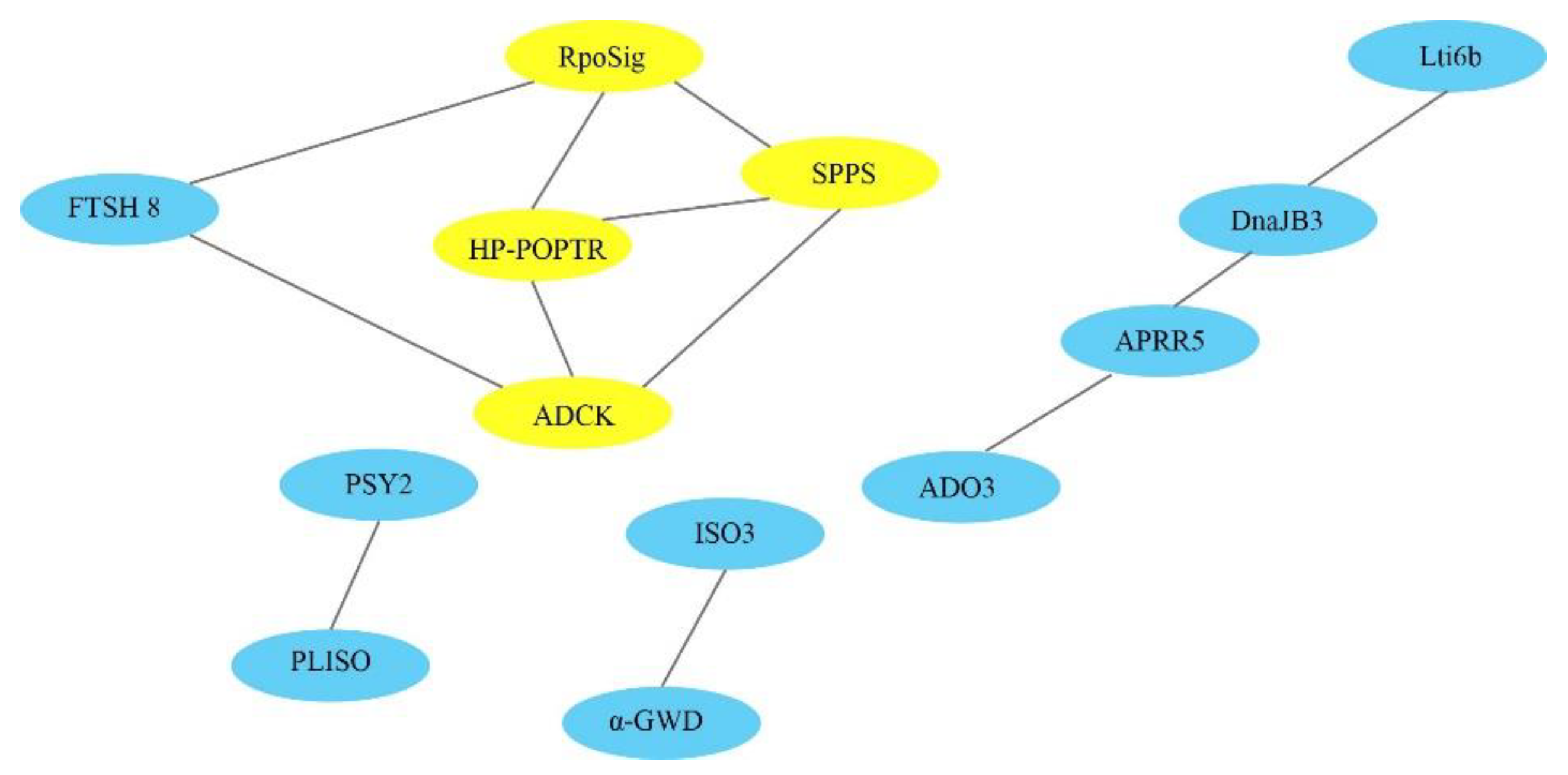
3.6. Selection of Core Candidate Genes
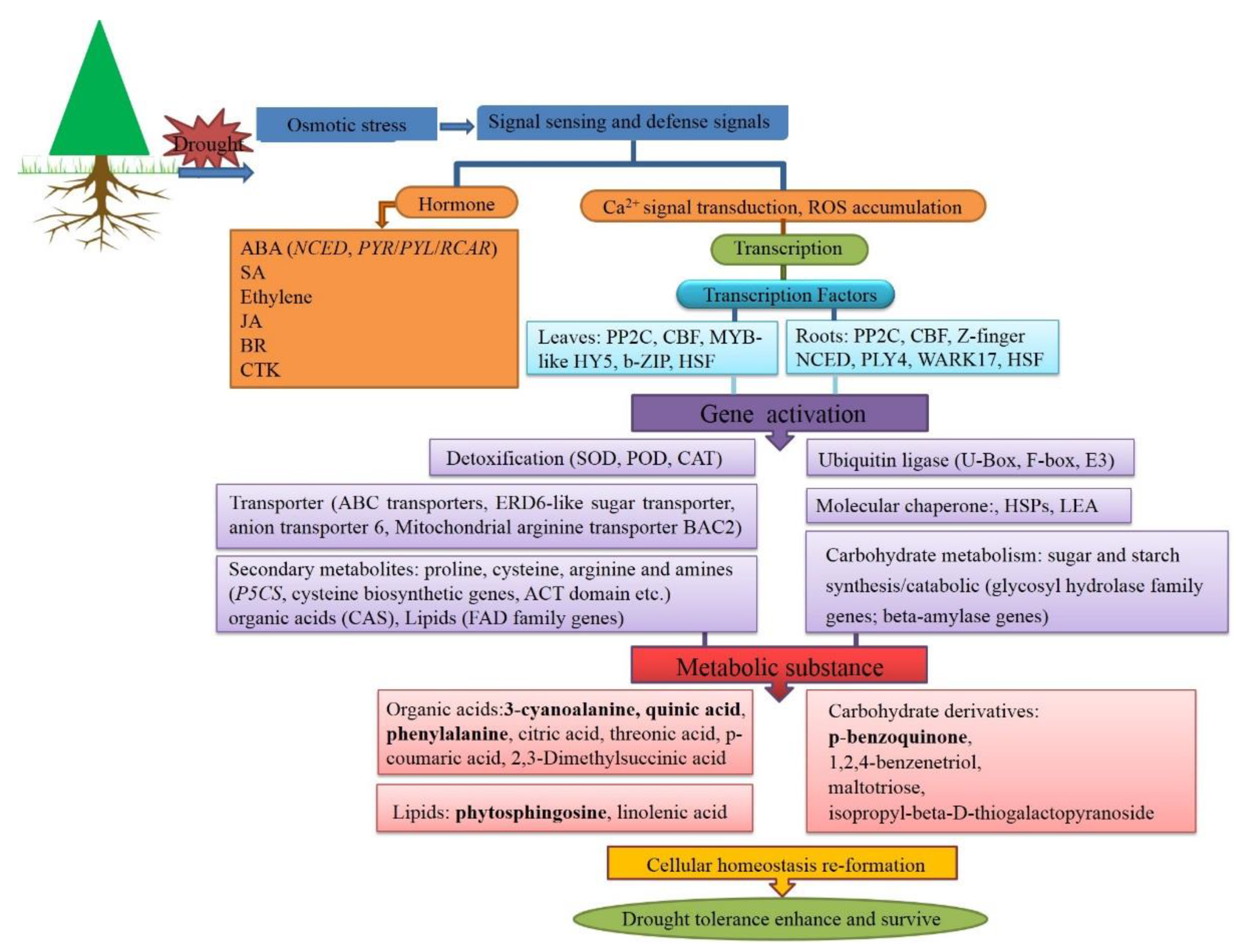
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DT | Drought-tolerant cherry rootstock |
| DS | Drought-susceptible cherry rootstock |
| TCA cycle | Tricarboxylic acid cycle |
| DEGs | Differentially expressed genes |
| PPI | Protein−Protein Interaction |
| ABA | Abscisic acid |
| SG | Stress group |
| CG | Control group |
| LWC | Relative water content |
| FPKM | Fragments Per Kilobase of transcript per Million mapped fragments |
| GO | Gene Ontology |
| KOG/COG | euKaryotic 87tssOrthologous Groups/Clusters of Orthologous Groups |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| qRT-PCR | Quantitative real-time PCR |
| GC-MS | Gas chromatography-mass spectrometer |
| RI | Retention time index |
| QC | Quality control |
| PCA | Principal component analysis |
| OPLS-DA | Orthogonal projections to latent structures-discriminant analysis |
| VIP | Variable importance in the projection |
| CDS | Coding sequence |
| TFs | Transcription factors |
References
- Chaves, M.M.; Maroco, J.P.; Pereira, J.S. Understanding plant responses to drought—From genes to the whole plant. Funct. Plant Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef]
- Kucukyumuk, Z.; Erbas, S.; Erdal, I.; Baydar, H.; Eraslan, F. Effect of different nitrogen doses on plant growth, quality characteristics and nutrient concentrations of lavandin (Lavandula×intermedia Emeric ex Loisel. var. Super A). J. Essent. Oil Bear. Pl. 2015, 18, 36–43. [Google Scholar] [CrossRef]
- Sivritepe, N.; Erturk, U.; Yerlikaya, C.; Turkan, I.; Bor, M.; Ozdemir, F. Response of the cherry rootstock to water stress induced in vitro. Biol. Plant. 2008, 52, 573–576. [Google Scholar] [CrossRef]
- Cai, Y.; Zhao, X.; Hrotkó, K. Development of cherry growing in Shaanxi province of PR China. Acta Hortic. 2019, 1235, 239–244. [Google Scholar] [CrossRef]
- Morandi, B.; Manfrini, L.; Lugli, S.; Tugnoli, A.; Boini, A.; Perulli, G.D.; Bresilla, K.; Venturi, M.; Corelli Grappadelli, L.J. Sweet cherry water relations and fruit production efficiency are affected by rootstock vigor. J. Plant Phys. 2019, 237, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Ranney, T.G.; Bassuk, N.L.; Whitlow, T.H. Osmotic adjustment and solute constituents in leaves and roots of water-stressed cherry (Prunus) trees. J. Am. Soc. Hortic. Sci. 1991, 116, 684–688. [Google Scholar] [CrossRef]
- Guo, Y.; Yu, H.; Kong, D.; Yan, F.; Liu, D.; Zhang, Y. Effects of gradual soil drought stress on the growth, biomass partitioning, and chlorophyll fluorescence of Prunus mongolica seedlings. Turk. J. Biol. 2015, 39, 532–539. [Google Scholar] [CrossRef] [Green Version]
