Haslea silbo, A Novel Cosmopolitan Species of Blue Diatoms
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Culture of Algae
2.2. Microscopy
2.3. Pigment Extraction and Purification
2.4. UV–Visible Spectrophotometry
2.5. Raman Spectrometry
2.6. Induction of Auxosporulation and Reproductive Behaviour
2.7. Molecular Barcoding
2.8. Next Generation Sequencing
2.9. Phylogenetic Analysis
3. Results
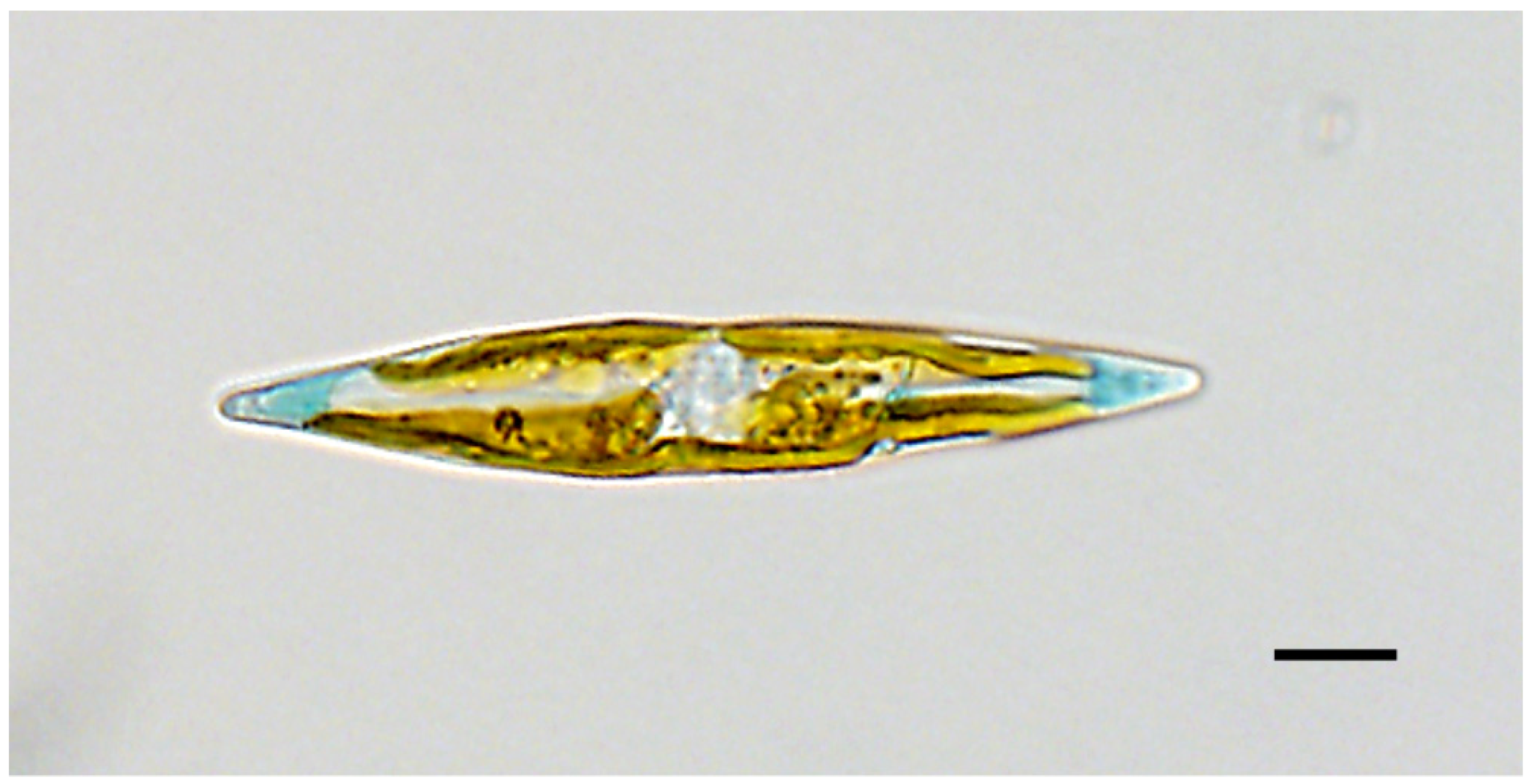
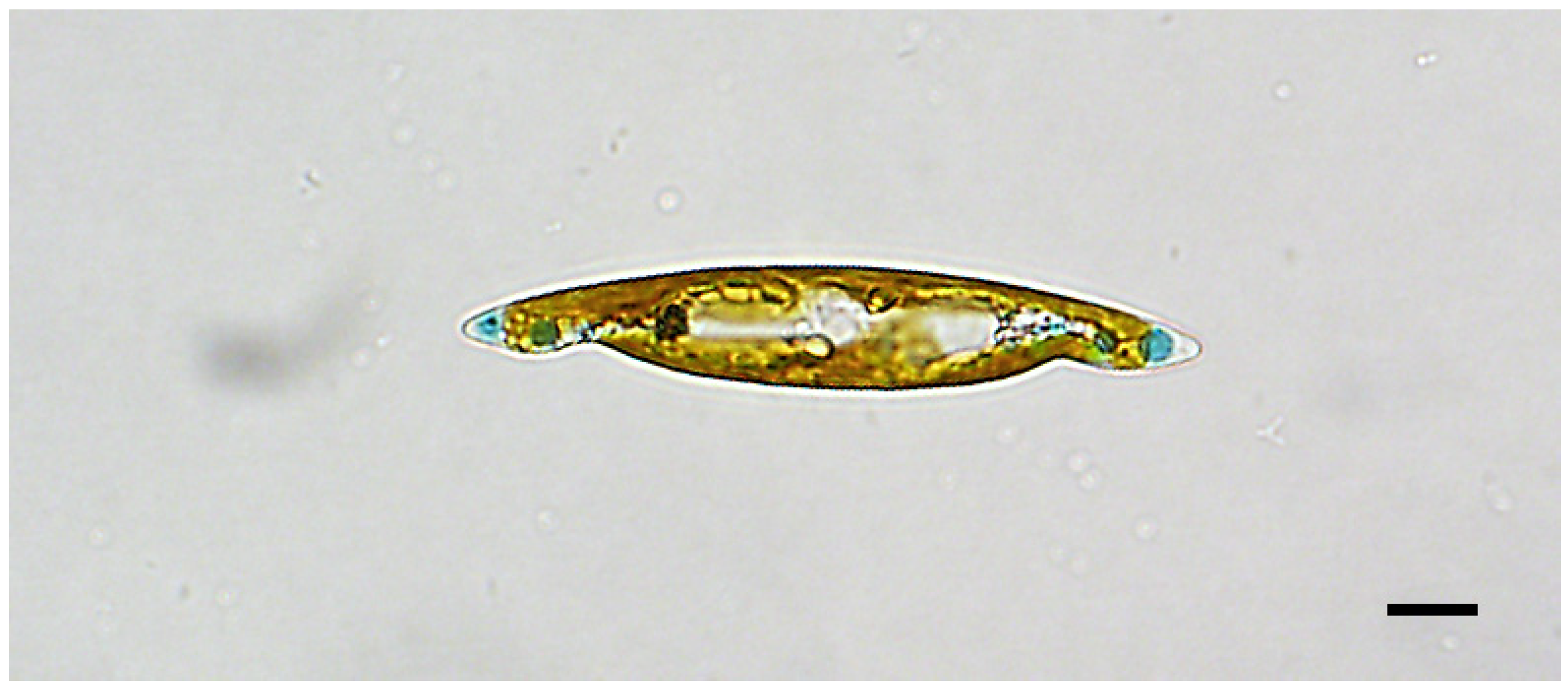
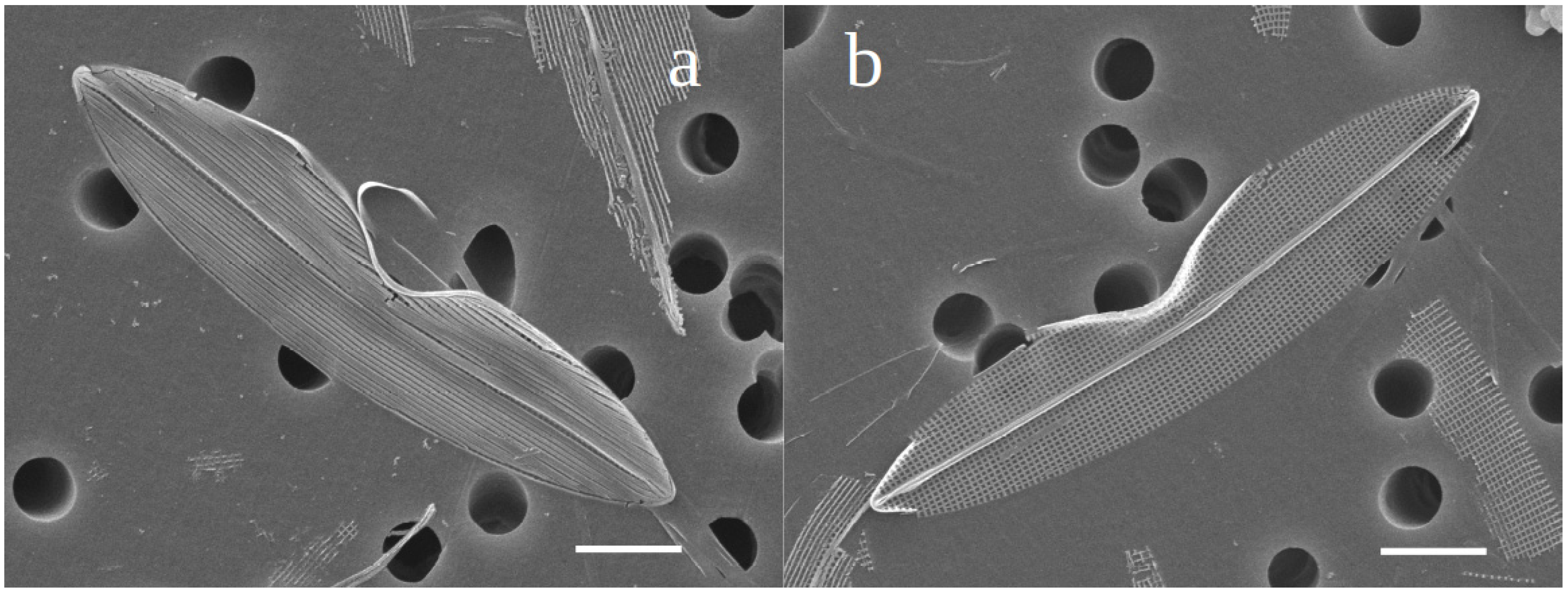
3.1. Description
3.1.1. Additional Morphometric and Morphological Data
3.1.2. Holotype
3.1.3. Type Locality
3.1.4. Etymology
3.1.5. Ecology
3.1.6. Molecular Signature
3.1.7. Differential Diagnosis
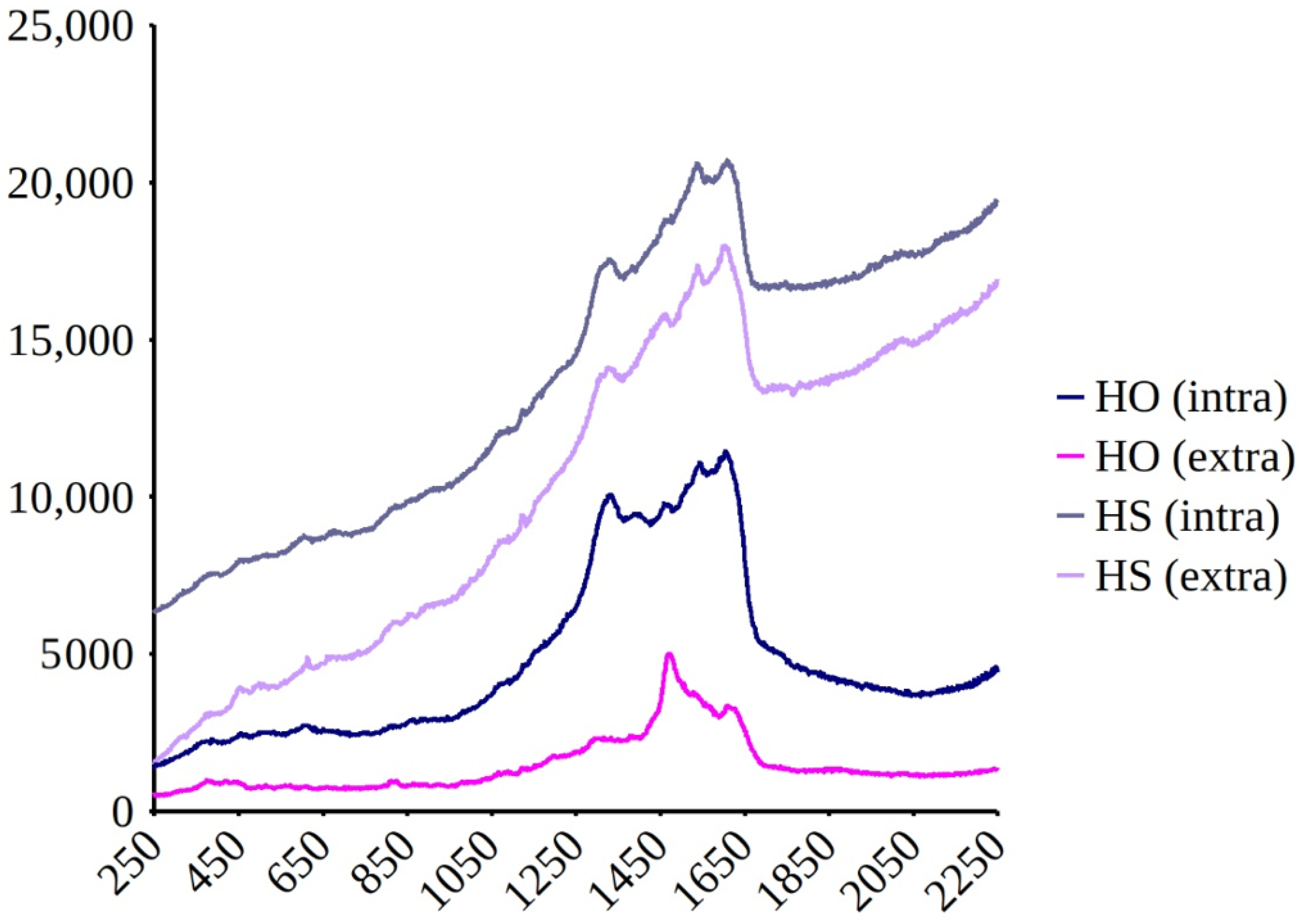
3.2. Spectrophotometric Analyses of H. silbo’s Blue Pigment
3.3. Reproductive Behaviour
3.4. Genomics
3.4.1. NCC456 Mitochondrial Genome
3.4.2. SZCZMV2009 Mitochondrial Genome
3.4.3. Comparison with The Mitochondrial Genome of H. nusantara
3.4.4. NCC456 Plastid Genome
3.4.5. SZCZMV2009 Plastid Genome
3.4.6. Comparison with The Plastid Genome of H. nusantara
3.4.7. NCC456 Plasmids
3.4.8. SZCZMV2009 Plasmids
3.4.9. Phylogeny
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simonsen, R. The diatom plankton of the Indian Ocean expedition of RV “Meteor” 1964–1965. Meteor Forschungsergebnisse Reihe D 1974, 9, 1–66. [Google Scholar]
- Poulin, M.; Méléder, V.; Mouget, J.-L. Typification of the first recognized blue pigmented diatom, Haslea ostrearia (Bacillariophyceae). Plant Ecol. Evol. 2019, 152, 402–408. [Google Scholar] [CrossRef]
- Gastineau, R.; Turcotte, F.; Pouvreau, J.-B.; Morançais, M.; Fleurence, J.; Windarto, E.; Semba Prasetiya, F.; Arsad, S.; Jaouen, P.; Babin, M.; et al. Marennine, promising blue pigments from a widespread Haslea diatom species complex. Mar. Drugs 2014, 12, 3161–3189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sprat, T. The history of the generation and ordering of green Oysters, commonly called Colchester Oysters. In The History of the Royal-Society of London for the Improving of Natural Knowledge by Tho. Sprat; Royal Society: London, UK, 1669; pp. 307–319. [Google Scholar]
- Gaillon, B. Des huîtres vertes, et des causes de cette coloration. J. Phys. Chim. Hist. Nat. Arts 1820, 91, 222–225. [Google Scholar]
- Dyer, W.T.T. Greening of oysters. Nature 1877, 16, 397. [Google Scholar] [CrossRef] [Green Version]
- Lankester, E.R. On green oysters. Q. J. Microsc. Sci. 1886, 26, 71–94. [Google Scholar] [CrossRef]
- Pouvreau, J.B.; Morançais, M.; Taran, F.; Rosa, P.; Dufossé, L.; Guérard, F.; Pin, S.; Fleurence, J.; Pondaven, P. Antioxidant and free radical scavenging properties of marennine, a blue-green polyphenolic pigment from the diatom Haslea ostrearia (Gaillon/Bory) Simonsen responsible for the natural greening of cultured oysters. J. Agric. Food Chem. 2008, 56, 6278–6286. [Google Scholar] [CrossRef]
- Gastineau, R.; Pouvreau, J.-B.; Hellio, C.; Morançais, M.; Fleurence, J.; Gaudin, P.; Bourgougnon, N.; Mouget, J.-L. Biological activities of purified marennine, the blue pigment responsible for the greening of oysters. J. Agric. Food Chem. 2012, 60, 3599–3605. [Google Scholar] [CrossRef]
