Insight into Liver lncRNA and mRNA Profiling at Four Developmental Stages in Ningxiang Pig
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. RNA Isolation, Library Construction, and RNA-seq
2.3. Identification and Classification of LncRNAs
2.4. Differential Expression Analysis and Functional Enrichment
2.5. Time-Series Analysis
2.6. Co-Expression Networks
2.7. Quantitative Real-Time PCR Analysis
3. Results
3.1. Identification and Classification of lncRNAs in Ningxiang Pig Liver
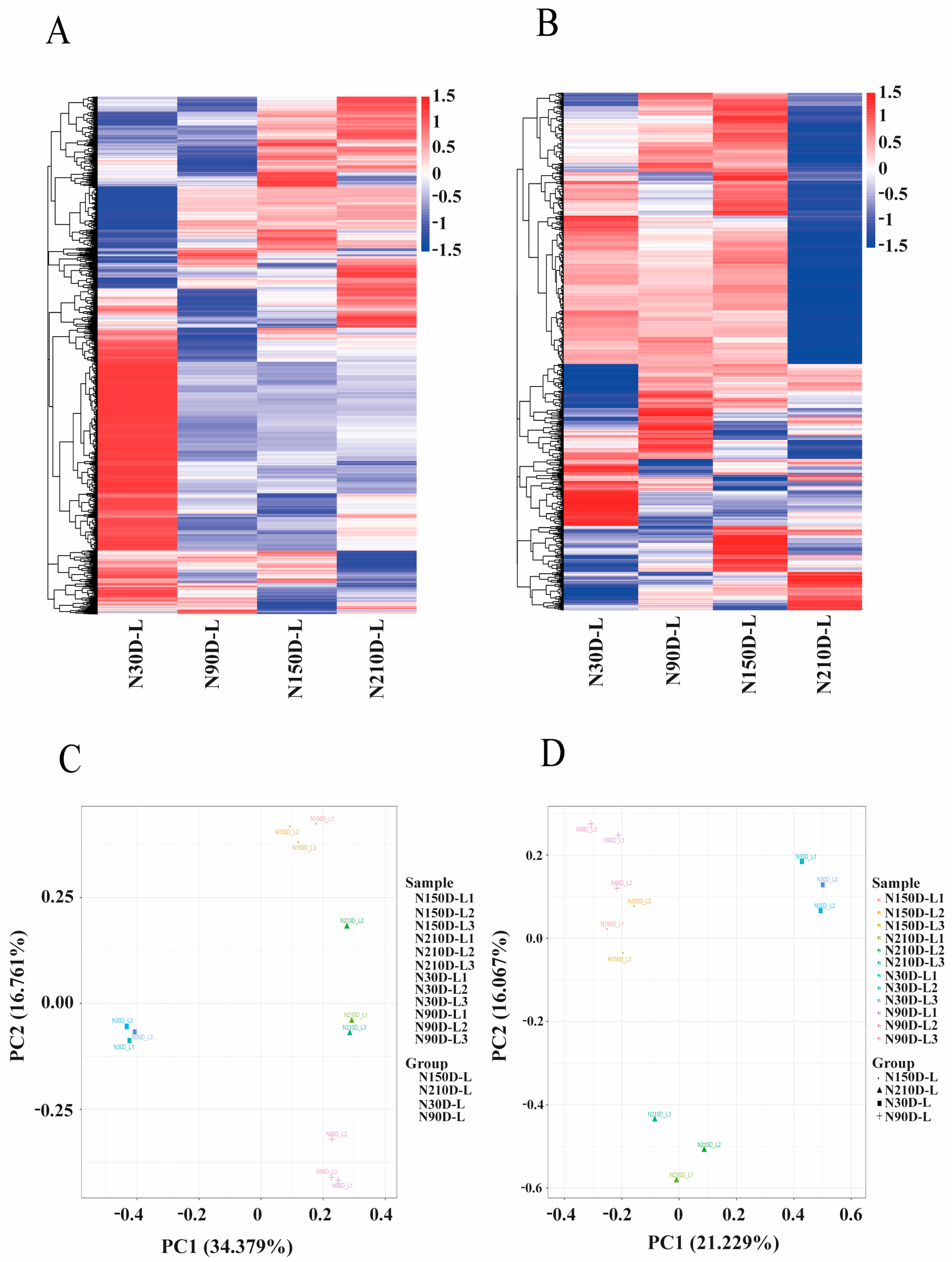
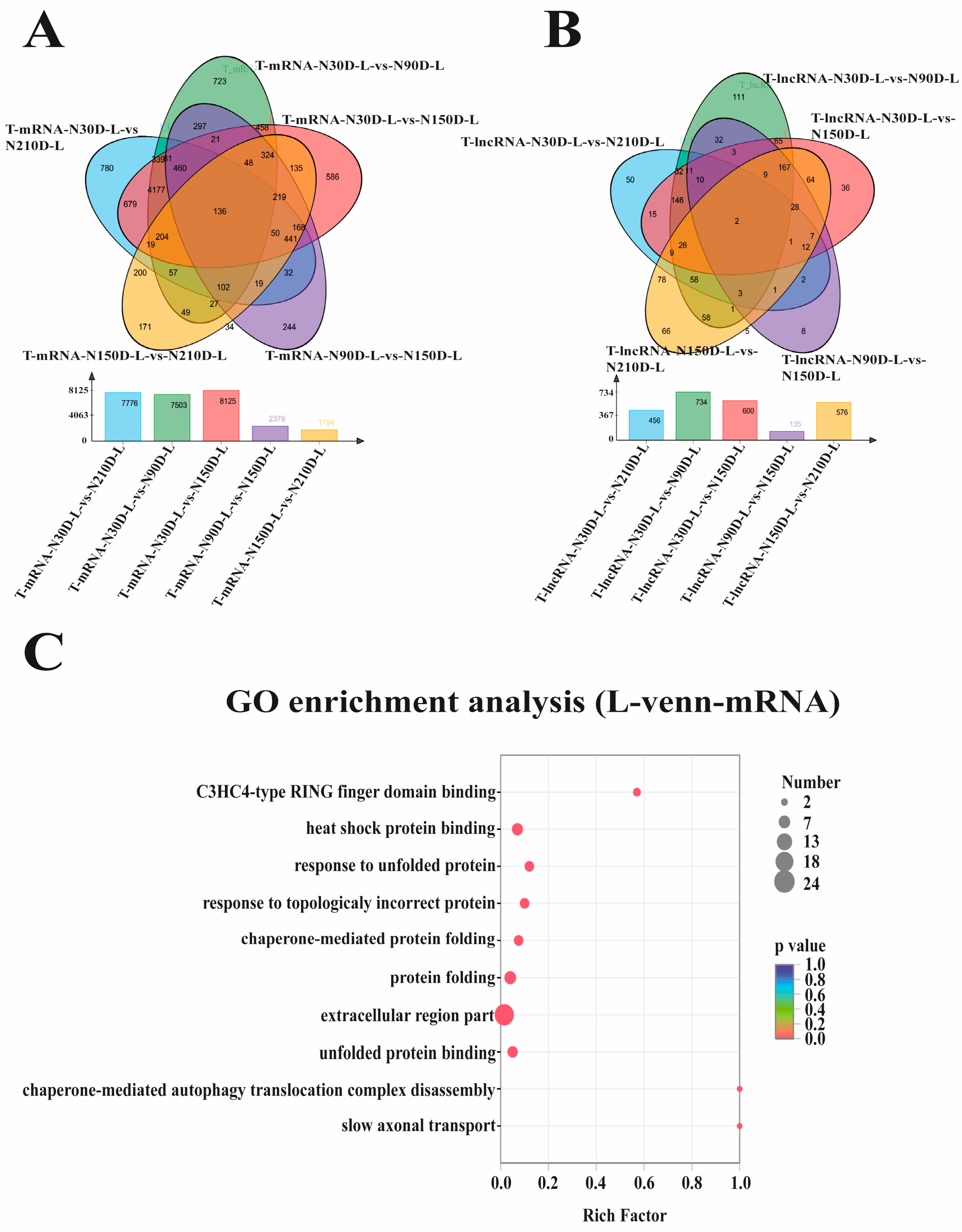
3.2. Identification of Differentially Expressed Protein-Coding Genes and lncRNAs
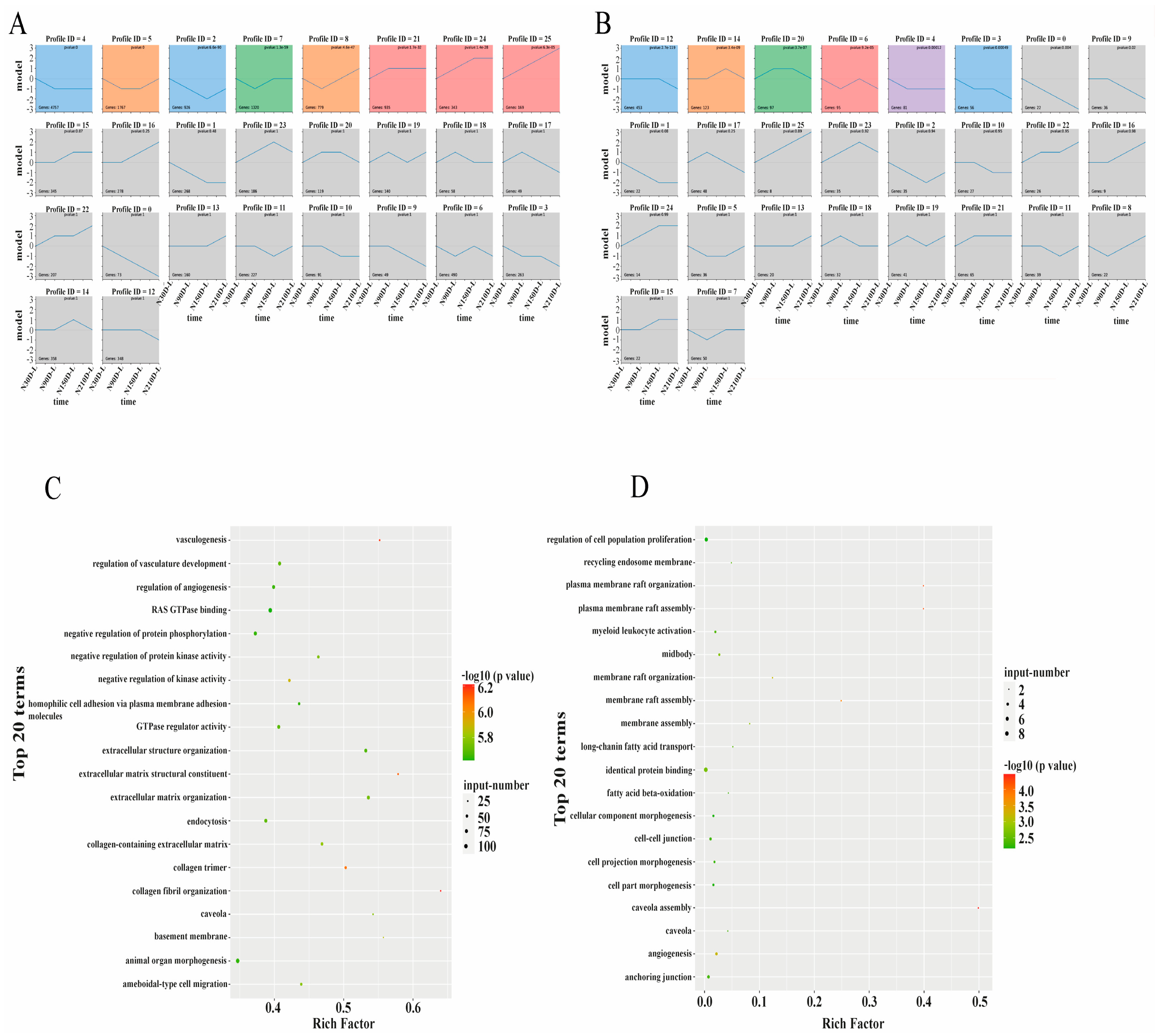
3.3. Time-Series Analysis of Protein-Coding Genes and LncRNAs
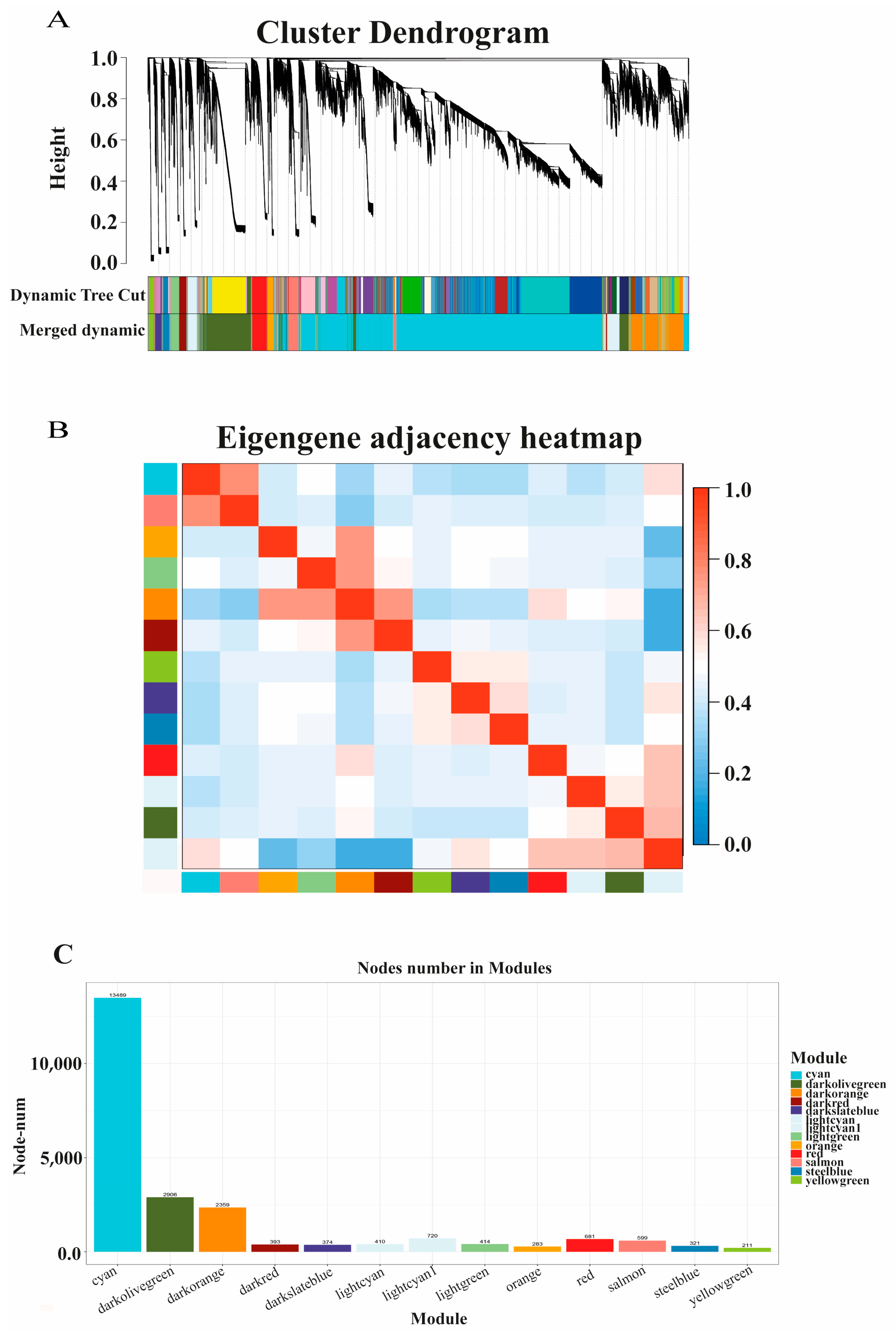
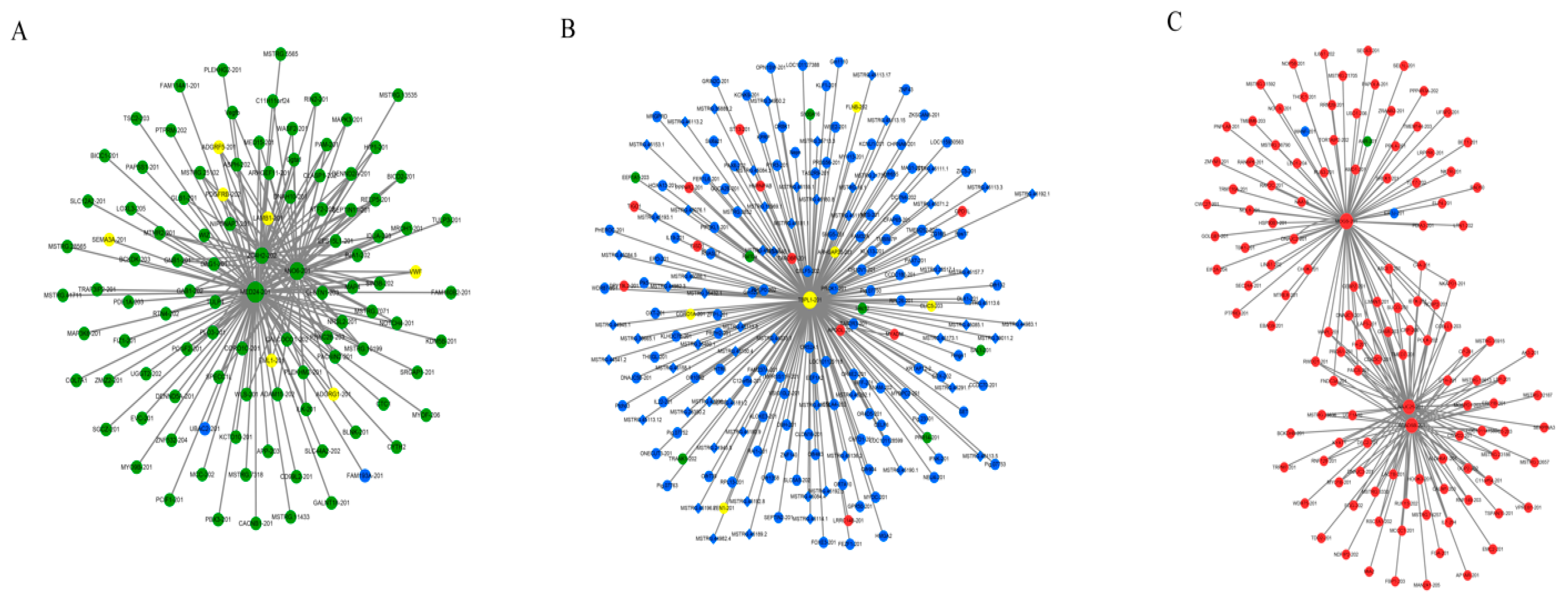
3.4. Co-Expression Network of Protein-Coding Genes and LncRNAs
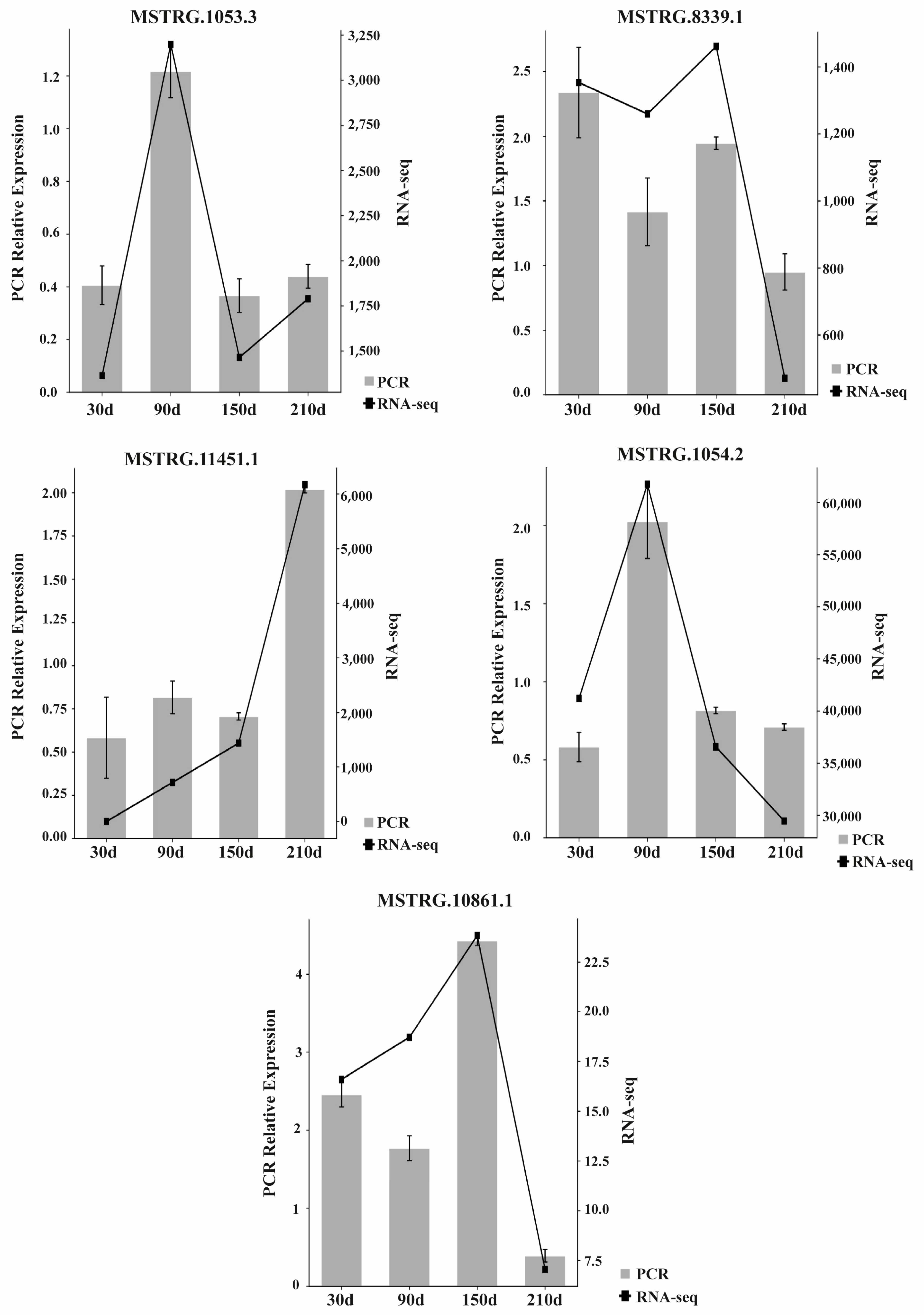
3.5. RT-qPCR Quantification of LncRNAs
4. Discussion
4.1. Differentially Expressed Protein-Coding Genes and lncRNAs
4.2. Time-Series Analysis of Protein-Coding Genes and LncRNAs
4.3. Co-Expression Network of Protein-Coding Genes and LncRNAs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Craig Venter, J.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef] [PubMed]
- Dunham, I.; Kundaje, A.; Aldred, S.F.; Collins, P.J.; Davis, C.A.; Doyle, F.; Epstein, C.B.; Frietze, S.; Harrow, J.; Kaul, R.; et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Necsulea, A.; Kaessmann, H. Evolutionary dynamics of coding and non-coding transcriptomes. Nat. Rev. Genet. 2014, 15, 734–748. [Google Scholar] [CrossRef]
- Bunch, H. Gene regulation of mammalian long non-coding RNA. Mol. Genet. Genom. 2018, 293, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Shields, E.J.; Petracovici, A.F.; Bonasio, R. LncRedibly versatile: Biochemical and biological functions of long noncoding RNAs. Biochem. J. 2019, 476, 1083–1104. [Google Scholar] [CrossRef]
