A Current Update on the Distribution, Morphological Features, and Genetic Identity of the Southeast Asian Mahseers, Tor Species
Abstract
:Simple Summary
Abstract
1. Introduction
2. The Genus Tor
3. Geographical Distribution of Tor Species in the SE Asian Region
3.1. Malaysia
3.2. Thailand
3.3. Vietnam
3.4. Myanmar
3.5. Lao PDR
3.6. Cambodia
3.7. Indonesia
3.8. Brunei, Singapore, Timor-Leste, and the Philippines
4. The Morphological Features of SE Asia’s Tor Species
4.1. T. ater (Robert, 1999)
4.2. T. dongnaiensis (Hoang et al., 2015)
4.3. T. douronensis (Valenciennes, 1842)
4.4. T. laterivittatus (Zhou & Cui, 1996)
4.5. T. mekongensis (Hoang et al., 2015)
4.6. T. mosal (Hamilton, 1822)
4.7. T. putitora (Hamilton, 1822)
4.8. T. sinensis (Wu, 1977)
4.9. T. tambra (Valenciennes, 1842)
4.10. T. tambroides (Bleeker, 1854)
4.11. T. tor (Hamilton, 1822)
4.12. T. yingjiangensis (Chen and Yang, 2004)
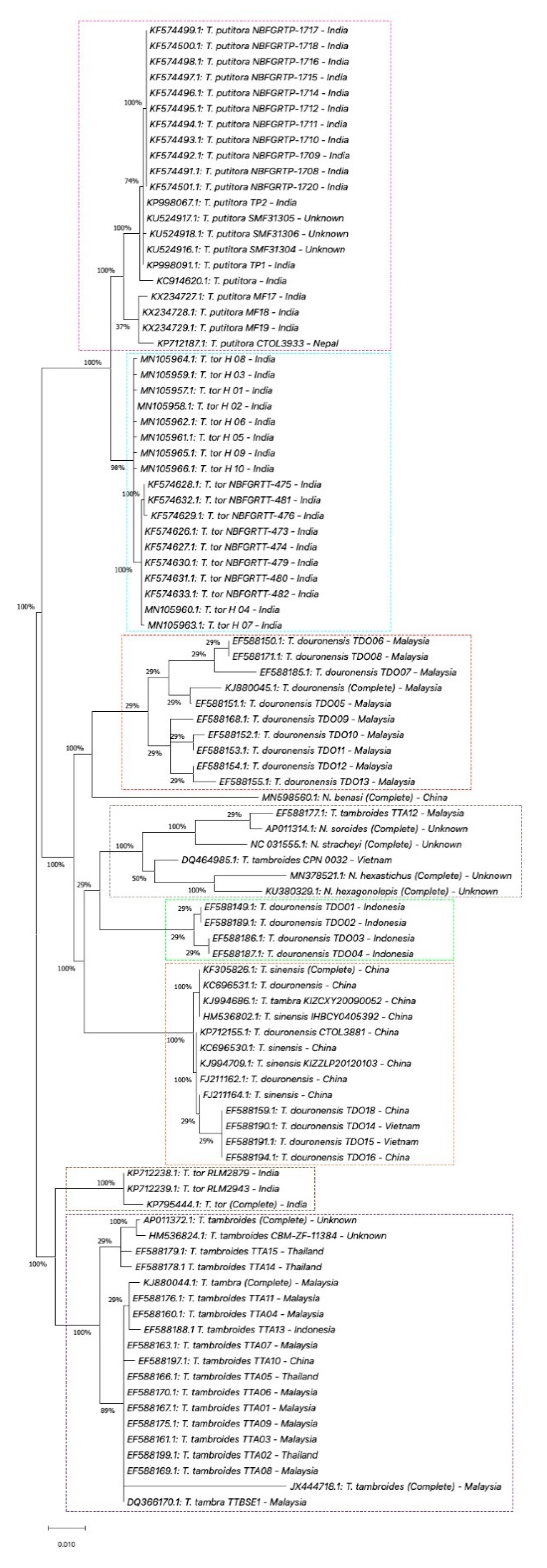
5. Genetic Identification of SE Asia’s Tor Species
5.1. Mitochondrial DNA
5.1.1. Cytochrome c Oxidase Subunit I (COX1) Gene
5.1.2. Cytochrome b (Cyt b) and ATPase 6/8 Gene
5.1.3. 16S rRNA Gene
5.2. Microsatellite DNA Marker
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Nguyen, T.T.; Na-Nakorn, U.; Sukmanomon, S.; ZiMing, C. A study on phylogeny and biogeography of mahseer species (Pisces: Cyprinidae) using sequences of three mitochondrial DNA gene regions. Mol. Phylogenet. Evol. 2008, 3, 1223–1231. [Google Scholar] [CrossRef]
- Ng, C.K. Kings of the Rivers: Mahseer in Malaysia and the Region; Inter Sea Fishery: Selangor, Malaysia, 2004. [Google Scholar]
- Pinder, A.C.; Britton, J.R.; Harrison, A.J.; Nautiyal, P.; Bower, S.D.; Cooke, S.J.; Lockett, S.; Everard, M.; Katwate, U.; Ranjeet, K.; et al. Mahseer (Tor spp.) fishes of the world: Status, challenges and opportunities for conservation. Rev. Fish Biol. Fish. 2019, 29, 417–452. [Google Scholar] [CrossRef] [Green Version]
- FAO. The State of World Fisheries and Aquaculture 2020. In brief. Sustainability in Action; FAO: Rome, Italy, 2020. [Google Scholar]
- Pinder, A.C.; Raghavan, R. Conserving the endangered mahseers (Tor spp.) of India: The positive role of recreational fisheries. Curr. Sci. 2013, 104, 1472–1475. [Google Scholar]
- Sungan, S.; Tinggi, D.; Salam, N.; Sadi, C. Aspects of the biology and ecology of empurau (Tor tambroides) and semah (T. douronensis) in Sarawak, Malaysia. In Mahseer 2006, Proceedings of the International Symposium on the Mahseer, Kuala Lumpur, Malaysia, 29–30 March 2006; Siraj, S.S., Christianus, A., Ng, C.K., Silva, S.S.D., Eds.; Malaysian Fisheries Society: Kuala Lumpur, Malaysia, 2006; p. 3. [Google Scholar]
- Jha, B.; Rayamajhi, A.; Dahanukar, N.; Harrison, A.; Pinder, A.C. Tor putitora. The IUCN Red List of Threatened Species. 2018. Available online: https://dx.doi.org/10.2305/IUCN.UK.2018-2.RLTS.T126319882A126322226.en (accessed on 13 February 2021).
- Azuadi, N.; Siraj, S.; Daud, S.; Christianus, A.; Harmin, S.; Sungan, S.; Britin, R. Enhancing ovulation of Malaysian mahseer (Tor tambroides) in captivity by removal of dopaminergic inhibition. J. Fish. Aquat. Sci 2011, 6, 740–750. [Google Scholar] [CrossRef] [Green Version]
- Childress, E.S.; McIntyre, P.B. Life history traits and spawning behavior modulate ecosystem-level effects of nutrient subsidies from fish migrations. Ecosphere 2016, 7, e01301. [Google Scholar] [CrossRef] [Green Version]
- Tamario, C.; Sunde, J.; Petersson, E.; Tibblin, P.; Forsman, A. Ecological and evolutionary consequences of environmental change and management actions for migrating fish: A review. Front. Ecol. Evol. 2019, 7, 271. [Google Scholar] [CrossRef] [Green Version]
- IUCN. The IUCN Red List of Threatened Species. Version 2020-3. Available online: https://www.iucnredlist.org (accessed on 13 February 2021).