- Gastineau, R.; Hardivillier, Y.; Leignel, V.; Morançais, M.; Fleurence, J.; Hellio, C.; Bourgougnon, N.; Davidovich, N.A.; Tekaya, N.; Gaudin, P.; et al. Greening effect on oysters and biological activities of the blue pigment produced by the diatom Haslea karadagensis (Naviculaceae). Aquaculture 2012, 368–369, 61–67. [Google Scholar] [CrossRef]
- Gastineau, R.; Prasetiya, F.S.; Falaise, C.; Cognie, B.; Decottignies, P.; Morançais, M.; Méléder, V.; Davidovich, N.; Turcotte, F.; Tremblay, R.; et al. Marennine-Like Pigments: Blue Diatom or Green Oyster Cult? In Blue Biotechnology; Barre, S.L., Bates, S.S., Eds.; Wiley: Hoboken, NJ, USA, 2020. [Google Scholar] [CrossRef]
- Falaise, C.; François, C.; Travers, M.A.; Morga, B.; Haure, J.; Tremblay, R.; Turcotte, F.; Pasetto, P.; Gastineau, R.; Hardivillier, Y.; et al. Antimicrobial Compounds from Eukaryotic Microalgae against Human Pathogens and Diseases in Aquaculture. Mar. Drugs 2016, 14, 159. [Google Scholar] [CrossRef] [Green Version]
- Falaise, C.; Cormier, P.; Tremblay, R.; Audet, C.; Deschênes, J.S.; Turcotte, F.; François, C.; Seger, A.; Hallegraeff, G.; Lindquist, N.; et al. Harmful or harmless: Biological effects of marennine on marine organisms. Aquat. Toxicol. 2019, 209, 13–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falaise, C.; James, A.; Travers, M.-A.; Zanella, M.; Badawi, M.; Mouget, J.-L. Complex Relationships between the Blue Pigment Marennine and Marine Bacteria of the Genus Vibrio. Mar. Drugs 2019, 17, 160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasetiya, F.S.; Safitri, I.; Widowati, I.; Cognie, B.; Decottignies, P.; Gastineau, R.; Morançais, M.; Windarto, E.; Tremblay, R.; Mouget, J. Does allelopathy affect co-culturing Haslea ostrearia with other microalgae relevant to aquaculture? J. Appl. Phycol. 2016, 28, 2241–2254. [Google Scholar] [CrossRef]
- Prasetiya, F.S.; Decottignies, P.; Tremblay, R.; Mouget, J.L.; Cognie, B. Does culture supernatant of Haslea ostrearia containing marennine affect short-term physiological traits in the adult blue mussel Mytilus edulis? Aquac. Rep. 2019, 15. [Google Scholar] [CrossRef]
- Prasetiya, F.S.; Sunarto, S.; Bachtiar, E.; Agung, M.U.K.; Nathanael, B.; Pambudi, A.C.; Lestari, A.D.; Astuty, S.; Mouget, J.-L. Effect of the blue pigment produced by the tropical diatom Haslea nusantara on marine organisms from different trophic levels and its bioactivity. Aquac. Rep. 2020, 17. [Google Scholar] [CrossRef]
- Permatasari, I.; Agung, M.U.K.; Liviawaty, E.; Astuty, S.; Risjani, Y.; Arsad, S.; Mouget, J.-L.; Prasetiya, F.S. Antibacterial Activity of Haslea ostrearia Supernatant Adapted in Indonesia against Pathogenic Bacteria Relevant to Mariculture (In-Vitro Study). Omni Akuatika 2019, 15, 30–38. [Google Scholar] [CrossRef]
- Gastineau, R.; Davidovich, N.A.; Bardeau, J.F.; Caruso, A.; Leignel, V.; Hardivillier, Y.; Jacquette, B.; Davidovich, O.; Rince, Y.; Gaudin, P.; et al. Haslea karadagensis (Bacillariophyta): A second blue diatom, recorded from the Black Sea and producing a novel blue pigment. Eur. J. Phycol. 2012, 47, 469–479. [Google Scholar] [CrossRef] [Green Version]
- Gastineau, R.; Davidovich, N.A.; Hansen, G.; Rines, J.; Wulff, A.; Kaczmarska, I.; Ehrman, J.; Hermann, D.; Maumus, F.; Hardivillier, Y.; et al. Haslea ostrearia-like diatoms: Biodiversity out of the blue. Adv. Bot. Res. 2014, 71, 441–446. [Google Scholar] [CrossRef]