- Jiang, Q.; Li, C.; Yu, Y.; Xing, Y.; Xiao, D.; Zhang, B. Comparison of fatty acid profile of three adipose tissues in Ningxiang pigs. Anim. Nutr. 2018, 4, 256–259. [Google Scholar] [CrossRef]
- Lada, E.; Anna, M.; Patrik, M.; Zbynek, T.; Miroslav, J.; Hynek, M.; Richard, P.; Sarah, L.; Vaclav, L. Porcine Liver Anatomy Applied to Biomedicine. J. Surg. Res. 2020, 250, 70–79. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Ren, P.; Kong, X.; Wu, Y.; Wu, G.; Li, P.; Hao, F.; Tang, H.; Blachier, F.; Yin, Y. Comparison of serum metabolite compositions between obese and lean growing pigs using an NMR-based metabonomic approach. J. Nutr. Biochem. 2012, 23, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Wu, X.; Xie, C.; Xiao, D.; Zhang, B. Meat quality and fatty acid profiles of Chinese Ningxiang pigs following supplementation with N-carbamylglutamate. Animals 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nafikov, R.A.; Beitz, D.C. Carbohydrate and lipid metabolism in farm animals. J. Nutr. 2007, 137, 702–705. [Google Scholar] [CrossRef] [PubMed]
- Anker, R.M.; Denbow, D.M. Anatomy and Physiology of Domestic Animals; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, 1–13. [Google Scholar] [CrossRef]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 1–11. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Zhang, B.; Horvath, S. Defining clusters from a hierarchical cluster tree: The Dynamic Tree Cut package for R. Bioinformatics 2008, 24, 719–720. [Google Scholar] [CrossRef]
- Kern, C.; Wang, Y.; Chitwood, J.; Korf, I.; Delany, M.; Cheng, H.; Medrano, J.F.; Van Eenennaam, A.L.; Ernst, C.; Ross, P.; et al. Genome-wide identification of tissue-specific long non-coding RNA in three farm animal species 06 Biological Sciences 0604 Genetics. BMC Genom. 2018, 19, 1–14. [Google Scholar] [CrossRef]
- Ito, M.; Okano, H.J.; Darnell, R.B.; Roeder, R.G. The TRAP100 component of the TRAP/Mediator complex is essential in broad transcriptional events and development. EMBO J. 2002, 21, 3464–3475. [Google Scholar] [CrossRef]