- Desai, V. Synopsis of Biological Data on the Tor Mahseer Tor Tor (Hamilton, 1822); Food & Agriculture Organization: Roma, Italy, 2003. [Google Scholar]
- Walton, S.; Gan, H.; Raghavan, R.; Pinder, A.C.; Ahmad, A. Disentangling the taxonomy of the mahseers (Tor spp.) of Malaysia: An integrated approach using morphology, genetics and historical records. Rev. Fish. Sci. Aquac. 2017, 25, 171–183. [Google Scholar] [CrossRef] [Green Version]
- Kottelat, M. The fishes of the Nam Theun and Xe Bangfai drainages, Laos. Hydroécologie Appl. 2016, 19, 271–320. [Google Scholar] [CrossRef] [Green Version]
- Hoang, H.D.; Pham, H.M.; Durand, J.-D.; Tran, N.T.; Phan, P.D. Mahseers genera Tor and Neolissochilus (Teleostei: Cyprinidae) from southern Vietnam. Zootaxa 2015, 4006, 551–568. [Google Scholar] [CrossRef] [Green Version]
- Kottelat, M. Fishes of the Xe Kong Drainage in Laos, Especially from the Xe Kaman; WWF: Gland, Switzerland, 2011; pp. 1–29. [Google Scholar]
- Oo, W. Inland fisheries of the Union of Myanmar; FAO Fisheries Technical Paper; FAO: Rome, Italy, 2002; pp. 71–78. [Google Scholar]
- Shahi, N.; Mallik, S.K.; Sarma, D. Golden mahseer, Tor putitora—A possible candidate species for hill aquaculture. Aquac Asia 2014, 14, 22–28. [Google Scholar]
- Tamba, I.; Muhtadi, A.; Ariyanti, J.; Leidonald, R. Diversity and habitat condition of Tor Fish (Tor spp.) in the upstream of Wampu Waters, North Sumatra, Indonesia. In Proceedings of the International Conference on Agriculture, Environment, and Food Security 2018, Medan, Indonesia, 24–25 October 2018; IOP Publishing: Bristol, UK, 2019; Volume 260, p. 012102. [Google Scholar]
- Arunkumar, L.; Basudha, C. Tor Barakae, a New Species of Mahseer fish (Cyprinidae: Cyprininae) from Manipur, India. Aquacult 2003, 4, 271–276. [Google Scholar]
- Mani, I.; Kumar, R.; Singh, M.; Kushwaha, B.; Nagpure, N.; Srivastava, P.; Murmu, K.; Rao, D.; Lakra, W. Karyotypic diversity and evolution of seven mahseer species (Cyprinidae) from India. J. Fish Biol. 2009, 75, 1079–1091. [Google Scholar] [CrossRef]
- Chen, X.-Y.; Yang, J.-X.; Chen, Y.-R. A review of the cyprinoid fish genus Barbodes Bleeker, 1859, from Yunnan, China, with descriptions of two new species. Zool. Stud. Taipei 1999, 38, 82–88. [Google Scholar]
- Varma, V.; Raghavan, R.; Binoy, V. Big fish in small tanks: Stunting in the Deccan Mahseer, Tor khudree (Sykes 1849). bioRxiv 2020. [Google Scholar] [CrossRef]
- Iqbal, Z.; Pervaiz, K.; Javed, M.N. Population dynamics of Tor macrolepis (Teleostei: Cyprinidae) and other fishes of Attock region, Pakistan. Can. J. Pure Appl. Sci 2013, 7, 2195–2201. [Google Scholar]
- Chandhini, S.; Arjunan, V.; Ali, P.A.; Raghavan, R.; Pillai, D.; Kumar, V.R. Complete mitogenome analysis of endangered Malabar mahseer (Tor malabaricus). Conserv. Genet. Resour. 2019, 11, 185–189. [Google Scholar] [CrossRef]
- Qin, T.; Chen, Z.-Y.; Xu, L.-L.; Zaw, P.; Kyaw, Y.M.M.; Maung, K.W.; Chen, X.-Y. Five newly recorded Cyprinid fish (Teleostei: Cypriniformes) in Myanmar. Zool. Res. 2017, 38, 300. [Google Scholar]
- Sarkar, U.; Mahapatra, B.; Saxena, S.R.; Singh, A. Mahseer in India: An overview on research status and future priorities. J. Ecophysiol. Occup. Health 2015, 15, 45. [Google Scholar]
- ZiMing, C.; JunXing, Y. A new species of the genus Tor from Yunnan, China (Teleostei: Cyprinidae). Environ. Biol. Fishes 2004, 70, 185–191. [Google Scholar] [CrossRef]
- Ingram, B.; Sungan, S.; Gooley, G.; Sim, S.Y.; Tinggi, D.; De Silva, S.S. Induced spawning, larval development and rearing of two indigenous Malaysian mahseer, Tor tambroides and T. douronensis. Aquac. Res. 2005, 36, 983–995. [Google Scholar] [CrossRef]
- Nguyen, T.; Baranski, M.; Rourke, M.; McPARTLAN, H. Characterization of microsatellite DNA markers for a mahseer species, Tor tambroides (Cyprinidae) and cross-amplification in four congeners. Mol. Ecol. Notes 2007, 7, 109–112. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Ingram, B.; Sungan, S.; Gooley, G.; Sim, S.Y.; Tinggi, D.; De Silva, S.S. Mitochondrial DNA diversity of broodstock of two indigenous mahseer species, Tor tambroides and T. douronensis (Cyprinidae) cultured in Sarawak, Malaysia. Aquaculture 2006, 253, 259–269. [Google Scholar] [CrossRef]
- Fricke, R.; Eschmeyer, W.N.; Laan, R.V.D. (Eds.) Eschmeyer’s Catalog of Fishes: Genera, Species, References. 2021. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 2 March 2021).