- Arndt, S.K.; Wanek, W.; Clifford, S.C.; Popp, M. Contrasting adaptations to drought stress in field-grown Ziziphus mauritiana and Prunus persica trees: Water relations, osmotic adjustment and carbon isotope composition. Funct. Plant Biol. 2000, 27, 985–996. [Google Scholar] [CrossRef]
- Kalefetoğlu, T.; Ekmekçi, Y. The effects of drought on plants and tolerance mechanisms. Gazi Univ. J. Sci. 2005, 18, 723–740.10. [Google Scholar]
- Jiménez, S.; Dridi, J.; Gutiérrez, D.; Moret, D.; Irigoyen, J.J.; Moreno, M.A.; Gogorcena, Y. Physiological, biochemical and molecular responses in four Prunus rootstocks submitted to drought stress. Tree Physiol. 2013, 33, 1061–1075. [Google Scholar] [CrossRef]
- Lang, G.A. Precocious, dwarfing, and productive—How will new cherry rootstocks impact the sweet cherry industry? HortTechnology 2000, 10, 719–725. [Google Scholar] [CrossRef]
- Hrotko, K. Potentials in Prunus mahaleb L. for cherry rootstock breeding. Sci. Hortic. 2016, 205, 70–78. [Google Scholar] [CrossRef]
- Faust, M.; Surányi, D. Origin and dissemination of cherry. Hort. Rev. 1997, 19, 263–317. [Google Scholar] [CrossRef]
- Hrotko, K. Progress in cherry rootstock research. Acta Hortic. 2008, 795, 171–178. [Google Scholar] [CrossRef]
- Bagati, S.; Mahajan, R.; Nazir, M.; Dar, A.A.; Zargar, S.M. Omics: A gateway towards abiotic stress tolerance. In Abiotic Stress-Mediated Sensing and Signaling In plants: An Omics Perspective; Springer: Singapore, 2018; pp. 1–45. [Google Scholar]
- Arbona, V.; Manzi, M.; Ollas, C.; Gómez-Cadenas, A. Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 4885–4911. [Google Scholar] [CrossRef]
- Park, C.H.; Yeo, H.J.; Park, Y.E.; Baek, S.-A.; Kim, J.K.; Park, S.U. Transcriptome analysis and metabolic profiling of Lycoris radiata. Biology 2019, 8, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Rong, Z.; Bai, S.; Gao, X.; Min, L.; Wei, Y.; Chan, Z. Mongolian almond (Prunus mongolica Maxim): The morpho-physiological, biochemical and transcriptomic response to drought stress. PLoS ONE 2015, 10, e0124442. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.; Ma, K.; Lu, Z.; Lu, J.; Cui, J.; Wang, L.; Jin, B. Physiological, transcriptomic, and metabolic responses of Ginkgo biloba L. to drought, salt, and heat stresses. Biomolecules 2020, 10, 1635. [Google Scholar] [CrossRef] [PubMed]
- Campos, C.; Nobre, T.; Goss, M.J.; Faria, J.; Barrulas, P.; Carvalho, M. Transcriptome analysis of wheat roots reveals a differential regulation of stress responses related to arbuscular mycorrhizal fungi and soil disturbance. Biology 2019, 8, 93. [Google Scholar] [CrossRef] [Green Version]
- You, J.; Zhang, Y.; Liu, A.; Li, D.; Wang, X.; Dossa, K.; Zhou, R.; Yu, J.; Zhang, Y.; Wang, L.; et al. Transcriptomic and metabolomic profiling of drought-tolerant and susceptible sesame genotypes in response to drought stress. BMC Plant Biol. 2019, 19. [Google Scholar] [CrossRef] [Green Version]