- Kunzelmann, K.; Ousingsawat, J.; Benedetto, R.; Cabrita, I.; Schreiber, R. Contribution of anoctamins to cell survival and cell death. Cancers 2019, 11, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Vangeel, L.; Janssens, A.; Lemmens, I.; Lievens, S.; Tavernier, J.; Voets, T. The zinc-finger domain containing protein ZC4H2 interacts with TRPV4, enhancing channel activity and turnover at the plasma membrane. Int. J. Mol. Sci. 2020, 21, 3556. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, W.; Veenstra, G.J.C. TBP-related factors: A paradigm of diversity in transcription initiation. Cell Biosci. 2011, 1, 1–12. [Google Scholar] [CrossRef]
- Monticelli, M.; Ferro, T.; Jaeken, J.; dos Reis Ferreira, V.; Videira, P.A. Immunological aspects of congenital disorders of glycosylation (CDG): A review. J. Inherit. Metab. Dis. 2016, 39, 765–780. [Google Scholar] [CrossRef]
- Sass, J.O.; Ensenauer, R.; Röschinger, W.; Reich, H.; Steuerwald, U.; Schirrmacher, O.; Engel, K.; Häberle, J.; Andresen, B.S.; Mégarbané, A.; et al. 2-Methylbutyryl-coenzyme A dehydrogenase deficiency: Functional and molecular studies on a defect in isoleucine catabolism. Mol. Genet. Metab. 2008, 93, 30–35. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Burghardt, T.P.; Vockley, J. A novel approach to the characterization of substrate specificity in short/branched chain acyl-CoA dehydrogenase. J. Biol. Chem. 2003, 278, 37974–37986. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Jiang, W.; Han, D.; Yu, L. DNAJC25 is downregulated in hepatocellular carcinoma and is a novel tumor suppressor gene. Oncol. Lett. 2012, 4, 1274–1280. [Google Scholar] [CrossRef]
- Wang, J.; Chen, L.; Li, P.; Li, X.; Zhou, H.; Wang, F.; Li, D.; Yin, Y.; Wu, G. Gene expression is altered in piglet small intestine by weaning and dietary glutamine supplementation. J. Nutr. 2008, 138, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Li, L.; Huang, A.; Cui, L.; Zhang, Y.; Liu, Q.; Wang, X.; Wang, Y.; Liu, Z.; Yuan, Z.; et al. Molecular characterization and biological function of a novel lncRNA CRNG in swine. Front. Pharmacol. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Lundell, K. Cloning and expression of two novel pig liver and kidney fatty acid hydroxylases [cytochrome P450 (CYP)4A24 and CYP4A25]. Biochem. J. 2002, 363, 297–303. [Google Scholar] [CrossRef]