- Anabella, N.; Subagja, J.; Arifin, O.; Muhiardi, I.; Arief, M. The motility of Tor soro fish (Valenciennes, 1842) using post cryopreservation sperm: The effect of grape juice (Vitis vinifera) as a natural antioxidant. In Proceedings of the International Conference on Fisheries and Marine, North Maluku, Indonesia, 13–14 July 2020; IOP Publishing: Bristol, UK, 2020; Volume 584, p. 012063. [Google Scholar]
- Hamilton, F. An Account of the Fishes Found in the River Ganges and Its Branches; Archibald Constable: Edinburgh, UK, 1822; Volume 1. [Google Scholar]
- Monkolprasit, S.; Sontirat, S.; Vimollohakarn, S.; Songsirikul, T. Checklist of fishes in Thailand; Office of Environmental Policy and Planning: Bangkok, Thailand, 1997. [Google Scholar]
- Roberts, T.R. Fishes of the cyprinid genus Tor in the Nam Theun watershed (Mekong basin) of Laos, with description of a new species. Raffles Bull. Zool. 1999, 47, 225–236. [Google Scholar]
- Wei, Z.; Cui, G.-H. A review of Tor species from the Lancangjiang river (Upper Mekong river), China (Teleostei: Cyprinidae). Ichthyol. Explor. Freshw. 1996, 7, 131–142. [Google Scholar]
- Cuvier, G.b.; Valenciennes, M.A. Histoire Naturelle des Poissons; Nabu Press: Paris, France, 1842; Volume 16, pp. 465–487. [Google Scholar]
- Wu, H.-W.E. The Cyprinid Fishes of China; Technical Printing House: Shanghai, China, 1977; Volume 2, pp. 229–598. [Google Scholar]
- Bleeker, P. Overzigt der Ichtyologische Fauna van Sumatra: Met Beschrijving van Eenige Nieuwe Soorten; Natuurkundig Tijdschrift voor Nederlandsch Indië; National Library of the Netherlands: Hague, The Netherlands, 1854; Volume 7, pp. 9–108. [Google Scholar]
- Abdullah, S.A.; Nakagoshi, N. Changes in landscape spatial pattern in the highly developing state of Selangor, peninsular Malaysia. Landsc. Urban Plan. 2006, 77, 263–275. [Google Scholar] [CrossRef]
- Chowdhury, M.S.U.; Othman, F.; Jaafar, W.Z.W.; Mood, N.C.; Adham, M.I. Assessment of pollution and improvement measure of water quality parameters using scenarios modeling for Sungai Selangor Basin. Sains Malays. 2018, 47, 457–469. [Google Scholar] [CrossRef]
- Esa, Y.B.; Siraj, S.S.; Rahim, K.A.A.; Daud, S.K.; Chong, H.G.; Guan, T.S.; Syukri, M.F. Genetic characterization of two mahseer species (Tor douronensis and Tor tambroides) using microsatellite markers from other cyprinids. Sains Malays. 2011, 40, 1087–1095. [Google Scholar]
- Esa, Y.B.; Siraj, S.S.; Daud, S.K.; Rahim, K.A.A.; Japning, J.R.R.; Tan, S.G. Mitochondrial DNA diversity of Tor tambroides Valenciennes (Cyprinidae) from five natural populations in Malaysia. Zool. Stud. 2008, 47, 360–367. [Google Scholar]
- Ambak, M.A.; Zakaria, M.Z. Freshwater fish diversity in Sungai Kelantan. J. Sustain. Sci. Manag. 2010, 5, 13–20. [Google Scholar]
- Hashim, R.; Azlan, M.R.P.; Zainuddin, M.; Amzar, W.M.; Jusoh, S.A.; Sah, M.; Ruddin, A.S. Fish distribution and composition of Kelantan river systems, Kelantan, Malaysia. Appl. Mech. Mater. 2015, 747, 294–297. [Google Scholar] [CrossRef]
- Alias, M.I.M.; Hambali, K.; Amir, A.; Fauzi, N.; Hassin, H.; Yin, S.A. Checklist of Fishes at Pergau Lake, Jeli, Kelantan, Malaysia. Trop. Life Sci. Res. 2019, 30, 161. [Google Scholar] [CrossRef] [PubMed]
- Asaduzzaman, M.; Ikeda, D.; Kader, M.A.; Kinoshita, S.; Ghaffar, M.A.; Abol-Munafi, A.B. Cellular muscle growth and molecular cloning and expression of growth-related gene of Malaysian Mahseer Tor tambroides larvae fed with live and formulated feeds in indoor nursery rearing system. Aquac. Rep. 2017, 5, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Kottelat, M.; Pinder, A.; Harrison, A. Tor Tambroides. The IUCN Red List of Threatened Species. 2018. Available online: https://dx.doi.org/10.2305/IUCN.UK.2018-2.RLTS.T187939A91076554.en (accessed on 13 February 2021).
- Vidthayanon, C.; Karnasuta, J.; Nabhitabhata, J. Diversity of Freshwater Fishes in Thailand; Office of Environmental Policy and Planning: Bangkok, Thailand, 1997; pp. 61–62. [Google Scholar]
- Nguyen, T.T. Population structure in the highly fragmented range of Tor douronensis (Cyprinidae) in Sarawak, Malaysia revealed by microsatellite DNA markers. Freshw. Biol. 2008, 53, 924–934. [Google Scholar] [CrossRef]
- Kottelat, M. The fishes of the inland waters of Southeast Asia: A catalogue and core bibliography of the fishes known to occur in freshwaters, mangroves and estuaries. Raffles Bull. Zool. 2013, 27, 1–663. [Google Scholar]
- Roesma, D.I.; Chornelia, A.; Mursyid, A.; Kamsi, M. Fish diversity of the Batang Toru River System, South Tapanuli, North Sumatra. Biodiversitas J. Biol. Divers. 2016, 17. [Google Scholar] [CrossRef]
- Herawati, T. Fish Food Habits In Upstream Of Cimanuk, Garut District West-Java. GSJ 2019, 7, 384–393. [Google Scholar]
- Haryono, S.J. Populasi dan habitat ikan tambra, Tor tambroide (Bleeker, 1854) di perairan kawasan Pegunungan Muller Kalimantan Tengah. Biodiversitas 2008, 9, 306–309. [Google Scholar] [CrossRef]
- Akmal, Y.; Rahardjo, M. Morphology of appendicular skeleton of the Thai mahseer’s Tor tambroides (Bleeker, 1854). J. Iktiologi Indones. 2018, 18, 261–274. [Google Scholar] [CrossRef]
- Mohamad, A.; Sharir, S.; Farinordin, F.A.; Zakeyuddin, M.S.; Shukor, A.M.; Samat, A.; Nor, S.M. Length-Weight Relationship and Relative Condition Factor of Tor Tambra in Tembat Reservoir, Terengganu, Peninsular Malaysia: Indication of Environmental Health. In ICDSME 2019: ICDSME 2019 Proceedings of the International Conference on Dam Safety Management and Engineering; Springer: Singapore, 2019; pp. 447–456. [Google Scholar]
- Ahmad, S.A.; Sabullah, M.K.; Shamaan, N.A.; Shukor, M.Y.A.; Jirangon, H.; Khalid, A.; Syed, M.A. Evaluation of acetylcholinesterase source from fish, for detection of carbamate. J. Environ. Biol. 2016, 37, 479. [Google Scholar]
- Kunlapapuk, S.; Kulabtong, S. Breeding, Nursing and Biology of Thai Mahseer (Tor tamboides) in Malaysia: An Overview. J. Agric. Sci. Technol. A 2011, 1, 1214–1216. [Google Scholar]
- Kunlapapuk, S.; Kulabtong, S.; Saipattana, P. Updated checklist of freshwater and brackish fishes of Phetchaburi Basin, Northwest Gulf of Thailand Drainages. Biodivers. J. 2015, 6, 837–842. [Google Scholar]
- Duangjai, E.; Punroob, J. Growth Performance of Mahseer Fish (Tor tambroides) in Different Types of Cultured Environments. Eau Herit. J. Sci. Technol. 2018, 12, 225–235. [Google Scholar]
- Nguyen, T.T.; Davy, F.B.; Rimmer, M.A.; De Silva, S.S. Use and exchange of genetic resources of emerging species for aquaculture and other purposes. Rev. Aquac. 2009, 1, 260–274. [Google Scholar] [CrossRef]
- Sampantamit, T.; Ho, L.; Van Echelpoel, W.; Lachat, C.; Goethals, P. Links and trade-offs between fisheries and environmental protection in relation to the sustainable development goals in Thailand. Water 2020, 12, 399. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, Y.; Lee, H.S.; Trung, B.H.; Tran, H.-D.; Lall, M.K.; Kakar, K.; Xuan, T.D. Impacts of Mainstream Hydropower Dams on Fisheries and Agriculture in Lower Mekong Basin. Sustainability 2020, 12, 2408. [Google Scholar] [CrossRef] [Green Version]
- Chanlabut, U.; Gomontean, B.; Srifa, A. Soil Organic Carbon Stocks across Hydrologic Schemes in Freshwater Wetlands of the Chi River Basin, Northeast Thailand. Wetlands 2020, 40, 377–389. [Google Scholar] [CrossRef]
- Rainboth, W.J. Fishes of the Cambodian Mekong; Food & Agriculture Organization: Roma, Italy, 1996. [Google Scholar]
- Vidthayanon, C.; Pinder, A.C. Tor sinensis. The IUCN Red List of Threatened Species. 2018. Available online: https://dx.doi.org/10.2305/IUCN.UK.2018-2.RLTS.T187891A126322879.en (accessed on 13 February 2021).