- Gastineau, R.; Hansen, G.; Davidovich, N.A.; Davidovich, O.; Bardeau, J.F.; Kaczmarska, I.; Ehrman, J.M.; Leignel, V.; Hardivillier, Y.; Jacquette, B.; et al. A new blue-pigmented hasleoid diatom, Haslea provincialis, from the Mediterranean Sea. Eur. J. Phycol. 2016, 51, 156–170. [Google Scholar] [CrossRef] [Green Version]
- Prasetiya, F.S.; Gastineau, R.; Poulin, M.; Hardivillier, Y.; Syakti, A.D.; Lemieux, C.; Widowati, I.; Falaise, C.; Turmel, M.; Risjani, Y.; et al. Haslea nusantara, a new blue diatom from the Java Sea, Indonesia: Morphology, biometry and molecular characterizations. Plant Ecol. Evol. 2019, 152, 188–202. [Google Scholar] [CrossRef]
- Davidovich, N.A.; Gastineau, R.; Gaudin, P.; Davidovich, O.; Mouget, J.-L. Sexual reproduction in the newly-described blue diatom Haslea karadagensis. Fottea 2012, 12, 219–229. [Google Scholar] [CrossRef]
- Guillard, R.R.L.; Hargraves, P.E. Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia 1993, 32, 234–236. [Google Scholar] [CrossRef]
- Perkins, R.G.; Mouget, J.L.; Lefevbre, S.; Lavaud, J. Light response curve methodology and possible implications in the application of chlorophyll fluorescence to benthic diatoms. Mar. Biol. 2006, 149, 703–712. [Google Scholar] [CrossRef]
- Hendey, N.I. The permanganate method for cleaning freshly gathered diatoms. Microscopy 1974, 32, 423–426. [Google Scholar]
- Pouvreau, J.B.; Morançais, M.; Masse, G.; Rosa, P.; Robert, J.M.; Fleurence, J.; Pondaven, P. Purification of the blue-green pigment “marennine” from the marine tychopelagic diatom Haslea ostrearia (Gaillon/Bory) Simonsen. J. Appl. Phycol. 2006, 18, 769–781. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, D.; Abajian, C.; Green, P. Consed: A graphical tool for sequence finishing. Genome Res. 1998, 3, 195–202. [Google Scholar] [CrossRef] [Green Version]
- Turmel, M.; Otis, C.; Lemieux, C. Divergent copies of the large inverted repeat in the chloroplast genomes of ulvophycean green algae. Sci. Rep. 2017, 7, 994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 2016, 32, 1933–1942. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Davidovich, N.A.; Mouget, J.L.; Gaudin, P. Heterothallism in the pennate diatom Haslea ostrearia (Bacillariophyta). Eur. J. Phycol. 2009, 44, 251–261. [Google Scholar] [CrossRef]
- Mouget, J.L.; Gastineau, R.; Davidovich, O.; Gaudin, P.; Davidovich, N.A. Light is a key factor in triggering sexual reproduction in the pennate diatom Haslea ostrearia. FEMS Microbiol. Ecol. 2009, 69, 194–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, S.M.; Noh, J.H.; Choi, D.H.; Lee, J.H.; Yang, E.C. Repeat region absent in mitochondrial genome of tube-dwelling diatom Berkeleya fennica (Naviculales, Bacillariophyceae). Mitochondrial DNA Part A 2016, 27, 2137–2138. [Google Scholar] [CrossRef]
- Pogoda, C.S.; Keepers, K.G.; Hamsher, S.E.; Stepanek, J.G.; Kane, N.C.; Kociolek, J.P. Comparative analysis of the mitochondrial genomes of six newly sequenced diatoms reveals group II introns in the barcoding region of cox1. Mitochondrial DNA Part A 2018, 30, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Ruck, E.C.; Linard, S.R.; Nakov, T.; Theriot, E.C.; Alverson, A.J. Hoarding and horizontal transfer led to an expanded gene and intron repertoire in the plastid genome of the diatom, Toxarium undulatum (Bacillariophyta). Curr. Genet. 2016, 63, 499–507. [Google Scholar] [CrossRef]