- Rosenbaum, S.; Ringseis, R.; Most, E.; Hillen, S.; Becker, S.; Erhardt, G.; Reiner, G.; Eder, K. Genes involved in carnitine synthesis and carnitine uptake are up-regulated in the liver of sows during lactation. Acta Vet. Scand. 2013, 55, 24. [Google Scholar] [CrossRef]
- Lester, S.N.; Li, K. Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 2014, 426, 1246–1264. [Google Scholar] [CrossRef]
- Halley, P.; Kadakkuzha, B.M.; Faghihi, M.A.; Magistri, M.; Zeier, Z.; Khorkova, O.; Coito, C.; Hsiao, J.; Lawrence, M.; Wahlestedt, C. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014, 6, 222–230. [Google Scholar] [CrossRef]
- Drixler, T.A.; Vogten, M.J.; Ritchie, E.D.; Van Vroonhoven, T.J.M.V.; Gebbink, M.F.B.G.; Voest, E.E.; Borel Rinkes, I.H.M.; Neuhaus, P.; Drixler, T.; Clavien, P.A.; et al. Liver regeneration is an angiogenesis-associated phenomenon. Ann. Surg. 2002, 236, 703–712. [Google Scholar] [CrossRef]
- AlAbdi, L.; He, M.; Yang, Q.; Norvil, A.B.; Gowher, H. The transcription factor Vezf1 represses the expression of the antiangiogenic factor Cited2 in endothelial cells. J. Biol. Chem. 2018, 293, 11109–11118. [Google Scholar] [CrossRef]
- Van Der Leij, F.R.; Cox, K.B.; Jackson, V.N.; Huijkman, N.C.A.; Bartelds, B.; Kuipers, J.R.G.; Dijkhuizen, T.; Terpstra, P.; Wood, P.A.; Zammit, V.A.; et al. Structural and functional genomics of the CPT1B gene for muscle-type carnitine palmitoyltransferase I in mammals. J. Biol. Chem. 2002, 277, 26994–27005. [Google Scholar] [CrossRef]
- Gargalovic, P.; Dory, L. Caveolins and macrophage lipid metabolism. J. Lipid Res. 2003, 44, 11–21. [Google Scholar] [CrossRef] [PubMed]
- del Pozo, M.A.; Balasubramanian, N.; Alderson, N.B.; Kiosses, W.B.; Grande-García, A.; Anderson, R.G.W.; Schwartz, M.A. Phospho-caveolin-1 mediates integrin-regulated membrane domain internalization. Nat. Cell Biol. 2005, 7, 901–908. [Google Scholar] [CrossRef]