- Teerawoot, L.; Chantana, K. Diversity and Distribution of Cyprinid Fishes in Tapi Upstream System. Res. J. Rajamangala Univ. Technol. Thanyaburi 2016, 15, 57–63. (In Thai) [Google Scholar]
- Mai, D.Y.; Nguyen, V.T.; Le, H.Y.; Hua, B.L. Identification of Freshwater Fishes of southern Vietnam. Nha Xuat Ban Khoa Hoc Va Ky Thuat Ha Noi 1992, 73–77. (In Vietnamese) [Google Scholar]
- Phan, P.D. Literature Review for the Development of Guideline for Prioritisation of Barriers to Fish Passage in Irrigation Schemes in Vietnam. Res. Inst. Aquac. 2015, 31. [Google Scholar] [CrossRef]
- Phan, D.P.; Duong, T.P. Some biological characteristics of Tor tambroides (Bleeker, 1954) in Central Highlands, Vietnam. Sci. Technol. J. Agric. Rural Dev. Vietnam 2014, 2, 87–94. [Google Scholar]
- Phonvisay, S. An Introduction to the Fisheries of Lao PDR; Mekong River Commission: Phnom Penh, Cambodia, 2013; Volume 6. [Google Scholar]
- Vilain, C.; Baran, E.; Gallego, G.; Samadee, S. Fish and the nutrition of rural Cambodians. Asian J. Agric. Food Sci. 2016, 4, 26–34. [Google Scholar]
- Rainboth, W.J.; Vidthayanon, C.; Yen, M.D. Fishes of the Greater Mekong Ecosystem with Species List and Photographic Atlas; University of Michigan, Museum of Zoology: Ann Arbor, MI, USA, 2012. [Google Scholar]
- Hanebuth, T.J.; Voris, H.K.; Yokoyama, Y.; Saito, Y.; Okuno, J.I. Formation and fate of sedimentary depocentres on Southeast Asia’s Sunda Shelf over the past sea-level cycle and biogeographic implications. Earth-Sci. Rev. 2011, 104, 92–110. [Google Scholar] [CrossRef]
- De Silva, S.S.; Ingram, B.; Sungan, S.; Tinggi, D.; Gooley, G.; Sim, S.Y. Artificial propagation of the indigenous Tor species, empurau (T. tambroides) and semah (T. douronensis), Sarawak, East Malaysia. Aquac. Asia 2004, 9, 15–20. [Google Scholar]
- Khaironizam, M.; Akaria-Ismail, M.; Armbruster, J.W. Cyprinid fishes of the genus Neolissochilus in Peninsular Malaysia. Zootaxa 2015, 3962, 139–157. [Google Scholar] [CrossRef] [Green Version]
- Timorya, Y.; Abdullah, A.; Batubara, A.; Muchlisin, Z. Conservation and economic status fishes in the Krueng Sabee River, Aceh Jaya District, Aceh Province, Indonesia. In Proceedings of the International Conference on Fisheries, Aquatic, and Environmental Sciences (ICFAES 2018), Banda Aceh, Indonesia; IOP Publishing: Bristol, UK, 2018; Volume 216, p. 012044. [Google Scholar]
- Roesma, D.I.; Tjong, D.H.; Munir, W.; Agesi, A.V.; Chornelia, A. Genetic diversity of Tor douronensis (Pisces: Cyprinidae) in West Sumatra, Indonesia. Biodivers. J. Biol. Divers. 2017, 18, 1018–1025. [Google Scholar] [CrossRef]
- Roberts, T.R. The Freshwater Fishes of Java, as Observed by Kuhl and van Hasselt in 1820-23; Nationaal Natuurhistorisch Museum: Leiden, The Netherlands, 1993. [Google Scholar]
- Abidin, Z.; Marwoto, P.; Iswari, R. The influence of physical and chemical parameters on the extinction of the god fish (Tor duoronensis) species: Case study in Balong Dalem Kuningan district. In Proceedings of the International Conference on Mathematics, Science and Education 2017 (ICMSE2017), Semarang, Indonesia, 18–19 September 2017; IOP Publishing: Bristol, UK, 2018; Volume 983, p. 012201. [Google Scholar]
- Cahyanti, W.; Soelistyowati, D.T.; Carman, O.; Kristanto, A.H. Artificial spawning and larvae performance of three Indonesian mahseer species. Aquac. Aquar. Conserv. Legis. 2019, 12, 280–288. [Google Scholar]
- Sulaiman, Z.; Hui, T.H.; Lim, K.K.P. Annotated checklist of freshwater fishes from Brunei Darussalam, Borneo. Zootaxa 2018, 4379, 24–46. [Google Scholar] [CrossRef]
- Alfred, E.R. The Fresh-Water Fishes of Singapore; EJ Brill: Leiden, The Netherlands, 1966. [Google Scholar]
- Ng, P.K.; Lim, K.K. The diversity and conservation status of fishes in the nature reserves of Singapore. Gard. Bull. Singap. 1997, 49, 245–265. [Google Scholar]
- Karin, B.R.; Das, I.; Jackman, T.R.; Bauer, A.M. Ancient divergence time estimates in Eutropis rugifera support the existence of Pleistocene barriers on the exposed Sunda Shelf. PeerJ 2017, 5, e3762. [Google Scholar] [CrossRef] [Green Version]
- Arifin, O.Z.; Subagja, J.; Hadie, W. Karakterisasi biometrik tiga populasi ikan semah Tor douronensis^ Valenciennes, 1842) dalam mendukung konservasi sumber daya genetik [Biometric characterization three population of semah mahseer Tor douronensis (Valenciennes, 1842) in support to conservation of genetic resources]. J. Iktiologi Indones 2017, 15, 143–154. [Google Scholar]
- Muséum national d’Histoire naturelle, P.F. Tor laterivittatus zhou & cui, 1996,Collection: Ichthyology (IC), Specimen MNHN-IC-2004-0164. Available online: http://coldb.mnhn.fr/catalognumber/mnhn/ic/2004-0164 (accessed on 1 March 2021).