- Sterrenburg, F.A.S.; Tiffany, M.A.; Hinz, F.; Herwig, W.E.; Hargraves, P.E. Seven new species expand the morphological spectrum of Haslea. A comparison with Gyrosigma and Pleurosigma (Bacillariophyta). Phytotaxa 2015, 207, 143–162. [Google Scholar] [CrossRef] [Green Version]
- Poulin, M.; Massé, G.; Belt, S.T.; Delavault, P.; Rousseau, F.; Robert, J.M.; Rowland, S.J. Morphological, biochemical and molecular evidence for the transfer of Gyrosigma nipkowii Meister to the genus Haslea (Bacillariophyta). Eur. J. Phycol. 2004, 39, 181–196. [Google Scholar] [CrossRef]
- Li, Y.; Chen, X.; Sun, Z.; Xu, K. Taxonomy and molecular phylogeny of three marine benthic species of Haslea (Bacillariophyceae), with transfer of two species to Navicula. Diatom Res. 2017, 32, 451–463. [Google Scholar] [CrossRef]
- Lobban, C.S.; Perez, C.O.; Ashworth, M.P. Non-blue Haslea species (Bacillariophyceae: Naviculaceae) in the benthic marine flora of Guam (Mariana Islands, Western Pacific Ocean). Diatom Res. 2020, 35, 163–183. [Google Scholar] [CrossRef]
- Amato, A.; Kooistra, W.H.C.F.; Levialdi Ghiron, J.H.; Mann, D.G.; Pröschold, T.; Montresor, M. Reproductive isolation among sympatric cryptic species in marine diatoms. Protist 2007, 158, 193–207. [Google Scholar] [CrossRef]
- Beszteri, B.; Ács, É.; Medlin, L.K. Ribosomal DNA sequence variation among sympatric strains of the Cyclotella meneghiniana complex (Bacillariophyceae) reveals cryptic diversity. Protist 2005, 156, 317–333. [Google Scholar] [CrossRef]
- Kaczmarska, I.; Ehrman, J.M.; Moniz, M.B.J.; Davidovich, N. Phenotypic and genetic structure of interbreeding populations of the diatom Tabularia fasciculata (Bacillariophyta). Phycologia 2009, 48, 391–403. [Google Scholar] [CrossRef]
- Kaczmarska, I.; Mather, L.; Luddington, I.A.; Musie, F.; Ehrman, J.M. Cryptic diversity in a cosmopolitan diatom known as Asterionellopsis glacialis (Fragilariaceae): Implications for ecology, biogeography, and taxonomy. Am. J. Bot. 2014, 101, 267–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kermarrec, L.; Bouchez, A.; Rimet, F.; Humbert, J.F. First evidence of the existence of semi-cryptic species and of a phylogeographic structure in the Gomphonema parvulum (Kützing) Kützing complex (Bacillariophyta). Protist 2013, 164, 686–705. [Google Scholar] [CrossRef] [PubMed]
- Poulíčková, A.; Veselá, J.; Neustupa, J.; Škaloud, P. Pseudocryptic diversity versus cosmopolitanism in diatoms: A case study on Navicula cryptocephala Kütz. (Bacillariophyceae) and morphologically similar taxa. Protist 2010, 161, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Vanormelingen, P.; Chepurnov, V.A.; Mann, D.G.; Sabbe, K.; Vyverman, W. Genetic divergence and reproductive barriers among morphologically heterogeneous sympatric clones of Eunotia bilunaris sensu lato (Bacillariophyta). Protist 2008, 159, 73–90. [Google Scholar] [CrossRef]