- Mayoral, R.; Fernández-Martínez, A.; Roy, R.; Boscá, L.; Martín-Sanz, P. Dispensability and dynamics of caveolin-1 during liver regeneration and in isolated hepatic cells. Hepatology 2007, 46, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Frank, P.G.; Cheung, M.W.C.; Pavlides, S.; Llaverias, G.; Park, D.S.; Lisanti, M.P. Caveolin-1 and regulation of cellular cholesterol homeostasis. Am. J. Physiol. Heart Circ. Physiol. 2006, 291, H677–H686. [Google Scholar] [CrossRef]
- Sala-Vila, A.; Navarro-Lérida, I.; Sánchez-Alvarez, M.; Bosch, M.; Calvo, C.; López, J.A.; Calvo, E.; Ferguson, C.; Giacomello, M.; Serafini, A.; et al. Interplay between hepatic mitochondria-Associated membranes, lipid metabolism and caveolin-1 in mice. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Senju, Y.; Itoh, Y.; Takano, K.; Hamada, S.; Suetsugu, S. Essential role of PACSIN2/syndapin-II in caveolae membrane sculpting. J. Cell Sci. 2011, 124, 2032–2040. [Google Scholar] [CrossRef] [PubMed]
- Amado-Azevedo, J.; Reinhard, N.R.; Van Bezu, J.; De Menezes, R.X.; Van Beusechem, V.W.; Van Nieuw Amerongen, G.P.; Van Hinsbergh, V.W.M.; Hordijk, P.L. A CDC42-centered signaling unit is a dominant positive regulator of endothelial integrity. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Del Maldonado, M.M.; Medina, J.I.; Velazquez, L.; Dharmawardhane, S. Targeting Rac and Cdc42 GEFs in Metastatic Cancer. Front. Cell Dev. Biol. 2020, 8, 1–17. [Google Scholar] [CrossRef]
- Duman, J.G.; Mulherkar, S.; Tu, Y.K.; Cheng, J.X.; Tolias, K.F. Mechanisms for spatiotemporal regulation of Rho-GTPase signaling at synapses. Neurosci. Lett. 2015, 601, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fondell, J.D. Identification of mouse TRAP100: A transcriptional coregulatory factor for thyroid hormone and vitamin D receptors. Mol. Endocrinol. 1999, 13, 1130–1140. [Google Scholar] [CrossRef]