- Khare, P.; Mohindra, V.; Barman, A.S.; Singh, R.K.; Lal, K.K. Molecular evidence to reconcile taxonomic instability in mahseer species (Pisces: Cyprinidae) of India. Org. Divers. Evol. 2014, 14, 307–326. [Google Scholar] [CrossRef]
- Akmal, Y.; Dhamayanti, Y.; Paujiah, E. Osteocranium of Tor tambroides (Cypriniformes: Cyprinidae) from Tangse River, Aceh, Indonesia. Biodivers. J. Biol. Divers. 2020, 21. [Google Scholar] [CrossRef]
- Kottelat, M. Fishes of the Nam Theun and Xe Bangfai basins (Laos), with diagnoses of a new genus and twenty new species (Cyprinidae, Balitoridae, Cobitidae, Coiidae and Eleotrididae). Ichthyol. Explor. Freshw. 1998, 9, 1–198. [Google Scholar]
- Menon, A.K. Taxonomy of mahseer fishes of the genus Tor Gray with description of a new species from the Deccan. J. Bombay Nat. Hist. Soc. 1992, 89, 210–228. [Google Scholar]
- Haryono, H.; Tjakrawidjaja, A.H. Morphological study for identification improvement of Tambra fish (Tor spp.: Cyprinidae) from Indonesia. Biodivers. J. Biol. Divers. 2006, 7, 59–62. [Google Scholar]
- Ningsih, D.T.M.; Hudaidah, S.; Sunarno, M.T.D. EFFECTIVENESS Daphnia sp. WHICH IS PELET FEEDING TO THE GROWTH OF SEMAH’S LARVAE Tor douronensis (Valenciennes, 1842). J. Aquatropica Asia 2020, 5, 24–28. [Google Scholar] [CrossRef]
- JAKA, O.; Azrita, A.; Endiryeni, E. Karakteristik Morfologi ikangaring (Tor douronensis) Berdasarkan truss Morfometrik padatiga Perairan Yang Berbeda di Kabupaten Padangpariaman. Ph.D Thesis, Universitas Bung Hatta, Sumatera Barat, Indonesia, 2020. [Google Scholar]
- Mellisa, S.; Hasri, I.; Agung, W.; Nurfadillah, N.; Putra, D. Induction of gonadal maturation in mahseer fish (Tor douronensis) using the PMSG hormone, antidopamin. In Proceedings of the 2nd ICFAES 2019 International Conference on Fisheries, Aquatic and Environmental Sciences 2019 in Conjunctions with the 6th ASI 2019 Annual Conference of The Asian Society of Ichthyologist 2019, Banda Aceh, Aceh Province, Indonesia, 19–20 June 2019; IOP Publishing: Bristol, UK, 2019; Volume 348, p. 012101. [Google Scholar]
- Rahimi, S.; Nawan, A.; Hasri, I.; Putra, D.; Ichsan, R. Effect of PMSG+ AD hormone variation on the gonadal maturation of Pedih fish (Tor douronensis). In Proceedings of the 2nd ICFAES 2019 International Conference on Fisheries, Aquatic and Environmental Sciences 2019 In Conjunctions with The 6th ASI 2019 Annual Conference of The Asian Society of Ichthyologist 2019, Banda Aceh, Aceh Province, Indonesia, 19–20 June 2019; IOP Publishing: Bristol, UK, 2019; Volume 348, p. 012126. [Google Scholar]
- Rahayu, D.A.; Nugroho, E.D. DNA Barcode Dan Haplotype Network Ikan Lokal Dari Telaga Banyu Biru Kabupaten Pasuruan. In Seminar Nasional Ikan Ke-8; Masyarakat Iktiologi Indonesia: Bogor, Indonesia, 2015; Volume 2, pp. 67–75. [Google Scholar]
- Doi, A. A review of taxonomic studies of cypriniform fishes in Southeast Asia. Jpn. J. Ichthyol. 1997, 44, 1–33. [Google Scholar]
- Laskar, B.A.; Kumar, V.; Kundu, S.; Tyagi, K.; Chandra, K. Taxonomic quest: Validating two mahseer fishes (Actinopterygii: Cyprinidae) through molecular and morphological data from biodiversity hotspots in India. Hydrobiologia 2018, 815, 113–124. [Google Scholar] [CrossRef]
- Bhatt, J.P.; Pandit, M.K. Endangered Golden mahseer Tor putitora Hamilton: A review of natural history. Rev. Fish Biol. Fish. 2016, 26, 25–38. [Google Scholar] [CrossRef]
- Holopainen, I.; Aho, J.; Vornanen, M.; Huuskonen, H. Phenotypic plasticity and predator effects on morphology and physiology of crucian carp in nature and in the laboratory. J. Fish Biol. 1997, 50, 781–798. [Google Scholar] [CrossRef]
- Tave, D. Inbreeding and Brood Stock Management; Food & Agriculture Organization: Roma, Italy, 1999. [Google Scholar]
- Pavan-Kumar, A.; Raman, S.; Koringa, P.G.; Patel, N.; Shah, T.; Singh, R.K.; Krishna, G.; Joshi, C.; Gireesh-Babu, P.; Chaudhari, A. Complete mitochondrial genome of threatened mahseer Tor tor (Hamilton 1822) and its phylogenetic relationship within Cyprinidae family. J. Genet. 2016, 95, 853–863. [Google Scholar] [CrossRef]
- Pinder, A.C.; Manimekalan, A.; Knight, J.M.; Krishnankutty, P.; Britton, J.R.; Philip, S.; Dahanukar, N.; Raghavan, R. Resolving the taxonomic enigma of the iconic game fish, the hump-backed mahseer from the Western Ghats biodiversity hotspot, India. PLoS ONE 2018, 13, e0199328. [Google Scholar] [CrossRef]
- Sati, J.; Sah, S.; Pandey, H.; Ali, S.; Sahoo, P.K.; Pande, V.; Barat, A. Phylogenetic relationship and molecular identification of five Indian Mahseer species using COI sequence. J. Environ. Biol. 2013, 34, 933. [Google Scholar]
- Wibowo, A.; Kaban, S. GENETIC VARIATION OF Tor tambroides (Bleeker, 1854) ALONG BATANG TARUSAN RIVER, WEST SUMATERA: IMPLICATIONS FOR STOCK IDENTIFICATION. Indones. Fish. Res. J. 2015, 21, 61–66. [Google Scholar] [CrossRef] [Green Version]
- Sah, P.; Mandal, S.; Singh, R.K.; Kumar, R.; Pathak, A.; Dutta, N.; Srivastava, J.; Saini, V.P.; Lal, K.K.; Mohindra, V. Genetic structure of natural populations of endangered Tor mahseer, Tor tor (Hamilton, 1822) inferred from two mitochondrial DNA markers. Meta Gene 2020, 23, 100635. [Google Scholar] [CrossRef]
- Yang, L.; Tan, Z.; Wang, D.; Xue, L.; Guan, M.-x.; Huang, T.; Li, R. Species identification through mitochondrial rRNA genetic analysis. Sci. Rep. 2014, 4, 4089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeSalle, R.; Schierwater, B.; Hadrys, H. MtDNA: The small workhorse of evolutionary studies. Front. Biosci. 2017, 22, 873–887. [Google Scholar] [CrossRef] [Green Version]
- Esa, Y.; Rahim, A.; Adha, K. Genetic structure and preliminary findings of cryptic diversity of the Malaysian Mahseer (Tor tambroides Valenciennes: Cyprinidae) inferred from mitochondrial DNA and microsatellite analyses. Biomed Res. Int. 2013, 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norfatimah, M.; Teh, L.; Salleh, M.; Isa, M.M.; SitiAzizah, M. Complete mitochondrial genome of Malaysian Mahseer (Tor tambroides). Gene 2014, 548, 263–269. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021, 49, D10–D17. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. AP011372.1, Tor Tambroides Mitochondrial DNA, Complete Genome, Except for D-loop, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/AP011372.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. KJ880044.1, Tor Tambra Mitochondrion, Complete Genome, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KJ880044.1 (accessed on 30 January 2021).
- Kumar, R.; Goel, C.; Kumari Sahoo, P.; Singh, A.K.; Barat, A. Complete mitochondrial genome organization of Tor tor (Hamilton, 1822). Mitochondrial DNA Part A 2016, 27, 2541–2542. [Google Scholar] [CrossRef] [PubMed]
- Sati, J.; Goel, C.; Kumar, R.; Ali, S.; Patiyal, R.; Singh, V.K.; Sahoo, P.K.; Barat, A. Complete Mitochondrial Genome Organization of Tor Putitora; Taylor & Francis: Abingdon, UK, 2014. [Google Scholar]
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. KJ880045.1, Tor Douronensis Mitochondrion, Complete Genome, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KJ880045.1 (accessed on 30 January 2021).
- Huang, F.; Liu, M.; Ye, C.; Liu, S. The complete mitochondrial genome sequence of Tor sinensis (Cypriniformes, Cyprinidae). Mitochondrial DNA 2015, 26, 712–713. [Google Scholar] [CrossRef]
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. MN378521.1, Neolissochilus Hexastichus Voucher CIFEFGB-SERB-NH Mitochondrion, Complete Genome, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/MN378521.1 (accessed on 30 January 2021).