- Mann, D.G. Discovering diatom species: Is a long history of disagreements about species-level taxonomy now at an end? Plant Ecol. Evol. 2010, 143, 251–264. [Google Scholar] [CrossRef]
- Gastineau, R.; Leignel, V.; Jacquette, B.; Hardivillier, Y.; Wulff, A.; Gaudin, P.; Bendahmane, D.; Davidovich, N.A.; Kaczmarska, I.; Mouget, J.L. Inheritance of mitochondrial DNA in the pennate diatom Haslea ostrearia (Naviculaceae) during auxosporulation suggests a uniparental transmission. Protist 2013, 164, 340–351. [Google Scholar] [CrossRef]
- Fernandes, L.F.; Jardim, P.F.G. Morfologia da valva de algumas espécies de Haslea Simonsen (Bacillariophyta) do Sul do Brasil. Iheringia Série Botânica 2016, 70, 375–384. [Google Scholar]
- Karsten, G. Untersuchungen über Diatomeen. III. Flora 1897, 83, 203–221. [Google Scholar]
- Chepurnov, V.A. Polovoj protsess u dvudomnoj vodorosli Haslea subagnita (Pr.-Lavr.) Makar. et Kar. (Bacillariophyta). Algologiya 1993, 3, 37–40. [Google Scholar]
- Chepurnov, V.A.; Mann, D.G.; Sabbe, K.; Vyverman, W. Experimental studies on sexual reproduction in diatoms. Int. Rev. Cytol. 2004, 237, 91–154. [Google Scholar] [CrossRef]
- Van Den Heuvel, H.M.; Prud’homme Van Reine, W.F. Marine, mainly benthic, diatoms of the West Coast of the Island La Palma (Canary Islands). Vieraea 1984, 14, 11–31. [Google Scholar]
- Van Den Heuvel, H.M. Diatoms from El Golfo on Lanzarote (Canary Islands). Vieraea 1991, 20, 53–70. [Google Scholar]
- Moro Abad, L.; Martín Esquivel, J.L.; Garrido, M.J.; Izquierdo, J. (Eds.) Lista de Especies Marinas de Canarias (Algas, Hongos, Plantas y Animales); Consejería de Política Territorial y Medio Ambiente del Gobierno de Canarias: La Laguna, Tenerife, Canary Islands, Spain, 2003; 248p. [Google Scholar]
- Ojeda, A.; Gil-Rodríguez, M.C.; Moreira-Reyes, A. Aportaciones al conocimiento de diatomeas bentónicas y ticoplanctónicas del puerto de Santa Cruz de Tenerife (islas Canarias). Vieraea 2005, 33, 59–78. [Google Scholar]
- Afonso-Carrillo, J. Lista Actualizada de las Algas Marinas de las Islas Canarias, 2014; Las Palmas: Elaborada para la Sociedad Española de Ficología (SEF): La Laguna, Tenerife, Canary Islands, Spain, 2014; 64p. [Google Scholar]
- Aboal, M.; Alvarez-Cobelas, M.; Cambra, J.; Ector, L. Floristic list of non marine diatoms (Bacillariophyceae) of Iberian Peninsula, Balearic Islands, and Canary Islands. Updated taxonomy and bibliography. In Diatom Monograph; Witkowski, A., Ed.; A.R.G. Gantner Verlag, K.G.: Ruggell, Liechtenstein, 2003; Volume 4, 639p. [Google Scholar]
- Witkowski, A.; Lange-Bertalot, H.; Metzeltin, D. Diatom flora of marine coasts I. Iconogr. Diatomol. 2000, 7, 1–925. [Google Scholar]
- Hallegraeff, G.M.; Bolch, C.J. Transport of diatom and dinoflagellate resting spores in ships’ ballast water: Implications for plankton biogeography and aquaculture. J. Plankton Res. 1992, 14, 1067–1084. [Google Scholar] [CrossRef]
- Villac, M.C.; Kaczmarska, I.; Ehrman, J.M. The diversity of diatom assemblages in ships’ ballast sediments: Colonization and propagule pressure on Canadian ports. J. Plankton Res. 2013, 35, 1267–1282. [Google Scholar] [CrossRef] [Green Version]
- Denys, L. Morphology and taxonomy of epizoic diatoms (Epiphalaina and Tursiocola) on a sperm whale (Physeter macrocephalus) stranded on the coast of Belgium. Diatom Res. 1997, 12, 1–18. [Google Scholar] [CrossRef]