- Zhao, P.; Peng, X.; Luo, S.; Huang, Y.; Tan, L.; Shao, J.; He, X. Identification and characterization of novel mutations in MOGS in a Chinese patient with infantile spams. Neurogenetics 2020, 21, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Lallès, J.P.; Bosi, P.; Smidt, H.; Stokes, C.R. Weaning—A challenge to gut physiologists. Livest. Sci. 2007, 108, 82–93. [Google Scholar] [CrossRef]
- Wu, X.; Xie, C.; Yin, Y.; Li, F.; Li, T.; Huang, R.; Ruan, Z.; Deng, Z. Effect of L-arginine on HSP70 expression in liver in weanling piglets. BMC Vet. Res. 2013, 9, 2–7. [Google Scholar] [CrossRef]

| Name | Sequence (5’ to 3’) |
|---|---|
| MSTRG.1053.3-F | ACTTGGGAAGAAAGCAATTTTAAGA |
| MSTRG.1053.3-R | TGTAGTCCCAGCTACTCGGG |
| MSTRG.11451.1-F | AGACATCCGAGCCTGGGATA |
| MSTRG.11451.1-R | CGTTTCAGAAAGCGTTGGAAGT |
| MSTRG.8339.1-F | GGCATATGGAGGTTCCCAGG |
| MSTRG.8339.1-R | GCGCAGTGGTTAACGAATCC |
| MSTRG.10861.1-F | GAGCCTGATTCCTCCAGCTC |
| MSTRG.10861.1-R | CCAGCCACAGCAATCAGAGA |
| MSTRG.1054.2-F | TGTAGTCCCAGCTACTCGGG |
| MSTRG.1054.2-R | ACAGGGTCTCGCTATGTTGC |
| Sample | Raw Reads | Raw Bases | Clean Reads | Clean Bases | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|---|
| 30d-1 | 110,376,466 | 16,666,846,366 | 108,613,548 | 14,971,657,073 | 0.0241 | 98.25 | 95.24 | 55.15 |
| 30d-2 | 113,435,178 | 17,128,711,878 | 111,741,648 | 15,445,970,217 | 0.0241 | 98.27 | 95.16 | 54.05 |
| 30d-3 | 115,191,290 | 17,393,884,790 | 113,234,706 | 15,440,805,570 | 0.0242 | 98.22 | 95.10 | 54.40 |
| 90d-1 | 111,744,756 | 16,873,458,156 | 110,334,062 | 14,899,645,712 | 0.0237 | 98.48 | 95.60 | 51.03 |
| 90d-2 | 112,938,114 | 17,053,655,214 | 111,212,336 | 15,058,781,243 | 0.0237 | 98.47 | 95.59 | 52.15 |
| 90d-3 | 108,509,032 | 16,384,863,832 | 107,067,364 | 14,561,278,014 | 0.0237 | 98.48 | 95.53 | 51.38 |
| 150d-1 | 92,416,686 | 13,954,919,586 | 90,822,938 | 12,377,703,789 | 0.0239 | 98.38 | 95.32 | 50.39 |
| 150d-2 | 92,911,502 | 14,029,636,802 | 91,797,388 | 12,720,267,538 | 0.0238 | 98.44 | 95.40 | 49.73 |
| 150d-3 | 84,857,708 | 12,813,513,908 | 83,747,750 | 11,702,176,288 | 0.0241 | 98.34 | 95.16 | 49.99 |
| 210d-1 | 103,579,630 | 15,640,524,130 | 102,245,470 | 14,008,181,654 | 0.0237 | 98.51 | 95.54 | 49.84 |