- Gu, W.; Xu, G.; Huang, T.; Wang, B. The complete mitochondrial genome of Neolissochilus benasi (Cypriniformes: Cyprinidae). Mitochondrial DNA Part B 2020, 5, 463–464. [Google Scholar] [CrossRef]
- Miya, M. Whole Mitochondrial Genome Sequences in Cypriniformes; Unpublished Manuscript; Natural History Museum & Institute: Chiba, Japan, 2009. [Google Scholar]
- Zhou, C.; Yuan, D.; Zhu, C.; Lei, L.; Zhang, C.; Zhou, J.; Gong, J.; Zhu, L.; Li, B.; Wu, Q. The complete mitochondrion genome of the Barbodes hexagonolepis (Cypriniformes, cyprinidae). Mitochondrial DNA Part B 2016, 1, 158–159. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Yang, L.; Mayden, R.L.; Sado, T.; He, S.; Saitoh, K.; Miya, M. Molecular phylogeny of the fishes traditionally referred to Cyprinini sensu stricto (Teleostei: Cypriniformes). Zool. Scr. 2010, 39, 527–550. [Google Scholar] [CrossRef]
- Dahruddin, H.; Hutama, A.; Busson, F.; Sauri, S.; Hanner, R.; Keith, P.; Hadiaty, R.; Hubert, N. Revisiting the ichthyodiversity of Java and Bali through DNA barcodes: Taxonomic coverage, identification accuracy, cryptic diversity and identification of exotic species. Mol. Ecol. Resour. 2017, 17, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Nadiatul, H.; Daud, S.; Siraj, S.; Sungan, S.; Moghaddam, F. Genetic diversity of Malaysian indigenous Mahseer, Tor douronensis in Sarawak river basins as revealed by cytochrome c oxidase I gene sequences. Iran. J. Anim. Biosyst. 2011, 7, 119–127. [Google Scholar]
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. JF810674.1, Tor Douronensis Haplotype Hap01 Cytochrome Oxidase Subunit I (COI) Gene, Partial cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/JF810674.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. JF810675.1, Tor Douronensis Haplotype Hap02 Cytochrome Oxidase Subunit I (COI) Gene, Partial cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/JF810675.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. JF810676.1, Tor Douronensis Haplotype Hap03 Cytochrome Oxidase Subunit I (COI) Gene, Partial cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/JF810676.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No.JF810677.1, Tor Douronensis Haplotype Hap04 Cytochrome Oxidase Subunit I (COI) Gene, Partial cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/JF810677.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. ethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. JF810678.1, Tor Douronensis Haplotype Hap05 Cytochrome Oxidase Subunit I (COI) Gene, Partial cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/JF810678.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. JF810679.1, Tor Douronensis Haplotype Hap06 Cytochrome Oxidase Subunit I (COI) Gene, Partial cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/JF810679.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. JF810680.1, Tor Douronensis Haplotype Hap07 Cytochrome Oxidase Subunit I (COI) Gene, Partial cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/335346757 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. JF810681.1, Tor Douronensis Haplotype Hap08 Cytochrome Oxidase Subunit I (COI) Gene, Partial cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/JF810681.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. KF240784.1, Tor Douronensis Voucher DOFS/MT/7 Cytochrome Oxidase Subunit I (COI) Gene, Partial cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KF240784.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. KF240785.1, Tor Tambroides Voucher DOFS/MT/8 Cytochrome Oxidase Subunit I (COI) Gene, Partial cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KF240785.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. KT001033.1, Tor Tambroides Isolate KLHh29 Cytochrome Oxidase Subunit I (COI) Gene, Partial cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KT001033.1 (accessed on 30 January 2021).
- Sadi, A.; Biun, H. The ichtyofauna of Maliau Basin buzzer zone at Maliau Basin Conservation Area, Sabah, Malaysia. J. Trop. Biol. Conserv. JTBC 2012, 9, 105–113. [Google Scholar]
- Zheng, L.-P.; Yang, J.-X.; Chen, X.-Y. Molecular phylogeny and systematics of the Barbinae (Teleostei: Cyprinidae) in China inferred from mitochondrial DNA sequences. Biochem. Syst. Ecol. 2016, 68, 250–259. [Google Scholar] [CrossRef]
- Borkenhagen, K. Molecular phylogeny of the tribe Torini Karaman, 1971 (Actinopterygii: Cypriniformes) from the Middle East and North Africa. Zootaxa 2017, 4236. [Google Scholar] [CrossRef]
- Esa, Y.B.; Japning, J.R.R.; Rahim, K.A.A.; Siraj, S.; Daud, S.; Tan, S.G.; Sungan, S. Phylogenetic relationships among several freshwater fishes (Family: Cyprinidae) in Malaysia inferred from partial sequencing of the cytochrome b mitochondrial DNA (mtDNA) gene. Pertanika J. Trop. Agric. Sci. 2012, 35, 307–318. [Google Scholar]
- Guo, B.; Tong, C.; He, S. Sox genes evolution in closely related young tetraploid cyprinid fishes and their diploid relative. Gene 2009, 439, 102–112. [Google Scholar] [CrossRef]
- Lal, K.; Mohindra, V.; Singh, R.; Dhawan, S.; Chandra, S.; Gupta, B.; Dwivedi, A.; Jena, J. Molecular Characterization of Indian Freshwater Cyprinids Using Cytochrome C Oxidase I Sequences; National Bureau of Fish Conservation Division, NBFGR: Uttar Pradesh, India, 2014. [Google Scholar]
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. DQ464985.1, Tor Tambroides Voucher CPN 0032 Cytochrome B Gene, Partial Cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/DQ464985.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. KP998091.1, Tor Putitora Isolate TP1 Cytochrome b (cytb) Gene, Partial cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KP998091.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. KP998067.1, Tor Putitora Isolate TP2 Cytochrome b (cytb) Gene, Partial Cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KP998067.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. KX234729.1, Tor Putitora Voucher MF19 Cytochrome b (cytb) Gene, Partial Cds, Mitochondrial. DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KX234729.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. KX234728.1, Tor Putitora Voucher MF18 Cytochrome b (cytb) Gene, Partial Cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KX234728.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. KX234727.1, Tor Putitora Voucher MF17 Cytochrome b (cytb) Gene, Partial Cds, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KX234727.1 (accessed on 30 January 2021).
- Wang, J.; Wu, X.; Chen, Z.; Yue, Z.; Ma, W.; Chen, S.; Xiao, H.; Murphy, R.W.; Zhang, Y.; Zan, R. Molecular phylogeny of European and African Barbus and their West Asian relatives in the Cyprininae (Teleostei: Cypriniformes) and orogenesis of the Qinghai-Tibetan Plateau. Chin. Sci. Bull. 2013, 58, 3738–3746. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Sado, T.; Hirt, M.V.; Pasco-Viel, E.; Arunachalam, M.; Li, J.; Wang, X.; Freyhof, J.; Saitoh, K.; Simons, A.M. Phylogeny and polyploidy: Resolving the classification of cyprinine fishes (Teleostei: Cypriniformes). Mol. Phylogenet. Evol. 2015, 85, 97–116. [Google Scholar] [CrossRef] [PubMed]
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. DQ464914.1, Tor Tambroides Voucher CPN 0032 16S Ribosomal RNA Gene, Partial Sequence, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/DQ464914.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. DQ464925.1, Tor Douronensis Voucher CPN 0030 16S Ribosomal RNA Gene, Partial Sequence, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/DQ464925.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. JX090196.1, Tor Tor Cell-Line TTH 16S Ribosomal RNA Gene, Partial Sequence, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/JX090196.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. KJ869423.1, Tor Putitora Isolate TPF 16S Ribosomal RNA Gene, Partial Sequence, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KJ869423.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. KX925208.1, Tor Sinensis 16S Ribosomal RNA Gene, Partial Sequence, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KX925208.1 (accessed on 30 January 2021).
- Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. MG229064.1, Tor Putitora Isolate TP-01/16S/AJK/2017 16S Ribosomal RNA Gene, Partial Sequence, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/MG229064.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. MG229065.1, Tor Putitora Isolate TP-02/16S/AJK/2017 16S Ribosomal RNA Gene, Partial Sequence, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/MG229065.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. MG229066.1, Tor Putitora Isolate TP-03/16S/AJK/2017 16S Ribosomal RNA Gene, Partial Sequence, Mitochondrial, DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/MG229066.1 (accessed on 30 January 2021).
- Nucleotide_[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]–Accession No. MG229067.1, Tor Putitora Isolate TP-04/16S/AJK/2017 16S Ribosomal RNA Gene, Partial Sequence, Mitochondrial. DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/MG229067.1 (accessed on 30 January 2021).
- Pandey, H.; Barat, A.; Singh, B.; Sahoo, P.K.; Ali, S.; Mahanta, P. Comparative Phylogenetic Study of four Coldwater Fishes (Family Cyprinidae) Based on Targeted 16S RN A Mitochondrial Gene. J. Ecophysiol. Occup. Health 2013, 13, 47. [Google Scholar]
- Yadav, K.; Lakra, W.; Sharma, J.; Goswami, M.; Singh, A. Development and characterization of a cell line TTCF from endangered mahseer Tor tor (Ham.). Fish Physiol. Biochem. 2012, 38, 1035–1045. [Google Scholar] [CrossRef]
- Li, J.; Wang, X.; Kong, X.; Zhao, K.; He, S.; Mayden, R.L. Variation patterns of the mitochondrial 16S rRNA gene with secondary structure constraints and their application to phylogeny of cyprinine fishes (Teleostei: Cypriniformes). Mol. Phylogenet. Evol. 2008, 47, 472–487. [Google Scholar] [CrossRef]
- Kochzius, M.; Seidel, C.; Antoniou, A.; Botla, S.K.; Campo, D.; Cariani, A.; Vazquez, E.G.; Hauschild, J.; Hervet, C.; Hjörleifsdottir, S. Identifying fishes through DNA barcodes and microarrays. PLoS ONE 2010, 5, e12620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; Dewaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arif, I.; Khan, H. Molecular markers for biodiversity analysis of wildlife animals: A brief review. Anim. Biodivers. Conserv. 2009, 32, 9–17. [Google Scholar]
- Patwardhan, A.; Ray, S.; Roy, A. Molecular markers in phylogenetic studies—A review. J. Phylogenet. Evol. Biol. 2014, 2, 1–9. [Google Scholar] [CrossRef]
- Divya, P.; Gopalakrishnan, A.; Basheer, V.; Swaminathan, R.; Mohitha, C.; Joy, L.; Kumar, R.; Manoj, P.; Jena, J. Mitochondrial ATPase 6/8 genes to infer the population genetic structure of silver pomfret fish Pampus argenteus along the Indian waters. Mitochondrial Dna 2015, 26, 189–194. [Google Scholar] [CrossRef] [Green Version]
- Sarri, C.; Stamatis, C.; Sarafidou, T.; Galara, I.; Godosopoulos, V.; Kolovos, M.; Liakou, C.; Tastsoglou, S.; Mamuris, Z. A new set of 16S rRNA universal primers for identification of animal species. Food Control 2014, 43, 35–41. [Google Scholar] [CrossRef]
- Mohanty, M.; Jayasankar, P.; Sahoo, L.; Das, P. A comparative study of COI and 16 S rRNA genes for DNA barcoding of cultivable carps in India. Mitochondrial DNA 2015, 26, 79–87. [Google Scholar] [CrossRef]
- Liu, Z.J.; Cordes, J. DNA marker technologies and their applications in aquaculture genetics. Aquaculture 2004, 238, 1–37. [Google Scholar] [CrossRef]





| No. | Species | Species Status According to Eschmeyer’s Catalog of Fishes as of 1 March 2021 | Species Status According to Pinder et al. (2019) |
|---|---|---|---|
| 1. | Tor ater (Robert, 1999) | Valid | Valid |
| 2. | Tor barakae (Arunkumar and Basudha, 2003) | Valid | Valid |
| 3. | Tor chelynoides (McClelland, 1839) | Not Valid Valid as Naziritor chelynoides (McClelland, 1839) | Not Valid |
| 4. | Tor dongnaiensis (Hoang et al., 2015) | Valid | Valid |
| 5. | Tor douronensis (Valenciennes, 1842) | Not Valid. Valid as T. tambra (Valenciennes, 1842) | Not Valid |
| 6. | Tor hemispinus (Chen and Chu, 1985) | Not Valid Valid as Neolissochilus hemispinus (Chen and Chu, 1985) | Not Valid |
| 7. | Tor khudree (Sykes, 1839) | Valid | Valid |
| 8. | Tor kulkarnii (Menon, 1992) | Valid | Valid |
| 9. | Tor laterivittatus (Zhou and Cui, 1996) | Valid | Valid |
| 10. | Tor macrolepis (Heckel, 1838) | Not Valid Valid as T. putitora (Hamilton, 1822) | Not Valid |
| 11. | Tor malabaricus (Jerdon, 1849) | Valid | Valid |
| 12. | Tor mekongensis (Hoang et al., 2015)¶ | Valid | Not Valid Valid as T. tambra (Valenciennes, 1842) |
| 13. | Tor mosal (Hamilton, 1822) | Valid | Valid |
| 14. | Tor polylepis (Zhou and Cui, 1996) | Valid | Valid |
| 15. | Tor putitora (Hamilton, 1822) | Valid | Valid |
| 16. | Tor qiaojiensis (Wu, 1977) | Not Valid Valid as Neolissochilus qiaojiensis (Wu, 1977) | Not Valid |
| 17. | Tor remadevii (Madhusoodana Kurup and Radhakrishnan, 2011) | Valid | Valid |
| 18. | Tor sinensis (Wu, 1977) | Valid | Valid |
| 19. | Tor soro (Valenciennes, 1842) | Not Valid Valid as Neolissochilus soro (Valenciennes, 1842) | Not Valid |
| 20. | Tor tambra (Valenciennes, 1842) | Valid | Valid |
| 21. | Tor tambroides (Bleeker, 1854) | Valid | Valid |
| 22. | Tor tor (Hamilton, 1822) | Valid | Valid |
| 23. | Tor yingjiangensis (Chen and Yang, 2004) | Valid | Valid |
| 24. | Tor yunnanensis (Wang, Zhuang, and Gao, 1982) | Not Valid Valid as Folifer yunnanensis (Wang, Zhuang, and Gao, 1982) | Not Valid |
| No. | Species | Common Name(s) | Reported Geographical Distribution | Status (Based on Eschmeyer’s Catalog of Fishes as of 1 March 2021 and Pinder et al. (2019)) | IUCN Red List of Threatened SpeciesTM (Version 2020-3) [11] | References | |
|---|---|---|---|---|---|---|---|
| Species Status | Population Status | ||||||
| 1. | T. ater (Robert, 1999) | Laos | Valid | Near Threatened | Unknown | [36] | |
| 2. | T. dongnaiensis (Hoang et al., 2015) | Dongnai mahseer | Vietnam | Valid | Near Threatened | Unknown | [15] |
| 3. | T. douronensis¶ (Valenciennes, 1842) | Semah (Indonesia/Borneo Island), Pelian (Malaysia) | Malaysia, Indonesia | Not Valid/Valid as T. tambra/Unknown/Questionable/Data Deficient | Not Listed | Not Listed | [1,38] |
| 4. | T. laterivittatus (Zhou and Cui, 1996) | Laos, China | Valid | Data Deficient | Decreasing | [16,37] | |