- Pan, L.; Meng, C.; Wang, J.; Ma, X.; Fan, X.; Yang, Z.; Zhou, M.; Zhang, X. Integrated omics data of two annual ryegrass (Lolium multiflorum L.) genotypes reveals core metabolic processes under drought stress. BMC Plant Biol. 2018, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glaubitz, U.; Li, X.; Schaedel, S.; Erban, A.; Sulpice, R.; Kopka, J.; Hincha, D.K.; Zuther, E. Integrated analysis of rice transcriptomic and metabolomic responses to elevated night temperatures identifies sensitivity- and tolerance-related profiles. Plant Cell Environ. 2017, 40, 121–137. [Google Scholar] [CrossRef] [Green Version]
- Wedow, J.M.; Yendrek, C.R.; Mello, T.R.; Creste, S.; Martinez, C.A.; Ainsworth, E.A. Metabolite and transcript profiling of Guinea grass (Panicum maximum Jacq) response to elevated [CO2] and temperature. Metabolomics 2019, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirai, M.Y.; Yano, M.; Goodenowe, D.B.; Kanaya, S.; Kimura, T.; Awazuhara, M.; Arita, M.; Fujiwara, T.; Saito, K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. P. Natl. A Sci. India. B. 2004, 101, 10205–10210. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.; Li, Z.; Dai, S.; Ding, H.; Wang, Q.; Li, X.; Ding, G.; Wang, P.; Guan, Y.; Liu, W. Integrative analyses of transcriptomics and metabolomics upon seed germination of foxtail millet in response to salinity. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Yan, Z.; Zuo, J.; Zhou, F.; Shi, J.; Xu, D.; Hu, W.; Jiang, A.; Liu, Y.; Wang, Q. Integrated analysis of transcriptomic and metabolomic data reveals the mechanism by which LED light irradiation extends the postharvest quality of pak-choi (Brassica campestris L. ssp. chinensis (L.) Makino var. communis Tsen et Lee). Biomolecules 2020, 10, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pascual, J.; Cañal, M.J.; Escandón, M.; Meijón, M.; Weckwerth, W.; Valledor, L. Integrated physiological, proteomic, and metabolomic analysis of UV stress responses and adaptation mechanisms in Pinus radiata. Mol. Cell. Proteomics 2017, 16, 485–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirasawa, K.; Isuzugawa, K.; Ikenaga, M.; Saito, Y.; Yamamoto, T.; Hirakawa, H.; Isobe, S. The genome sequence of sweet cherry (Prunus avium) for use in genomics-assisted breeding. DNA Res. 2017, 24, 499–508. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Nat. Prec. 2010. [Google Scholar] [CrossRef]
- Feng, Y.; Liang, C.; Li, B.; Wan, T.; Liu, T.; Cai, Y. Differential expression profiles and pathways of genes in drought resistant tree species Prunus mahaleb roots and leaves in response to drought stress. Sci. Hortic. 2017, 226, 75–84. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Kumar, V.; Kohli, S.K.; Sidhu, G.P.S.; Bali, A.S.; Handa, N.; Kapoor, D.; Bhardwaj, R.; Zheng, B. Phytohormones regulate accumulation of osmolytes under abiotic stress. Biomolecules 2019, 9, 285. [Google Scholar] [CrossRef] [Green Version]