| 210d-2 | 101,318,164 | 15,299,042,764 | 99,445,274 | 13,643,601,019 | 0.0237 | 98.45 | 95.52 | 52.12 |
| 210d-3 | 106,488,022 | 16,079,691,322 | 104,848,812 | 14,255,190,000 | 0.0237 | 98.45 | 95.56 | 50.78 |
| Sample | Clean Reads | Mapped Reads | Mapping Rate (%) |
|---|---|---|---|
| 30d-1 | 108,613,548 | 100,520,492 | 92.55 |
| 30d-2 | 111,741,648 | 103,608,606 | 92.72 |
| 30d-3 | 113,234,706 | 104,826,456 | 92.57 |
| 90d-1 | 110,334,062 | 104,211,698 | 94.45 |
| 90d-2 | 111,212,336 | 105,230,225 | 94.62 |
| 90d-3 | 107,067,364 | 101,424,972 | 94.73 |
| 150d-1 | 90,822,938 | 85,849,715 | 94.52 |
| 150d-2 | 91,797,388 | 85,907,029 | 93.58 |
| 150d-3 | 83,747,750 | 78,654,116 | 93.92 |
| 210d-1 | 102,245,470 | 95,722,450 | 93.62 |
| 210d-2 | 99,445,274 | 92,960,760 | 93.48 |
| 210d-3 | 104,848,812 | 98,178,592 | 93.64 |
| Groups | Total DEmRNAs | Upregulated | Downregulated |
|---|---|---|---|
| 30 vs. 90 d | 7345 | 3473 | 3872 |
| 30 vs. 150 d | 7971 | 4039 | 3932 |
| 30 vs. 210 d | 7634 | 3761 | 3873 |
| 90 vs. 150 d | 2309 | 1434 | 873 |
| 90 vs. 210 d | 2754 | 1518 | 1236 |
| 150 vs. 210 d | 1717 | 479 | 1238 |
| Groups | Total DElncRNAs | Upregulated | Downregulated |
|---|---|---|---|
| 30 vs. 90 d | 734 | 646 | 88 |
| 30 vs. 150 d | 600 | 521 | 79 |
| 30 vs. 210 d | 456 | 195 | 261 |
| 90 vs. 150 d | 135 | 64 | 71 |
| 90 vs. 210 d | 671 | 29 | 642 |
| 150 vs. 210 d | 576 | 24 | 552 |
| mRNA | Module | Function of mRNA | Associated lncRNAs |
|---|---|---|---|
| MED24 | Molecular function | Interact with RNA polymerase II to promote formation of transcriptional pre-initiation complex to induce gene expression [17] | MSTRG.2158.2 MSTRG.34993.2 MSTRG.16183.1 MSTRG.17517.2 |
| ANO6 | Molecular function | Essential component for calcium-dependent exposure of phosphatidylserine on cell surface, essential for triggering clotting system and deposition in bone mineralization [18] | MSTRG.31383.3 MSTRG.9728.2 MSTRG.33542.1 MSTRG.25050.1 MSTRG.40598.2 MSTRG.18175.1 MSTRG.42141.2 MSTRG.860.1 MSTRG.24048.3 MSTRG.13083.1 |