| 5. | T. mekongensis¶ (Hoang et al., 2015) | Vietnam | Not Valid/Valid as T. tambra/Unknown/Questionable/Data Deficient | Not Listed | Not Listed | [15] | |
| 6. | T. mosal (Hamilton, 1822) | Mosal mahseer, Copper mahseer | Myanmar, India | Valid | Data Deficient | Unknown | [3,34] |
| 7. | T. putitora (Hamilton, 1822) | Himalayan mahseer, Golden mahseer, Putitora mahseer | Myanmar, Afghanistan, Bangladesh, Bhutan, India, Nepal, Pakistan | Valid | Endangered | Decreasing | [17] |
| 8. | T. sinensis (Wu, 1977) | Pba daeng (Laos), Red mahseer | Vietnam, Laos, China | Valid | Vulnerable | Unknown | [15,39] |
| 9. | T. soro (Valenciennes, 1842) | Kancra or Gemo fish | Indonesia | Not Valid/Valid as Other Species/Unknown/Questionable/Data Deficient | Not Listed | Not Listed | [33,38] |
| 10. | T. tambra (Valenciennes, 1842) | Keureling (Indonesia), Pba tohn (Laos) | Malaysia, Thailand, Indonesia | Valid | Data Deficient | Decreasing | [3,35,38] |
| 11. | T. tambroides (Bleeker, 1854) | Kelah, Empurau (Malaysia) Jurung (Indonesia) | Malaysia, Thailand, Indonesia | Valid | Data Deficient | Unknown | [3,35,40] |
| 12. | T. tor (Hamilton, 1822) | Myanmar, Pakistan, Nepal, Bhutan, India, Bangladesh | Valid | Data Deficient | Unknown | [17,34] | |
| 13. | T. yingjiangensis (Chen and Yang, 2004) | Myanmar, China | Valid | Data Deficient | Unknown | [26,28] | |
| Species | Reported Maximum Size (Standard Length for Adult, cm) | Mouth Position | Lower Median Lobe (Scale Bars Represent the Length of Lower Median Lobe) | Body Color | Lateral Scales | Distinctive Features | Reference |
|---|---|---|---|---|---|---|---|
| T. ater (Robert, 1999) | ~30 | Sub-terminal | Short | Dark brown with the presence of a dark longitudinal stripe | 30–31 | The smallest in body size among SE Asia’s Tor species. Short median lobe and longitudinal dark stripe on the body. | [52] |
| T. dongnaiensis (Hoang et al., 2015) | ~40 | Sub-terminal | Long | Silver gray and yellowish in sub-adult | 23–24 | Long lower median lobe without upper median projection. | [15] |
| T. douronensis¶ (Valenciennes, 1842) | ~100 | Sub-terminal | Medium | Silvery, darkish above and dark fins | 24–27 | Short lower median lobe and silvery and yellowish in color. | [2] |
| T. laterivittatus (Zhou and Cui, 1996) | ~60 | Sub-terminal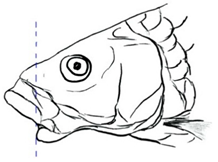 | Long | Dark green to silver with the presence of longitudinal stripe | 25–27 | Long lower median lobe without upper median projection and longitudinal dark brown stripe along the middle side of the adult body. | [91] |
| T. mekongensis¶ (Hoang et al., 2015) | ~33 | Sub-terminal | Medium | Silver gray | 23 | Short lower median lobe, blunt rostral hood, and silvery in color. | [15] |
| T. mosal (Hamilton, 1822) | ~270 | Terminal | Long | Delicate yellowish shade below, caudal reddish orange | 23–26 | Terminal mouth position with long lower median lobe, yellowish color on the lower body, and fin ray counts: 13 dorsal fin rays, 17 pectoral fin rays, and 8 anal fin rays. | [3] |
| T. putitora (Hamilton, 1822) | ~270 | Sub-terminal | Varying in length | Reddish sap green, light orange fading to silvery white | 23–28 | Blunt rostral hood and dark longitudinal stripe in the middle of the side of the body. The color of the caudal, pelvic, and anal fins is reddish gold. | [92] |
| T. sinensis (Wu, 1977) | ~46 | Sub-terminal | Long | Darkish on the back and brownish or bronzy beneath, a dark longitudinal stripe | 23–28 | Long lower median lobe without upper median projection, pointed rostral hood, and a slate gray longitudinal stripe along the side of the body from head to caudal base.  | [37] |
| T. tambra (Valenciennes, 1842) | ~100 | Sub-terminal | Short | Olive or dark olive, reddish | 22–24 | Short lower median lobe, blunt rostral hood, and reddish body color. | [13] |
| T. tambroides (Bleeker, 1854) | ~100 | Sub-terminal | Long | Silver bronze and reddish with dark fin | 23–26 | Long lower median lobe, present with upper median projection, pointed rostral hood, and reddish body color. | [93] |
| T. tor (Hamilton, 1822) | ~200 | Terminal | Varying in length | Grayish green | 22–28 | Small head and mouth, blunt rostral hood, reddish fin, silvery abdomen, and dark gray dorsal side. | [12] |
| T. yingjiangensis (Chen and Yang, 2004) | ~20 | Terminal | Short | Yellowish | 24–26 | Longer head, conical shape of the head, and a slightly convex body. All fins are yellowish, and there is no mid-lateral line. | [28] |
| No. | Species Name |
|---|---|
| 1. | T. ater (Robert, 1999) |
| 2. | T. douronensis (Valenciennes, 1842) |
| 3. | T. laterivittatus (Zhou and Cui, 1996) |
| 4. | T. mosal (Hamilton, 1822) |
| 5. | T. putitora (Hamilton, 1822) |
| 6. | T. sinensis (Wu, 1977) |
| 7. | T. tambra (Valenciennes, 1842) |
| 8. | T. tambroides (Bleeker, 1854) |
| 9. | T. tor (Hamilton, 1822) |
| 10. | T. yingjiangensis (Chen and Yang, 2004) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaafar, F.; Na-Nakorn, U.; Srisapoome, P.; Amornsakun, T.; Duong, T.-Y.; Gonzales-Plasus, M.M.; Hoang, D.-H.; Parhar, I.S. A Current Update on the Distribution, Morphological Features, and Genetic Identity of the Southeast Asian Mahseers, Tor Species. Biology 2021, 10, 286. https://doi.org/10.3390/biology10040286
Jaafar F, Na-Nakorn U, Srisapoome P, Amornsakun T, Duong T-Y, Gonzales-Plasus MM, Hoang D-H, Parhar IS. A Current Update on the Distribution, Morphological Features, and Genetic Identity of the Southeast Asian Mahseers, Tor Species. Biology. 2021; 10(4):286. https://doi.org/10.3390/biology10040286
Chicago/Turabian StyleJaafar, Faizul, Uthairat Na-Nakorn, Prapansak Srisapoome, Thumronk Amornsakun, Thuy-Yen Duong, Maria Mojena Gonzales-Plasus, Duc-Huy Hoang, and Ishwar S. Parhar. 2021. "A Current Update on the Distribution, Morphological Features, and Genetic Identity of the Southeast Asian Mahseers, Tor Species" Biology 10, no. 4: 286. https://doi.org/10.3390/biology10040286
APA StyleJaafar, F., Na-Nakorn, U., Srisapoome, P., Amornsakun, T., Duong, T.-Y., Gonzales-Plasus, M. M., Hoang, D.-H., & Parhar, I. S. (2021). A Current Update on the Distribution, Morphological Features, and Genetic Identity of the Southeast Asian Mahseers, Tor Species. Biology, 10(4), 286. https://doi.org/10.3390/biology10040286