- Santisree, P.; Bhatnagar-Mathur, P.; Sharma, K.K. NO to drought-multifunctional role of nitric oxide in plant drought: Do we have all the answers? Plant Sci. (Amsterdam, Neth.) 2015, 239, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.R.; Chaitanya, K.V.; Vivekanandan, M. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J. Plant Physiol. 2004, 161, 1189–1202. [Google Scholar] [CrossRef]
- Rathinasabapathi, B. Metabolic engineering for stress tolerance: Installing osmoprotectant synthesis pathways. Ann. Bot. 2000, 86, 709–716. [Google Scholar] [CrossRef]
- Hura, T.; Grzesiak, S.; Hura, K.; Thiemt, E.; Tokarz, K.; Wedzony, M. Physiological and biochemical tools useful in drought-tolerance detection in genotypes of winter triticale: Accumulation of ferulic acid correlates with drought tolerance. Ann. Bot. 2007, 100, 767–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Xia, H.; Liu, Y.; Wei, H.; Zheng, X.; Song, C.; Chen, L.; Liu, H.; Luo, L. Transcriptomic and metabolomic studies disclose key metabolism pathways contributing to well-maintained photosynthesis under the drought and the consequent drought-tolerance in rice. Front. Plant 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, M.; Kumar, J.; Singh, S.; Singh, V.P.; Prasad, S.M. Roles of osmoprotectants in improving salinity and drought tolerance in plants: A review. Rev. Environ. Sci. Bio. 2015, 14, 407–426. [Google Scholar] [CrossRef]
- Bohnert, H.J.; Jensen, R.G. Strategies for engineering water-stress tolerance in plants. Trends Biotechnol. 1996, 14, 89–97. [Google Scholar] [CrossRef]
- Ashraf, M.; Foolad, M.R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Sieciechowicz, K.A.; Joy, K.W.; Ireland, R.J. The metabolism of asparagine in plants. Phytochemistry 1988, 27, 663–671. [Google Scholar] [CrossRef]
- Lea, P.J.; Sodek, L.; Parry, M.A.J.; Shewry, P.R.; Halford, N.G. Asparagine in plants. Ann. Appl. Biol. 2007, 150, 1–26. [Google Scholar] [CrossRef]
- Toumi, I.; Gargouri, M.; Nouairi, I.; Moschou, P.N.; Salem-Fnayou, A.B.; Mliki, A.; Zarrouk, M.; Ghorbel, A. Water stress induced changes in the leaf lipid composition of four grapevine genotypes with different drought tolerance. Biol. Plant. 2008, 52, 161–164. [Google Scholar] [CrossRef]
- Scotti-Campos, P.; Pham-Thi, A.-T.; Semedo, J.; Pais, I.; Ramalho, J.; Matos, M. Physiological responses and membrane integrity in three Vigna genotypes with contrasting drought tolerance. Emir. J. Food Agric. 2013, 25, 1002. [Google Scholar] [CrossRef] [Green Version]