| ZC4H2 | Molecular function | ZC4H2 may improve channel activity and turnover of plasma membrane and is identified as potential candidate for X-linked mental retardation [19] | MSTRG.33542.1 MSTRG.40598.2 MSTRG.11929.4 MSTRG.3259.4 |
| TBPL1 | Cellular component | TBPL1 may play an important role in transcription by RNA polymerase II as a component of the transcription factor complex [20] | None |
| MOGS | Biological process | MOGS may cleave distal alpha-1,2-linked glucose residue from Glc(3)-Man(9)-GlcNAc(2) oligosaccharide precursor [21] | MSTRG.3283.1 |
| ACADSB | Biological process | Catalyze dehydrogenation of short branched chain acyl-CoA derivatives in metabolism of fatty acids [22,23] | MSTRG.6256.1 MSTRG.22802.1 MSTRG.11451.1 MSTRG.24004.1 MSTRG.26481.9 MSTRG.32831.1 MSTRG.29185.1 MSTRG.3444.1 |
| DNAJC25 | Biological process | DNAJC25 may play an important role in cell protection, protein folding, refolding, aggregation and degradation, and protein translocation [24] | MSTRG.3827.1 MSTRG.8329.3 MSTRG.20562.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, Y.; Zhang, Y.; Li, B.; Xiao, Y.; Zeng, Q.; Xu, K.; Duan, Y.; He, J.; Ma, H. Insight into Liver lncRNA and mRNA Profiling at Four Developmental Stages in Ningxiang Pig. Biology 2021, 10, 310. https://doi.org/10.3390/biology10040310
Gong Y, Zhang Y, Li B, Xiao Y, Zeng Q, Xu K, Duan Y, He J, Ma H. Insight into Liver lncRNA and mRNA Profiling at Four Developmental Stages in Ningxiang Pig. Biology. 2021; 10(4):310. https://doi.org/10.3390/biology10040310
Chicago/Turabian StyleGong, Yan, Yuebo Zhang, Biao Li, Yu Xiao, Qinghua Zeng, Kang Xu, Yehui Duan, Jianhua He, and Haiming Ma. 2021. "Insight into Liver lncRNA and mRNA Profiling at Four Developmental Stages in Ningxiang Pig" Biology 10, no. 4: 310. https://doi.org/10.3390/biology10040310
APA StyleGong, Y., Zhang, Y., Li, B., Xiao, Y., Zeng, Q., Xu, K., Duan, Y., He, J., & Ma, H. (2021). Insight into Liver lncRNA and mRNA Profiling at Four Developmental Stages in Ningxiang Pig. Biology, 10(4), 310. https://doi.org/10.3390/biology10040310