- Majewska, R.; Santoro, M.; Bolaños, F.; Chaves, G.; De Stefano, M. Diatoms and other epibionts associated with Olive Ridley (Lepidochelys olivacea) Sea Turtles from the Pacific coast of Costa Rica. PLoS ONE 2015, 10, e0130351. [Google Scholar] [CrossRef]
- Kaleli, A.; Krzywda, M.; Witkowski, A.; Riaux-Gobin, C.; Nadir Solak, C.; Zgłobicka, I.; Płociński, T.; Grzonka, J.; Kurzydłowski, K.J.; Car, A.; et al. A new sediment dwelling and epizoic species of Olifantiella (Bacillariophyceae), with an account on the genus ultrastructure based on Focused Ion Beam nanocuts. Fottea 2018, 18, 212–226. [Google Scholar] [CrossRef] [Green Version]
- Masó, M.; Garcés, E.; Pagès, F.; Camp, J. Drifting plastic debris as a potential vector for dispersing harmful algae bloom (HAB) species. Sci. Mar. 2003, 67, 107–111. [Google Scholar] [CrossRef] [Green Version]
- Welch, W.R. The European oyster, Ostrea edulis, in Maine. Proc. Natl. Shellfish. Assoc. 1966, 54, 7–23. [Google Scholar]
- Ruesink, J.L.; Lenihan, H.S.; Trimble, A.C.; Heiman, K.W.; Micheli, F.; Byers, J.E.; Kay, M.C. Introduction of non-native oysters: Ecosystem effects and restoration implications. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 643–689. [Google Scholar] [CrossRef] [Green Version]
- Wolff, W.J.; Reise, K. Oyster imports as a vector for the introduction of alien species into Northern and Western European coastal waters. In Invasive Aquatic Species of Europe. Distribution, Impact and Management; Leppäkoski, E., Gollasch, S., Olenin, S., Eds.; Springer Science+Business Media: Dordrecht, The Netherlands; Berlin/Heidelberg, Germany, 2002; pp. 193–205. [Google Scholar]
- Massé, G.; Rincé, Y.; Cox, E.J.; Allarda, G.; Belt, S.T.; Rowland, S.J. Haslea salstonica sp. nov. and Haslea pseudostrearia sp. nov. (Bacillariophyta), two new epibenthic diatoms from the Kingsbridge estuary, United Kingdom. Comptes Rendus de l’Académie des Sciences de Paris Sciences de la vie/Life Sci. 2001, 324, 617–626. [Google Scholar] [CrossRef]
- Brembu, T.; Winge, P.; Tooming-Klunderud, A.; Nederbragt, A.J.; Jakobsen, K.S.; Bones, A.M. The chloroplast genome of the diatom Seminavis robusta: New features introduced through multiple mechanisms of horizontal gene transfer. Mar. Genom. 2014, 16, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Ruck, E.C.; Nakov, T.; Jansen, R.K.; Theriot, E.C.; Alverson, A.J. Serial gene losses and foreign DNA underlie size and sequencevariation in the plastid genomes of diatoms. Genome Biol. Evol. 2014, 6, 644–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imanian, B.; Pombert, J.F.; Keeling, P.J. The complete plastid genomes of the two ‘dinotoms’ Durinskia baltica and Kryptoperidinium foliaceum. PLoS ONE 2010, 19, e10711. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Gastineau, R.; Turmel, M.; Witkowski, A.; Otis, C.; Car, A.; Lemieux, C. Complete chloroplast genome of the tiny marine diatom Nanofrustulum shiloi (Bacillariophyta) from the Adriatic Sea. Mitochondrial DNA Part B 2019, 4, 3374–3376. [Google Scholar] [CrossRef] [Green Version]