- Scotti-Campos, P.; Pais, I.P.; Partelli, F.L.; Batista-Santos, P.; Ramalho, J.C. Phospholipids profile in chloroplasts of Coffea spp. genotypes differing in cold acclimation ability. J. Plant Physiol. 2014, 171, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Partelli, F.L.; Vieira, H.D.; Pais, I.P.; Quartin, V.L.; Ramalho, J.C. Chloroplast membrane lipids from Coffea sp. under low positive temperatures. In Proceedings of the 22nd International Conference on Coffee Science, Campinas, Brazil, 14–19 September 2009; pp. 891–898. [Google Scholar]
- Scotti-Campos, P.; Pham-Thi, A.-T. Correlation between total lipids, linolenic acid and membrane injury under PEG-induced dehydration in leaves of Vigna genotypes differing in drought resistance. Emir. J. Food Agric. 2016, 28, 485. [Google Scholar] [CrossRef]
- Xu, F.; Zhang, D.; Zhu, F.; Tang, H.; Lv, X.; Cheng, J.; Xie, H.; Lin, H. A novel role for cyanide in the control of cucumber (Cucumis sativus L.) seedlings response to environmental stress. Plant Cell Environ. 2012, 35, 1983–1997. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, B.; Preston, G.M.; Sweetlove, L.J. Increased β-cyanoalanine nitrilase activity improves cyanide tolerance and assimilation in Arabidopsis. Mol. Plant 2014, 7, 231–243. [Google Scholar] [CrossRef]
- Yip, W.-K.; Yang, S.F. Cyanide metabolism in relation to ethylene production in plant tissues. Plant Physiol. 1988, 88, 473–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Chen, G.; Zhao, P.; Zhou, Q.; Zhao, X. The abundance of certain metabolites responds to drought stress in the highly drought tolerant plant Caragana korshinskii. Acta Physiol. Plant. 2017, 39, 116. [Google Scholar] [CrossRef]
- Barnaby, J.Y.; Fleisher, D.H.; Singh, S.K.; Sicher, R.C.; Reddy, V.R. Combined effects of drought and CO2 enrichment on foliar metabolites of potato (Solanum tuberosum L.) cultivars. J. Plant Interact. 2019, 14, 110–118. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.; Bano, A.; Rahman, M.A.; Rathinasabapathi, B.; Babar, M.A. UPLC-HRMS-based untargeted metabolic profiling reveals changes in chickpea (Cicer arietinum) metabolome following long-term drought stress. Plant Cell Environ. 2019, 42, 115–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabella, E.; Luvisi, A.; Aprile, A.; Negro, C.; Vergine, M.; Nicolì, F.; Miceli, A.; De Bellis, L. Xylella fastidiosa induces differential expression of lignification related-genes and lignin accumulation in tolerant olive trees cv. Leccino. J. Plant Physiol. 2018, 220, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Peltier, J.P.; Marigo, D.; Marigo, G. Involvement of malate and mannitol in the diurnal regulation of the water status in members of Oleaceae. Trees 1997, 12, 27–34. [Google Scholar] [CrossRef]
- Rahmati, M.; Vercambre, G.; Davarynejad, G.; Bannayan, M.; Azizi, M.; Génard, M. Water scarcity conditions affect peach fruit size and polyphenol contents more severely than other fruit quality traits. J. Sci. Food Agric. 2015, 95, 1055–1065. [Google Scholar] [CrossRef] [PubMed]