- Hildebrand, M.; Hasegawa, P.; Ord, R.W.; Thorpe, V.S.; Glass, C.A.; Volcani, B.E. Nucleotide sequence of diatom plasmids: Identification of open reading frames with similarity to site-specific recombinases. Plant Mol. Biol. 1992, 19, 759–770. [Google Scholar] [CrossRef]
- Jacobs, J.D.; Ludwig, J.R.; Hildebrand, M.; Kukel, A.; Feng, T.Y.; Ord, R.W.; Volcani, B.E. Characterization of two circular plasmids from the marine diatom Cylindrotheca fusiformis: Plasmids hybridize to chloroplast and nuclear DNA. Mol. Gen. Genet. 1992, 233, 302–310. [Google Scholar] [CrossRef]
- Yu, M.; Ashworth, M.P.; Hajrah, N.H.; Khiyami, M.A.; Sabir, M.J.; Alhebshi, A.M.; Al-Malki, A.L.; Sabir, J.S.M.; Theriot, E.C.; Jansen, R.K. Evolution of the plastid genomes in diatoms. Adv. Bot. Res. 2018, 85, 129–155. [Google Scholar] [CrossRef]
- Oudot-Le Secq, M.-P.; Green, B.R. Complex repeat structures and novel features in the mitochondrial genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana. Gene 2011, 476, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Ravin, N.V.; Galachyants, Y.P.; Mardanov, A.V.; Beletsky, A.V.; Petrova, D.P.; Sherbakova, T.A.; Zakharova, Y.R.; Likhoshway, Y.V.; Skryabin, K.G.; Grachev, M.A. Complete sequence of the mitochondrial genome of a diatom alga Synedra acus and comparative analysis of diatom mitochondrial genomes. Curr. Genet. 2010, 56, 215–223. [Google Scholar] [CrossRef]
- Villain, A.; Kojadinovic, M.; Puppo, C.; Prioretti, L.; Hubert, P.; Zhang, Y.; Grégori, G.; Roulet, A.; Roques, C.; Claverie, J.-M.; et al. Complete mitochondrial genome sequence of the freshwater diatom Asterionella formosa. Mitochondrial DNA Part B 2017, 2, 97–98. [Google Scholar] [CrossRef]
- An, S.M.; Noh, J.H.; Lee, H.R.; Choi, D.H.; Lee, J.H.; Yang, E.C. Complete mitochondrial genome of biraphid benthic diatom, Navicula ramosissima (Naviculales, Bacillariophyceae). Mitochondrial DNA Part B 2016, 1, 549–550. [Google Scholar] [CrossRef] [PubMed]
- Bensasson, D.; Zhang, D.X.; Hartl, D.L.; Hewitt, G.M. Mitochondrial pseudogenes: Evolution’s misplaced witnesses. Trends Ecol. Evol. 2001, 16, 314–321. [Google Scholar] [CrossRef]
- Raboin, M.J.; Timko, A.F.; Howe, D.K.; Marie-Anne Félix, M.-A.; Denver, D.R. Evolution of Caenorhabditis mitochondrial genome pseudogenes and Caenorhabditis briggsae natural isolates. Mol. Biol. Evol. 2010, 27, 1087–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eimanifar, A.; Kimball, R.T.; Braun, E.L.; Ellis, J.D. The complete mitochondrial genome of the Cape honey bee, Apis mellifera capensis Esch. (Insecta: Hymenoptera: Apidae). Mitochondrial DNA Part B 2016, 1, 817–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mabuchi, K. Complete mitochondrial genome of the parrotfish Calotomus japonicus (Osteichthyes: Scaridae) with implications based on the phylogenetic position. Mitochondrial DNA Part B 2016, 1, 643–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Xu, X.; Yu, Z.; Wei, Z.; Xia, J. Comparison of seven Crassostrea mitogenomes and phylogenetic analyses. Mol. Phylogenet. Evol. 2010, 57, 448–454. [Google Scholar] [CrossRef]
- Wu, X.; Li, X.; Li, L.; Xu, X.; Xia, X.; Yu, Z. New features of Asian Crassostrea oyster mitochondrial genomes: A novel alloacceptor tRNA gene recruitment ant two novels ORFs. Gene 2012, 507, 112–118. [Google Scholar] [CrossRef]