- Larher, F.R.; Lugan, R.; Gagneul, D.; Guyot, S.; Monnier, C.; Lespinasse, Y.; Bouchereau, A. A reassessment of the prevalent organic solutes constitutively accumulated and potentially involved in osmotic adjustment in pear leaves. Environ. Exp. Bot. 2009, 66, 230–241. [Google Scholar] [CrossRef]
- Gargallo-Garriga, A.; Sardans, J.; Pérez-Trujillo, M.; Oravec, M.; Urban, O.; Jentsch, A.; Kreyling, J.; Beierkuhnlein, C.; Parella, T.; Peñuelas, J. Warming differentially influences the effects of drought on stoichiometry and metabolomics in shoots and roots. New Phytol. 2015, 207, 591–603. [Google Scholar] [CrossRef] [PubMed]
- Passarinho, J.A.P.; Lamosa, P.; Baeta, J.P.; Santos, H.; Ricardo, C.P.P. Annual changes in the concentration of minerals and organic compounds of Quercus suber leaves. Physiol. Plant 2006, 127, 100–110. [Google Scholar] [CrossRef]
- Kamzolova, S.V.; Vinokurova, N.G.; Dedyukhina, E.G.; Samoilenko, V.A.; Lunina, J.N.; Mironov, A.A.; Allayarov, R.K.; Morgunov, I.G. The peculiarities of succinic acid production from rapeseed oil by Yarrowia lipolytica yeast. Appl. Microbiol. Biotechnol. 2014, 98, 4149–4157. [Google Scholar] [CrossRef]
- Morgunov, I.G.; Kamzolova, S.V.; Dedyukhina, E.G.; Chistyakova, T.I.; Lunina, J.N.; Mironov, A.A.; Stepanova, N.N.; Shemshura, O.N.; Vainshtein, M.B. Application of organic acids for plant protection against phytopathogens. Appl. Microbiol. Biotechnol. 2016, 101, 921–932. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, H. Method for Regulating the Growth of Plants. U.S. Patent US3326659A, 20 June 1967. [Google Scholar]
- Chung, N.; Mao, C.; Heitman, J.; Hannun, Y.A.; Obeid, L.M. Phytosphingosine as a specific inhibitor of growth and nutrient import in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 35614–35621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peer, M.; Stegmann, M.; Mueller, M.J.; Waller, F. Pseudomonas syringaeinfection triggers de novo synthesis of phytosphingosine from sphinganine in Arabidopsis thaliana. FEBS Lett. 2010, 584, 4053–4056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaelson, L.V.; Napier, J.A.; Molino, D.; Faure, J.-D. Plant sphingolipids: Their importance in cellular organization and adaption. Biochim. Biophys. Acta 2016, 1861, 1329–1335. [Google Scholar] [CrossRef] [PubMed]
- Niinemets, Ü. Uncovering the hidden facets of drought stress: Secondary metabolites make the difference. Tree Physiol. 2016, 36, 129–132. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Liu, W.; Su, Y.; Cui, J.; Zhang, Z.; Yuan, M.; Zhang, H.; Yuan, S. Different response of photosystem II to short and long-term drought stress in Arabidopsis thaliana. Physiol. Plant 2016, 158, 225–235. [Google Scholar] [CrossRef]
- Pommerrenig, B.; Ludewig, F.; Cvetkovic, J.; Trentmann, O.; Klemens, P.A.; Neuhaus, H.E. In concert: Orchestrated changes in carbohydrate homeostasis are critical for plant abiotic stress tolerance. Plant Cell Physiol. 2018, 59, 1290–1299. [Google Scholar] [CrossRef] [PubMed]
- Janska, A.; Marsík, P.; Zelenková, S.; Ovesná, J. Cold stress and acclimation—What is important for metabolic adjustment? Plant Biol. 2010, 12, 395–405. [Google Scholar] [CrossRef] [PubMed]


| Material | Tissue | Group | Samples for RNA-seq | Samples for GC-MS Tests |
|---|---|---|---|---|
| DT | Leaves | Drought stress | CDR-LS1, CDR-LS2, CDR-LS3 | CDR-LS1, CDR-LS2, CDR-LS3 CDR-LS4, CDR-LS5, CDR-LS6 |
| Control | CDR-LCK1, CDR-LCK2, CDR-LCK3 | CDR-LCK1, CDR-LCK2, CDR-LCK3 CDR-LCK4, CDR-LCK5, CDR-LCK6 | ||
| Root tip tissues | Drought stress | CDR-RS1, CDR-RS2, CDR-RS3 | - | |
| Control | CDR-RCK1, CDR-RCK2, CDR-RCK3 | - | ||
| DS | Leaves | Drought stress | G5-LS1, G5-LS2, G5-LS3 | G5-LS1, G5-LS2, G5-LS3 G5-LS4, G5-LS5, G5-LS6 |
| Control | G5-LCK1, G5-LCK2, G5-LCK3 | G5-LCK1, G5-LCK2, G5-LCK3 G5-LCK4, G5-LCK5, G5-LCK6 | ||
| Root tip tissues | Drought stress | G5-RS1, G5-RS2, G5-RS3 | - | |
| Control | G5-RCK1, G5-RCK2, G5-RCK3 | - |
| Metabolite | Mean CCL | Mean CSL | FC | VIP | p | Mean GCL | Mean GSL | FC | VIP | p |
|---|---|---|---|---|---|---|---|---|---|---|
| Proline | 0.8516 | 0.9086 | 1.1 | 1.034 | 0.048 | 0.2199 | 0.4656 | 2.1 | 1.096 | 0.042 |
| Asparagine | 0.0321 | 0.2258 | 7.0 | 1.141 | 0.049 | 0.1021 | 0.2609 | 2.6 | 1.273 | 0.022 |
| Quinic acid | 0.0005 | 0.0985 | 182.6 | 2.381 | 0.031 | 0.1715 | 0.2519 | 1.5 | 1.282 | 0.048 |
| Purine riboside | 0.0196 | 0.0785 | 4.0 | 1.228 | 0.039 | 0.1414 | 0.1594 | 1.1 | 1.333 | 0.028 |
| Glutamine | 0.0349 | 0.1036 | 3.0 | 1.416 | 0.014 | 0.0111 | 0.0917 | 8.3 | 1.085 | 0.010 |
| 3-cyanoalanine | 8.5523 × 10−9 | 0.0352 | 4,111,229.0 | 2.902 | 0.012 | 0.0141 | 0.0292 | 2.1 | 1.032 | 0.028 |
| L-allothreonine | 0.0130 | 0.0245 | 1.9 | 1.358 | 0.014 | 0.0114 | 0.0206 | 1.8 | 1.233 | 0.026 |
| Phenylalanine | 0.0110 | 0.0146 | 1.3 | 1.165 | 0.040 | 0.0090 | 0.0109 | 1.2 | 1.080 | 0.049 |
| Fucose | 0.0045 | 0.0075 | 1.7 | 1.519 | 0.027 | 0.0049 | 0.0079 | 1.6 | 1.567 | 0.046 |
| Salicylic acid | 0.0037 | 0.0059 | 1.6 | 1.000 | 0.027 | 0.0040 | 0.0074 | 1.8 | 1.264 | 0.008 |
| Sulfuric acid | 0.0035 | 0.0056 | 1.6 | 1.000 | 0.032 | 0.0028 | 0.0060 | 2.2 | 1.081 | 0.006 |
| Phytosphingosine | 0.0016 | 0.0118 | 7.3 | 1.295 | 0.044 | 0.0216 | 0.0359 | 1.7 | 1.085 | 0.032 |
| Biotin | 0.0013 | 0.0002 | 0.2 | 1.032 | 0.047 | 0.0001 | 0.0002 | 2.0 | 1.021 | 0.048 |
| D-glucoheptose | 0.0016 | 0.0033 | 2.1 | 1.565 | 0.031 | 0.0010 | 0.0042 | 4.4 | 1.256 | 0.000 |
| Gene ID | Putative Function | Number of Amino Acids | Theoretical pI | Putative Subcellular Localization |
|---|---|---|---|---|
| Pav_sc0006061.1_g110.1.mk | Beta-amylase 3, chloroplastic [Prunus avium] | 547 | 9.01 | Chloroplasts, nucleus, mitochondria |