- Gastineau, R.; Nguyễn, D.H.; Lemieux, C.; Turmel, M.; Tremblay, R.; Nguyễn, V.D.; Widowati, I.; Witkowski, A.; Mouget, J.-L. The complete mitochondrial DNA of the tropical oyster Crassostrea belcheri from the Cần Giò’ mangrove in Vietnam. Mitochondrial DNA Part B 2018, 3, 462–463. [Google Scholar] [CrossRef] [Green Version]
- Gastineau, R.; Wawrzyniak-Wydrowska, B.; Lemieux, C.; Turmel, M. Complete mitogenome of the invasive bivalve Rangia cuneata. Mitochondrial DNA Part B 2019, 4, 2794–2795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Xue, J.Y.; Li, L.; Liu, L.; Qiu, Y.L. The complete mitochondrial genome sequence of the liverwort Pleurozia purpurea reveals extremely conservative mitochondrial genome evolution in liverworts. Curr. Genet. 2009, 55, 601–609. [Google Scholar] [CrossRef]
- Xue, J.Y.; Liu, Y.; Li, L.; Wang, B.; Qiu, Y.L. The complete mitochondrial genome sequence of the hornwort Phaeoceros laevis: Retention of many ancient pseudogenes and conservative evolution of mitochondrial genomes in hornworts. Curr. Genet. 2010, 56, 53–61. [Google Scholar] [CrossRef]
- Choi, M.N.; Han, M.; Lee, H.; Park, H.S.; Kim, M.Y.; Kim, J.S.; Na, Y.J.; Sim, S.W.; Park, E.J. The complete mitochondrial genome sequence of Populus davidiana Dode. Mitochondrial DNA Part B 2017, 2, 113–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Liu, N.; Yin, H.; Liu, C.; Zhang, L.; Jin, Y.; Wang, H.; Chi, S.; Liu, T. Complete sequences of the mitochondrial DNA of the Petalonia binghamiae. Mitochondrial DNA Part B 2018, 3, 95–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goruynov, D.V.; Goryunova, S.V.; Kuznetsova, O.I.; Logacheva, M.D.; Milyutina, I.A.; Fedorova, A.V.; Gnatov, M.S.; Troitsky, A.V. Complete mitochondrial genome sequence of the “copper moss” Mielichhoferia elongata reveals independent nad7 gene functionality loss. PeerJ 2018, 6, e4350. [Google Scholar] [CrossRef] [PubMed] [Green Version]














Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gastineau, R.; Hansen, G.; Poulin, M.; Lemieux, C.; Turmel, M.; Bardeau, J.-F.; Leignel, V.; Hardivillier, Y.; Morançais, M.; Fleurence, J.; et al. Haslea silbo, A Novel Cosmopolitan Species of Blue Diatoms. Biology 2021, 10, 328. https://doi.org/10.3390/biology10040328
Gastineau R, Hansen G, Poulin M, Lemieux C, Turmel M, Bardeau J-F, Leignel V, Hardivillier Y, Morançais M, Fleurence J, et al. Haslea silbo, A Novel Cosmopolitan Species of Blue Diatoms. Biology. 2021; 10(4):328. https://doi.org/10.3390/biology10040328
Chicago/Turabian StyleGastineau, Romain, Gert Hansen, Michel Poulin, Claude Lemieux, Monique Turmel, Jean-François Bardeau, Vincent Leignel, Yann Hardivillier, Michèle Morançais, Joël Fleurence, and et al. 2021. "Haslea silbo, A Novel Cosmopolitan Species of Blue Diatoms" Biology 10, no. 4: 328. https://doi.org/10.3390/biology10040328
APA StyleGastineau, R., Hansen, G., Poulin, M., Lemieux, C., Turmel, M., Bardeau, J. -F., Leignel, V., Hardivillier, Y., Morançais, M., Fleurence, J., Gaudin, P., Méléder, V., Cox, E. J., Davidovich, N. A., Davidovich, O. I., Witkowski, A., Kaczmarska, I., Ehrman, J. M., Soler Onís, E., ... Mouget, J. -L. (2021). Haslea silbo, A Novel Cosmopolitan Species of Blue Diatoms. Biology, 10(4), 328. https://doi.org/10.3390/biology10040328