| Pav_sc0005750.1_g010.1.br | Beta-glucosidase 11-like | 508 | 6.25 | Vacuole, chloroplasts, extracellular membrane |
| Pav_sc0000004.1_g040.1.mk | ABC transporter G family member 22-like isoform X3 [Prunus avium] | 603 | 8.23 | Plasma membrane, vacuole |
| Pav_sc0004467.1_g120.1.mk | Sugar transporter ERD6-like 16 | 376 | 6.84 | Plasma membrane, nucleus |
| Pav_sc0000491.1_g270.1.mk | Stachyose synthase-like, raffinose synthase or seed imbibition protein | 867 | 5.66 | Chloroplasts, cytoplasm, and nucleus |
| Pav_sc0001335.1_g550.1.mk | RNA polymerase sigma factor sigE, chloroplastic/mitochondrial [Prunus avium] | 550 | 10.00 | Nucleus, chloroplasts, nucleus, mitochondria |
| Pav_sc0002893.1_g310.1.mk | Alpha-glucan water dikinase, chloroplastic isoform X1 [Glycine max] | 1468 | 6.40 | Chloroplasts |
| Pav_sc0001335.1_g050.1.mk | Probable protein phosphatase 2C 51 | 395 | 7.62 | Chloroplasts, nucleus, and cytoplasm |
| Pav_sc0001405.1_g970.1.mk | Temperature-induced lipocalin-1 [Prunus avium] | 185 | 5.97 | Nucleus and cytoplasm |
| Pav_sc0002493.1_g100.1.mk | Early light-induced protein 1, chloroplastic-like [Prunus avium] | 94 | 10.16 | Chloroplasts, nucleus |
| Pav_sc0000138.1_g610.1.mk | Two-component response regulator-like APRR5 | 691 | 5.96 | Nucleus |
| Pav_sc0001305.1_g820.1.mk | Late embryogenesis abundant protein 1-like | 164 | 9.40 | Mitochondria, chloroplasts, cytoplasm |
| Pav_sc0000719.1_g800.1.mk | E3 ubiquitin-protein ligase At4g11680 isoform X1 | 330 | 8.94 | Plasma membrane, vacuole, endoplasmic reticulum, and Golgi apparatus |
| Pav_sc0000464.1_g820.1.mk | U-box domain-containing protein 4 | 383 | 6.03 | Nucleus, cytoplasm |
| Pav_sc0000067.1_g380.1.mk | F-box/LRR-repeat MAX2 homolog A-like | 654 | 5.86 | Nucleus, cytoplasm |
| Pav_sc0000586.1_g780.1.mk | Homeobox-leucine zipper protein ATHB-12-like | 322 | 4.58 | Nucleus |
| Pav_sc0004290.1_g270.1.mk | Zinc finger CCCH domain-containing protein 23-like [Prunus avium] | 111 | 4.60 | Nucleus, chloroplasts, extracellular membrane |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, T.; Feng, Y.; Liang, C.; Pan, L.; He, L.; Cai, Y. Metabolomics and Transcriptomics Analyses of Two Contrasting Cherry Rootstocks in Response to Drought Stress. Biology 2021, 10, 201. https://doi.org/10.3390/biology10030201
Wan T, Feng Y, Liang C, Pan L, He L, Cai Y. Metabolomics and Transcriptomics Analyses of Two Contrasting Cherry Rootstocks in Response to Drought Stress. Biology. 2021; 10(3):201. https://doi.org/10.3390/biology10030201
Chicago/Turabian StyleWan, Tian, Ying Feng, Chenglin Liang, Liuyi Pan, Ling He, and Yuliang Cai. 2021. "Metabolomics and Transcriptomics Analyses of Two Contrasting Cherry Rootstocks in Response to Drought Stress" Biology 10, no. 3: 201. https://doi.org/10.3390/biology10030201






